Abstract
The mammalian target of rapamycin complex 1 (mTORC1) is a critical regulator of G1 cell cycle progression. Two key substrates of mTORC1 are ribosomal subunit S6 kinase (S6K) and eukaryotic initiation factor 4E (eIF4E) binding protein-1 (4E-BP1). We reported previously that simultaneous knockdown of S6K and eIF4E causes a transforming growth factor-β (TGF-β)-dependent G1 cell cycle arrest in MDA-MB-231 human breast cancer cells. Rapamycin inhibits the phosphorylation of S6K at nano-molar concentrations in MDA-MB-231 cells; however, micro-molar concentrations of rapamycin are required to inhibit phosphorylation of 4E-BP1 – the phosphorylation of which, liberates eIF4E to initiate translation. Micro-molar doses of rapamycin are required for complete G1 cell cycle arrest – indicating that 4E-BP1 is a critical target of mTOR for promoting cell cycle progression. Data are provided demonstrating that G1 cell cycle arrest induced by rapamycin is due to up-regulation of TGF-β signaling and down-regulation of Rb phosphorylation via phosphorylation of the mTORC1 substrates S6K and 4E-BP1 respectively. These findings enhance the current understanding of the cytostatic effects of mTORC1 suppression with therapeutic implications.
Keywords: mTOR, rapamycin, Rb, TGF-β, eIF4E
1. Introduction
Understanding control of G1 cell cycle progression has central position in the search for therapeutic options for cancer and other proliferative disorders. This is due to the finding that a majority of the driver mutations in cancer cells are to genes that encode proteins involved in the control of G1 cell cycle progression [1]. A key signaling node for the control of G1 cell cycle progression is the mammalian/mechanistic target of rapamycin (mTOR) complex 1 (mTORC1). It has been suggested that signals that regulate mTOR are the most commonly dysregulated signals in cancer [2, 3]. Although activating gain-of-function mTOR mutations have been reported in human cancers [4], more commonly there are mutations in genes encoding proteins that regulate mTOR activity. There are two key downstream substrates of mTORC1 – ribosomal subunit S6 kinase (S6K) and eukaryotic initiation factor (eIF4E) binding protein-1 (4E-BP1). Both S6K and 4E-BP1/eIF4E have been implicated in rapamycin-induced retardation of G1 cell cycle progression [5]. While the phosphorylation of S6K by mTORC1 is suppressed by conventional nano-molar doses of rapamycin, 4E-BP1 phosphorylation is not generally affected at these lower concentrations [6–8]. However, micro-molar concentrations of rapamycin do suppress phosphorylation of 4E-BP1 in MDA-MB-231 breast cancer cells, and it is at these higher doses that rapamycin induces complete cell cycle arrest in these cells [7] – suggesting that suppression of 4E-BP1 phosphorylation is also important for complete G1 cell cycle arrest. The cell cycle arrest induced by rapamycin was dependent on TGF-β signaling, which was elevated in response to rapamycin [9–11]. However, stimulating TGF-β signals could be achieved with nano-molar concentrations of rapamycin in MDA-MB-231 cells [10]. Thus, there is something in addition to stimulating TGF-β signaling mediated by 4E-BP1/eIF4E that is also responsible for the complete G1 cell cycle arrest caused by inhibition of mTORC1.
In this report, we provide evidence that suppression 4E-BP1 phosphorylation with rapamycin is required for the suppression of Rb phosphorylation; and that it is the suppression of Rb phosphorylation along with elevated TGF-β signals that causes complete G1 arrest.
2. Materials and methods
2.1. Cells and cell culture conditions
The human cancer cell lines MDA-MB-231 and MCF-7 cells were obtained from the American Tissue Type Culture Collection (ATCC) and cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Sigma, Saint Louis, MO, D6429) supplemented with 10% Fetal Bovine Serum (Sigma F4135).
2.2. Antibodies and reagents
The following antibodies were used: Cleaved PARP (9541), P-S6KT389 (9205), S6K (9202), P-4E-BP1T37/46 (9459), 4E-BP1 (9452), eIF4E (9742), Smad2 (5339), Smad3 (9523), Smad4 (9515), P-RbS780 (9307), Rb (9309), Cyclin D1 (2978) and α-Actin (8457) (Cell Signaling); P-Smad2S465/467(Millipore 04-953); p-Smad3S423/425 (Abcam ab52903). Negative control scrambled siRNA (Dharmacon), siRNAs targeted against S6K (sc-36165), eIF4E (sc-35284), Smad4 (sc-29484) and Rb (sc-29468) (Santa Cruz Biotechnology) were purchased. Lipofectamine RNAiMax (Invitrogen, 56532) were used for transient transfections. Rapamycin (R-5000) was obtained from LC Laboratories and the TGF-β inhibitor SB-431542 (S4317) was obtained from Sigma.
2.3. Western blot analysis
Extraction of proteins from cultured cells and Western blot analysis of extracted proteins was performed using the ECL system (Thermo Scientific, 34080) as described previously [7, 12].
2.4. Transient transfections
Cells were plated in 6-well plates in medium containing 10% FBS. The next day (30% confluence), transfections with siRNAs (100nM) in Lipofectamine RNAiMAX were performed. After 6 hours, reagents were replaced with fresh 10% FBS and cells were allowed to incubate for an additional 48 hours.
2.5. Flow cytometric analysis
Cells were washed and trypsinized. The cell suspensions were recovered and resuspended in the following fixing solution: 7 ml 1× phosphate buffer saline, 2% bovine serum albumin, 5mM EDTA, 0.1% NaN3. 3 ml of 100% ethanol was added drop wise. Fixed cells were centrifuged, washed and then resuspended in 500µl sorting buffer: 1× phosphate buffered saline, 0.1% Triton-X 100, 2% bovine serum albumin, 5mM EDTA, 40µg/ml propidium iodide, 100µg/ml RNAse A, and incubated at 37C for 30 min. The cells were filtered through 70 micrometer mesh to remove all cell aggregates. The DNA content was analyzed by flow cytometry (FACSCalibur; Becton Dickinson), and percentages of cells within each phase of the cell cycle was determined using Win Cycle software (Phoenix Flow Systems).
3. Results
3.1. Low dose of rapamycin and suppression of S6K elevate TGF-β signals
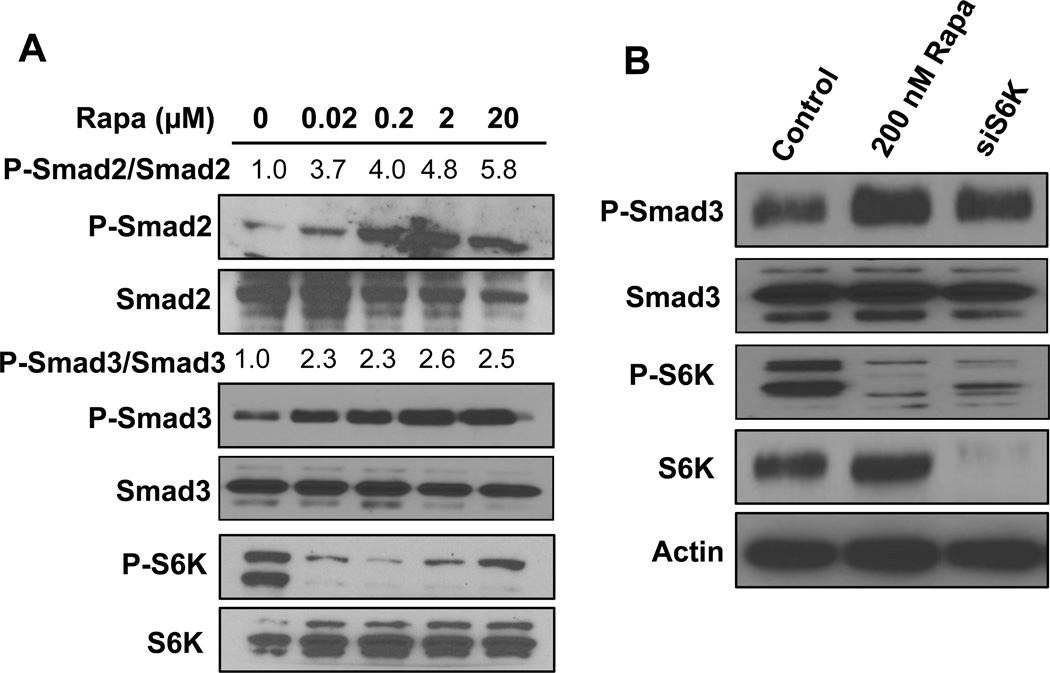
We previously reported that rapamycin caused a TGF-β-dependent G1 cell cycle arrest in MDA-MB-231 [11]. We also demonstrated that while G1 cell cycle progression in MDA-MB-231 breast cancer cells could be somewhat retarded by conventional nano-molar doses of rapamycin, complete G1 arrest required micro-molar doses of rapamycin [7]. TGF-β signaling is suppressed by mTORC1 and therefore is activated in response to rapamycin treatment [9, 13]. We therefore investigated the dose of rapamycin required to induce TGF-β signaling in MDA-MB-231 cells. Following stimulation with TGF- β, Smad2 and Smad3 become phosphorylated at carboxyl terminal serine residues (Ser465 and 467 on Smad2; Ser423 and 425 on Smad3) by TGF- β receptor I [14]. As shown in Fig. 1A, the induction of both Smad2 and Smad3 phosphorylation with rapamycin could be achieved between 20 and 200 nM in MDA-MB-231 cells. This induction correlated with the loss of S6K phosphorylation. We therefore examined whether suppression of S6K expression could elevate Smad phosphorylation. MDA-MB-231 cells were treated with siRNA directed against S6K and the level of phosphorylated Smad3 was determined. As shown in Fig. 1B, suppressing S6K expression, like rapamycin, elevated the level of phosphorylated Smad3. These data demonstrate that suppression of S6K phosphorylation is sufficient to elevate TGF-β signaling and establish a correlation between the suppression of S6K phosphorylation and increased TGF-β signaling.
Fig. 1.
Low dose of rapamycin and suppression of S6K causes Smad phosphorylation. (A) MDA-MB-231 cells were plated at a density of 400,000/60 mm dish in complete medium containing 10% serum overnight. Cells were treated with rapamycin for 24 hrs at the indicated doses. Cells were then harvested and the levels of P-Smad2S465/467, Smad2, P-Smad3S423/425, Smad3, P-S6KT389 and S6K were determined by Western blot analysis. The levels of P-Smad 2 and P-Smad 3 relative to the levels of Smad2 and Smad3 respectively were determined by LI-COR-Image studio lite and normalized to the levels observed in the absence of rapamycin treatment. (B) MDA-MB-231 cells were plated at a density of 300,000 in a 6 well plate in media containing 10% serum and no antibiotics overnight. Cells were transfected with negative control siRNA or siRNA for S6K as indicated. Six hr later the cells were shifted to regular media. 24 hr after transfection, cells were treated with rapamycin (200 nM) where indicated. After another 24hr, the level of P-Smad3, Smad3, P-S6K, S6K and actin were determined by Western blot analysis. All data are representative of at least two independent experiments.
3.2. High dose rapamycin inhibits Rb phosphorylation and suppresses cyclin D1 expression
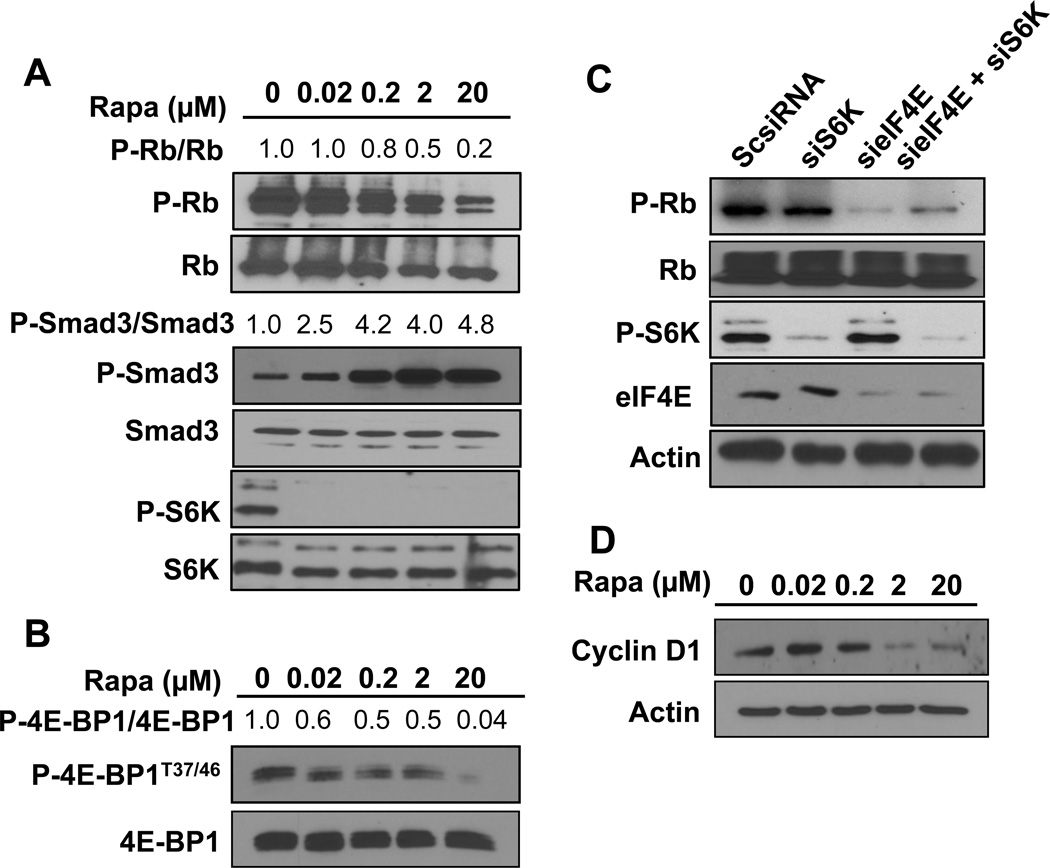
While suppression of S6K phosphorylation is apparently sufficient to de-repress TGF-β signaling, the low dose rapamycin treatments capable of suppressing S6K phosphorylation are not sufficient to induce complete G1 cell cycle arrest in MDA-MB-231 cells [7]. TGF-β suppresses G1 cell cycle progression by elevating the levels of p21 and p27 – factors that suppresses cyclin E/cyclin-dependent kinase (CDK) 2 [15]. Rapamycin causes a late G1 cell cycle arrest that occurs at a point in G1 that depends upon the phosphorylation of Rb by cyclin D-CDK4/6 and cyclin E-CDK2 [1, 16]. Since Rb phosphorylation is critical for late G1 cell cycle progression, we examined the effect of rapamycin on Rb phosphorylation. As shown in Fig. 2A, rapamycin inhibited Rb phosphorylation in MDA-MB-231. Importantly, the effect required the micro-molar doses needed to suppress 4E-BP1 phosphorylation (Fig. 2B) [7]. The effect of rapamycin on 4E-BP1 phosphorylation was examined at 4 hr in Fig. 2B as opposed to the 24 hr time point used in Fig. 2A. This was due to a stronger effect of rapamycin at the 4 hr time point for suppression of 4E-BP1, which becomes re-phosphorylated at the 24 time point – an effect that we and others have observed previously [8, 16]. These data reveal a correlation between suppression of Rb phosphorylation and the complete G1 cell cycle arrest induced by micro-molar doses of rapamycin.
Fig. 2.
High dose rapamycin inhibits Rb phosphorylation and suppresses cyclin D1 expression. (A) MDA-MB-231 cells were plated as in Fig. 1A. Cells were then treated with rapamycin for 24 hr at the indicated doses. Cells were then harvested and the levels of P-RbS780, Rb, P-Smad3, Smad3, P-S6KT389 and S6K were determined by Western blot analysis. The levels of P-Smad3 relative to Smad3 and P-Rb to Rb were determined as in Fig. 1A. (B) MDA-MB-231 cells were plated as in Fig. 1A. Cells were then treated with rapamycin with the indicated doses for 4hr and the levels of P-4E-BP1T37/46 and total 4E-BP1 was determined by Western blot analysis. The levels of P-4E-BP1 relative to 4EBP1 were determined as in (A). (C) MDA-MB-231 cells were plated as in Fig. 1B. Cells were transfected with negative control siRNA, siRNA for eIF4E or S6K as indicated. 6 hrs later the cells were shifted to regular media. 48 hrs later, the level of P-Rb, Rb, eIF4E, S6K and actin was determined by Western blot analysis. (D) MDA-MB-231 cells were plated as in Fig. 1A. Cells were treated with rapamycin for 24 hrs at indicated doses. Cells were then harvested and the levels of cyclin D1 and actin were determined by Western blot analysis. All data are representative of at least two independent experiments.
To further establish that the effect of rapamycin is due to inhibition of 4E-BP1 phosphorylation, we used an siRNA knockdown approach. The phosphorylation of 4E-BP1 by mTORC1 results in the liberation of eIF4E, which can then facilitate cap-dependent translation of RNAs that encode proteins critical for cell cycle progression [17]. We demonstrated previously that suppressing expression of both S6K and eIF4E resulted in G1 cell cycle arrest [11]. To determine whether the phosphorylation of Rb stimulated by high dose rapamycin was dependent on the inhibition of 4E-BP1 phosphorylation, we investigated whether suppressing eIF4E expression would suppress Rb phosphorylation. As shown in Fig. 2C, suppressing eIF4E expression with siRNA suppressed the phosphorylation Rb in MDA-MB-231 cells. Suppression of S6K expression did not significantly affect Rb phosphorylation. These data indicate that the suppression of Rb phosphorylation by high dose rapamycin is due to inhibition of 4E-BP1 phosphorylation and the consequent sequestration of eIF4E.
Proud and colleagues [18] reported previously that regulation of cyclin D1 expression by mTOR was mediated at the translational level and required eIF4E as evidenced by the loss of an effect of rapamycin when 4E-BP1 expression was suppressed. We therefore looked at the dose response to rapamycin of cyclin D1 expression. As shown in Fig. 2D, rapamycin suppressed cyclin D1 expression at micro-molar doses that correlated well with the rapamycin dose needed to suppress the phosphorylation of Rb. These data further support the hypothesis that the high dose rapamycin requirement for complete G1 arrest involves the suppression of 4E-BP1 phosphorylation and the subsequent suppression of Rb phosphorylation.
3.3. MCF7 breast cancer cells display a greater sensitivity to rapamycin
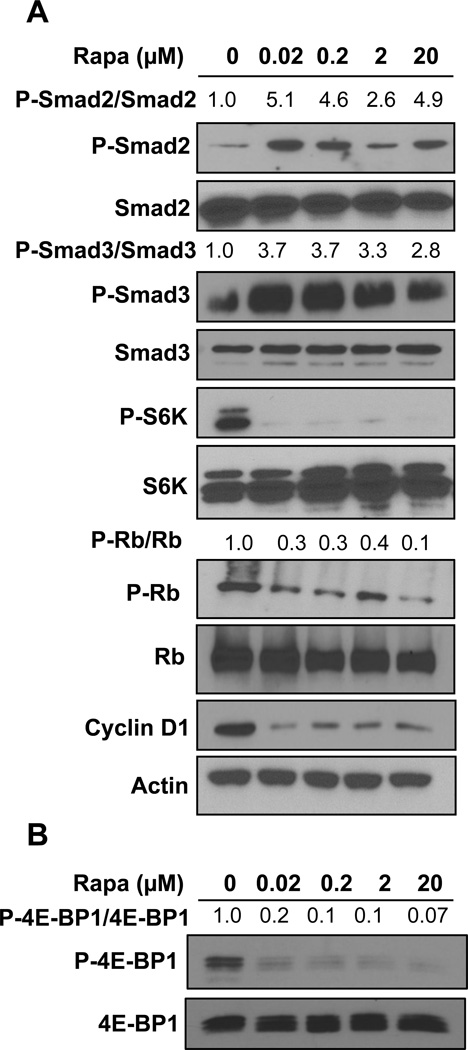
The data in Figs. 1 and 2 reveal that different doses of rapamycin that suppress TGF-β signals are at least an order of magnitude less than that needed to suppress Rb phosphorylation and cyclin D1. This reflects the differential sensitivity of mTORC1 to rapamycin for different substrates – with S6K phosphorylation being sensitive to low nano-molar concentrations, and 4E-BP1 needing micro-molar concentrations to suppress phosphorylation [7]. We demonstrated previously that the difference was due to partial, as opposed to complete, dissociation of mTOR from the substrate-recognizing mTORC1 subunit Raptor [7]. Another critical factor relating to rapamycin dose is that in different cancer cell lines, different doses of rapamycin are required to suppress phosphorylation of the same substrate. We previously reported that while suppression of S6K phosphorylation in MDA-MB-231 required around 50 nano-molar rapamycin, whereas in MCF7 breast cancer cells, S6K phosphorylation could be suppressed at 1 nano-molar [19]. This was due to elevated levels of phospholipase D activity in MDA-MB-231 cells [19]. Phospholipase D generates the metabolite phosphatidic acid, which interacts with and activates mTOR in a manner that is competitive with rapamycin [20]. We therefore investigated the effect of rapamycin on the parameters that control cell cycle progression in response to rapamycin in MCF7 cells. As shown in Fig. 3A, it can be seen that the levels of rapamycin needed to suppress Smad2 and Smad3 phosphorylation were somewhat less than that needed to suppress the phosphorylation of Smad2 and Smad3 in MDA-MB-231 cells (Fig. 1 and 2). More significantly, Rb phosphorylation and cyclin D1 expression in MCF7 cells (Fig. 3A) were sensitive to substantially lower doses of rapamycin than that needed in MDA-MB-231 cells (Fig. 2A). As with MDA-MB-231 cells long-term rapamycin treatment was less effective at suppressing 4E-BP1 than was a 4-hr treatment. However, as shown, rapamycin could suppress 4E-BP1 phosphorylation at the same dose that suppressed Rb phosphorylation and cyclin D1 expression (Figs. 3A and 3B). Thus, while MCF7 cells are more sensitive to rapamycin than MDA-MB-231 cells, they also display the differential sensitivity to rapamycin for TGF-β signaling and Rb phosphorylation.
Fig. 3.
MCF-7 cells display greater sensitivity to rapamycin. (A) MCF-7 cells were plated as in Fig. 1A. Cells were then treated with rapamycin for 24 hrs at indicated doses. Cells were then harvested and the levels of P-Smad2, Smad2, P-Smad3, Smad3, P-S6K, S6K, P-Rb, Rb and cyclin D1 were determined by Western blot analysis. All data are representative of at least two independent experiments. (B) MCF-7 cells were plated as in Fig. 1A. Cells were then treated with rapamycin with the indicated doses for 4hr and the levels of P-4EBP1T37/46 and total 4EBP1 was determined by Western blot analysis. The levels of P-Smad2 and P-Smad3 relative to the levels of Smad2 and Smad3 respectively and the levels of P-Rb relative to Rb were determined as in Figs. 1 and 2.
3.4. High dose rapamycin employs both TGF-β and Rb to cause G1 arrest
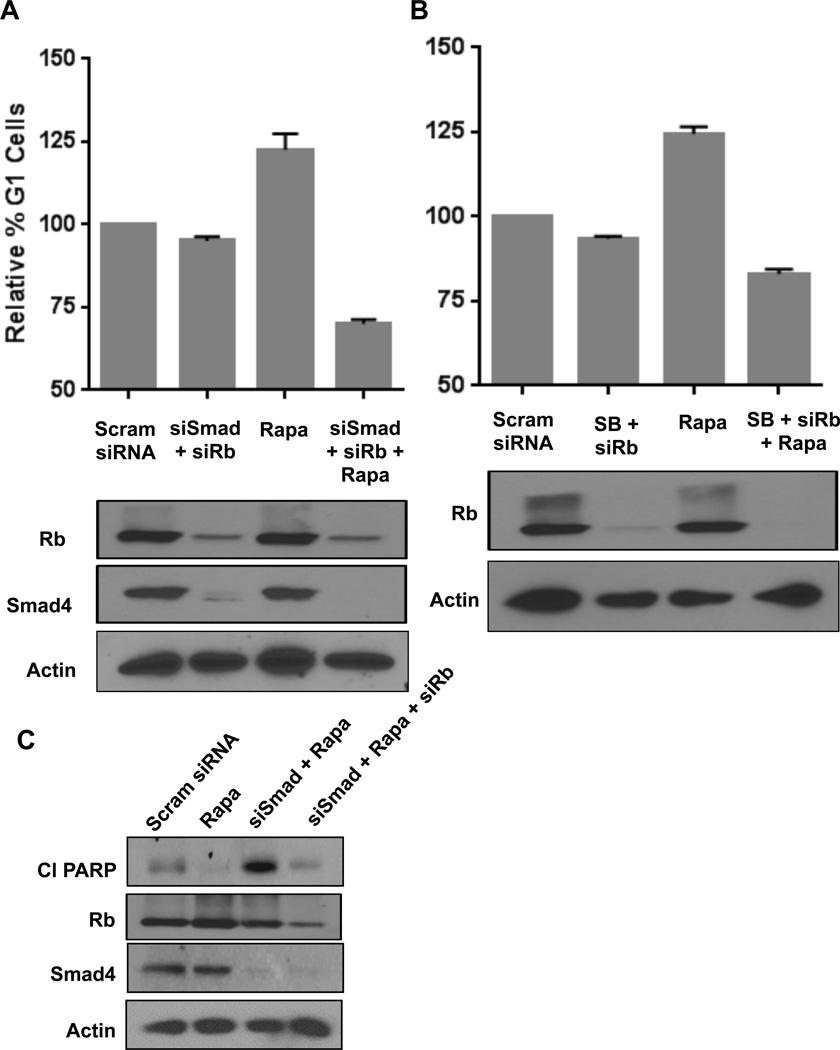
To further establish that the G1 cell cycle arrest caused by high dose rapamycin was due to inhibition of both S6K and 4E-BP1 phosphorylation, we examined the effect of high dose rapamycin on cell cycle progression in MDA-MB-231 cells where we suppressed TGF-β signaling along with suppression of Rb expression. As shown in Fig. 4A, high dose rapamycin increased the percentage of cells in G1 as reported previously [7, 11]. Upon phosphorylation, Smad2 or Smad3 combines with Smad4 and migrates to the nucleus where it acts as a transcription factor [15]. We therefore suppressed the expression Smad4 along with Rb using siRNAs. As shown in Fig. 4A, suppression of both Smad4 and Rb expression did not significantly alter the percentage of cells in G1; however, under these conditions, rapamycin was no longer able to increase the percentage of cells in G1. The same results were obtained using SB-431542 (Fig. 4B), a compound that inhibits the TGF-β receptor [21]. The observation that under conditions where both TGF-β signals and Rb are compromised rapamycin actually suppresses the percentage of cells in G1 is likely a reflection that there is a higher percentage of cells in S-phase (not shown) – indicating that rapamycin slows progression through S-phase in the absence of TGF-β signals and Rb. These data demonstrate that the ability of high dose rapamycin to arrest MDA-MB-231 cells in G1 is dependent on both TGF-β and Rb.
Fig. 4.
Rapamycin employs TGF-β and Rb pathway to cause cell cycle arrest. (A) MDA-MB-231 cells were plated as in Fig. 1B. The cells were transfected with scrambled (Scram) siRNA, siRNA for Rb or Smad4 as indicated. 6hrs later, the cells were shifted to a 10 cm dish containing regular media. 24 hrs after transfection rapamycin (20µM) was added where indicated. After an additional 48hrs the cells were collected for flow cytometry and Western blot analysis. (B) MDA-MB-231 cells were plated as in Fig. 1B. The cells were transfected with scrambled siRNA or siRNA for Rb where indicated. 6hrs later, the cells were shifted to 10 cm dish containing regular media. 24 hrs after transfection rapamycin (20µM) or SB-431542 (SB) (10µM) was added where indicated. After an additional 48hrs the cells were collected for FACS and Western blot analysis. Error bars represent the standard deviation for at least two independent experiments. (C) MDA-MB-231 cells were plated as in Fig. 1B. Cells were transfected with negative control siRNA, siRNA for Smad4 or Rb as indicated. 6 hrs later the cells were shifted to fresh regular media. 24 hrs after transfection, cells were treated with rapamycin (20µM) where indicated. After another 18hr, the levels of cleaved PARP (Cl PARP), Smad4, Rb and actin were determined by Western blot analysis. The data are representative of experiments repeated at least two times.
We reported previously that in the absence of TGF-β, high dose rapamycin treatment results in apoptotic cell death in MDA-MB-231 cells [7, 9, 11, 22]. As expected, rapamycin induced cleavage of the caspase 3 substrate poly-ADP ribose polymerase (PARP) in MDA-MB-231 cells that had been treated with siRNA for the co-Smad - Smad4. However, as shown in Fig. 4C, the loss of both TGF-β signals and Rb did not result in apoptosis as indicated by the lack of PARP cleavage, which was observed with the combination rapamycin and suppression of Smad4 expression. Apparently, the loss of Rb and the concomitant release of E2F family transcription factors prevented the apoptosis observed with rapamycin treatment in cells with defective TGF-β signaling.
4. Discussion
In this report, we have identified a missing factor in rapamycin-induced TGF-β-dependent G1 cell cycle arrest. We previously reported that complete G1 cell cycle arrest in MDA-MB-231 cells required high micro-molar doses of rapamycin, whereas low nano-molar doses were sufficient for activating TGF-β signals [7, 11]. Rapamycin suppresses S6K phosphorylation at nano-molar doses, however micro-molar doses are required for suppression of 4E-BP1 phosphorylation. In this report, we have provided evidence that complete G1 cell cycle arrest requires suppression of both S6K-dependent inhibition of TGF-β signals and 4E-BP1-dependent suppression of Rb phosphorylation. While there are other substrates of mTOR that are sensitive to the lower doses of rapamycin that could also be involved, the higher doses appear to be restricted to suppressing phosphorylation of 4E-BP1 and Rb.
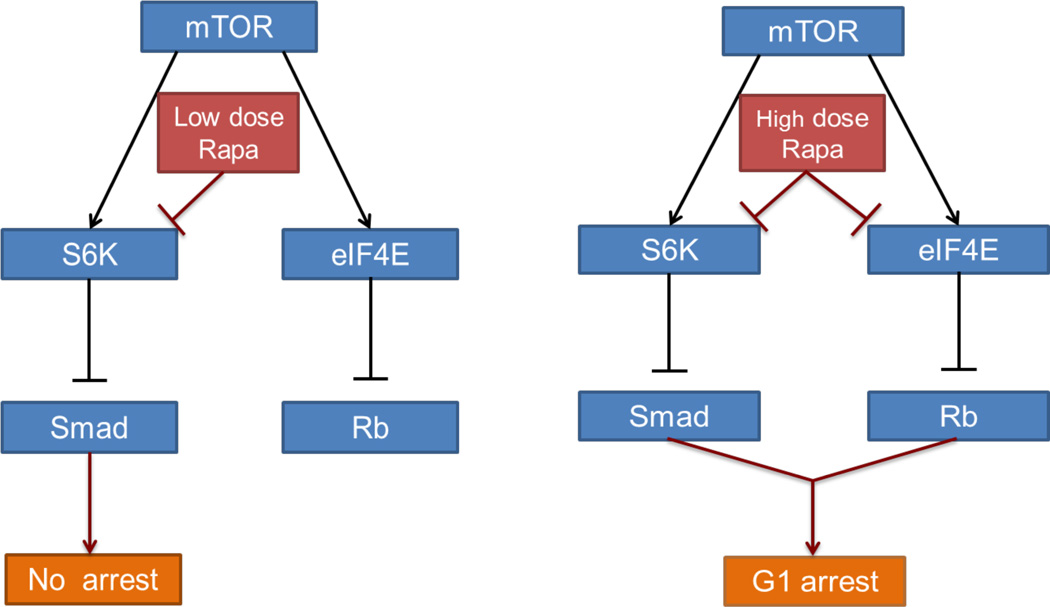
The integration of TGF-β signals with Rb highlights the significance of cyclin D-CDK4/6 and cyclin E-CDK2, which phosphorylates Rb in late G1 to promote cell cycle progression into S-phase [1]. Rb becomes hyper-phosphorylated by cyclin E/CDK2 and dissociates from E2F family transcription factors allowing E2F to stimulate the expression of genes critical for progression from G1 into S-phase [23, 24]. TGF-β stimulates the expression of the cyclin E-CDK2 inhibitors p21Cip1 and p27Kip1 [25]. Thus, in principle, elevating TGF-β could suppress Rb phosphorylation. However, as shown in Fig. 2, nano-molar doses of rapamycin could completely suppress S6K phosphorylation, but only partially inhibit Rb phosphorylation. This is consistent with a previous report by Blenis and colleagues where nano-molar doses of rapamycin slowed G1 progression, but did not arrest cells in G1 [5]. The high doses of rapamycin that completely block G1 cell cycle progression [7, 16] caused a more significant drop in Rb phosphorylation that was dependent on suppression of 4E-BP1 phosphorylation, which results in the sequestration of eIF4E. Thus, while de-repression of TGF-β signaling by low dose rapamycin is necessary for blocking G1 cell cycle progression, suppression of eIF4E, which requires higher doses of rapamycin, is also required. These data indicate a more complex regulation of the cyclin E-CDK2 cell cycle checkpoint that involve signal input to both TGF-β signals and Rb that involve cyclin D-CDK4/6 and cyclin E-CDK2. This is shown schematically in Fig. 5.
Fig. 5.
Model for the differential effects of low and high dose rapamycin. (A) Low dose rapamycin inhibits S6K and thereby up-regulates TGF-β signaling. Up regulation of TGF-β signaling is insufficient to cause cell cycle arrest. (B) High dose rapamycin inhibits S6K and eIF4E and thus activates TGF-β signaling and Rb, both of which are required for causing cell cycle arrest
While it is not clear how eIF4E contributes to the suppression of Rb phosphorylation, it was reported that a constitutively active 4EBP-1, which suppresses eIF4E, caused a cell cycle arrest in MCF7 cells that correlated with decreased cyclin E-CDK2 activity and increased association of CDK2 with p27Kip1 [26]. It was also reported that rapamycin can suppress cyclin D levels in a manner that could be reversed with suppression of 4E-BP1 – indicating that the effects of rapamycin were mediated by suppression of 4E-BP1 phosphorylation [18]. While the concentrations of rapamycin used (100 nM) are not sufficient to suppress 4E-BP1 phosphorylation in most cells [7], the effect was only observed in MCF7 cells, which are much more sensitive to rapamycin than most cell lines [19]. However, if mTORC1 was suppressed by amino acid withdrawal, which inhibited 4E-BP1 phosphorylation, then cyclin D levels were reduced in HEK-293 cells that were resistant to 100 nM rapamycin [18]. Data provided here (Figs. 2 and 3) show that cyclin D levels and Rb phoaphorylation were sensitive to nano-molar levels of rapamycin in MCF7 cells (Fig. 3) and micro-molar levels in MDA-MB-231 cells (Fig. 2).
Since cyclin D-CDK4/6 mono-phosphorylates Rb, which is required for the phosphorylation of Rb by cyclin E/CDK2, [27] suppression of cyclin D levels would also suppress the ability of cyclin E/CDK2 to phosphorylate Rb. In that cyclin D-CDK4/6 complexes bind p27 and prevent it from binding to cyclin E-CDK2 where it is inhibitory, [27] reduced cyclin D levels could lead to elevated levels of p27 that could also contribute to the suppression of cyclin E/CDK2 activity. We demonstrated here that cyclin D1 levels were suppressed by doses of rapamycin needed to suppress 4E-BP1 phosphorylation. Thus, it is likely that the key effect of high dose rapamycin treatment on G1 cell cycle progression was to prevent eIF4E-dependent translation of cyclin D1 as was reported by Rosen and colleagues [28]. Consistent with this hypothesis, it was reported that rapamycin inhibited cyclin D1 expression at the level of translation [18, 28]. It is also of interest that rapamycin treatment was associated with decreased cyclin D1 levels in rapamycin-sensitive cells but not in rapamycin-resistant cells [29] – further supporting the importance of suppressing cyclin D1 for the efficacy of rapamycin as a suppressor of G1 cell cycle progression.
An intriguing finding here was that suppressing both TGF-β signaling and Rb expression did not result in cell death with high dose rapamycin treatment – as was observed with suppression of TGF-β signaling alone [9]. This finding suggests that rapamycin-induced suppression of Rb phosphorylation – and the sequestration of E2F family transcription factors – is responsible for the cell death observed with the high dose rapamycin treatment that inhibits 4E-BP1 phosphorylation. The data are consistent with a model whereby the lack of TGF-β signals allows cells to progress into S-phase, but with suppression of Rb phosphorylation, expression of E2F is suppressed and the genes needed for S-phase are not expressed. Since the cell cycle is not reversible at this point, the cells undergo apoptosis. Thus, Rb-null cancer cells are not likely to be vulnerable to therapeutic strategies that target mTOR.
Highlights.
Rapamycin-induced G1 cell cycle arrest is dependent on TGF-β.
Rapamycin-induced TGF-β signals requires nano-molar doses that suppress the phosphorylation of S6 kinase.
Complete G1 arrest requires micro-molar doses of rapamycin that suppress phosphorylation of 4E-BP1.
The effect of suppressing 4E-BP1 phosphorylation on G1 cell cycle progression is the inhibition of Rb phosphorylation.
This study reveals the integration of TGF-β signals with Rb signals to promote complete G1 cell cycle progression.
Acknowledgements
This study was supported by National Institute of Health grants R01-CA046677 and R01-CA179542 and a pilot project award from the Research Centers in Minority Institutions award RP-03037 from the National Center for Research Resources of the National Institute of Health.
Abbreviations
- 4E-BP1
eIF4E-binding protein-1
- CDK
cyclin-dependent kinase
- eIF4E
eukaryotic initiation factor 4E
- mTOR
mammalian target of rapamycin
- mTORC1
mTOR complex 1
- S6K
S6 kinase
- TGF-β
transforming growth factor-β
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
The authors declare no conflicts of interest.
References
- 1.Foster DA, Yellen P, Xu L, Saqcena M. Regulation of G1 cell cycle progression: Distinguishing the restriction point from a nutrient-sensing cell growth checkpoint(s) Genes Cancer. 2010;1:1124–1131. doi: 10.1177/1947601910392989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blagosklonny MV. Molecular damage in cancer: an argument for mTOR-driven aging. Aging. 2011;3:1130–1141. doi: 10.18632/aging.100422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hardt M, Chantaravisoot N, Tamanoi F. Activating mutations of TOR (target of rapamycin) Genes Cells. 2011;16:141–151. doi: 10.1111/j.1365-2443.2010.01482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fingar DC, Richardson CJ, Tee AR, Cheatham L, Tsou C, Blenis J. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mo Cell Biol. 2004;24:200–216. doi: 10.1128/MCB.24.1.200-216.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choo AY, Blenis J. Not all substrates are treated equally: implications for mTOR, rapamycin-resistance and cancer therapy. Cell Cycle. 2009;8:567–572. doi: 10.4161/cc.8.4.7659. [DOI] [PubMed] [Google Scholar]
- 7.Yellen P, Saqcena M, Salloum D, Feng J, Preda A, Xu L, Rodrik-Outmezguine V, Foster DA. High-dose rapamycin induces apoptosis in human cancer cells by dissociating mTOR complex 1 and suppressing phosphorylation of 4E-BP1. Cell Cycle. 2011;10:3948–3956. doi: 10.4161/cc.10.22.18124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci USA. 2008;105:17414–17419. doi: 10.1073/pnas.0809136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gadir N, Jackson DN, Lee E, Foster DA. Defective TGF-β signaling sensitizes human cancer cells to rapamycin. Oncogene. 2008;27:1055–1062. doi: 10.1038/sj.onc.1210721. [DOI] [PubMed] [Google Scholar]
- 10.Gadir N, Lee E, Garcia A, Toschi A, Foster DA. Suppression of TGF-β signaling by phospholipase D. Cell Cycle. 2007;6:2840–2845. doi: 10.4161/cc.6.22.4921. [DOI] [PubMed] [Google Scholar]
- 11.Yellen P, Chatterjee A, Preda A, Foster DA. Inhibition of S6 kinase suppresses the apoptotic effect of eIF4E ablation by inducing TGF-beta-dependent G1 cell cycle arrest. Cancer Lett. 2013;333:239–243. doi: 10.1016/j.canlet.2013.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toschi A, Lee E, Xu L, Garcia A, Gadir N, Foster DA. Regulation of mTORC1 and mTORC2 complex assembly by phosphatidic acid: competition with rapamycin. Mol Cell Biol. 2009;29:1411–1420. doi: 10.1128/MCB.00782-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Law BK, Chytil A, Dumont N, Hamilton EG, Waltner-Law ME, Aakre ME, Covington C, Moses HL. Rapamycin potentiates transforming growth factor β-induced growth arrest in nontransformed, oncogene-transformed, and human cancer cells. Mol Cell Biol. 2002;22:8184–8198. doi: 10.1128/MCB.22.23.8184-8198.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abdollah S, Macias-Silva M, Tsukazaki T, Hayashi H, Attisano L, Wrana JL. TbetaRI phosphorylation of Smad2 on Ser465 and Ser467 is required for Smad2–Smad4 complex formation and signaling. J Biol Chem. 1997;272:27678–27685. doi: 10.1074/jbc.272.44.27678. [DOI] [PubMed] [Google Scholar]
- 15.Massague J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saqcena M, Menon D, Patel D, Mukhopadhyay S, Chow V, Foster DA. Amino acids and mTOR mediate distinct metabolic checkpoints in mammalian G1 cell cycle. PLoS One. 2013;8:e74157. doi: 10.1371/journal.pone.0074157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sonenberg N. eIF4E, the mRNA cap-binding protein: from basic discovery to translational research. Biochem Cell Biol. 2008;86:178–183. doi: 10.1139/O08-034. [DOI] [PubMed] [Google Scholar]
- 18.Averous J, Fonseca BD, Proud CG. Regulation of cyclin D1 expression by mTORC1 signaling requires eukaryotic initiation factor 4E-binding protein 1. Oncogene. 2008;27:1106–1113. doi: 10.1038/sj.onc.1210715. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y, Zheng Y, Foster DA. Phospholipase D confers rapamycin resistance in human breast cancer cells. Oncogene. 2003;22:3937–3942. doi: 10.1038/sj.onc.1206565. [DOI] [PubMed] [Google Scholar]
- 20.Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 2001;294:1942–1945. doi: 10.1126/science.1066015. [DOI] [PubMed] [Google Scholar]
- 21.Inman GJ, Nicolas FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, Laping NJ, Hill CS. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- 22.Le Gendre O, Sookdeo A, Duliepre SA, Utter M, Frias M, Foster DA. Suppression of AKT phosphorylation restores rapamycin-based synthetic lethality in SMAD4-defective pancreatic cancer cells. Mol Cancer Res. 2013;11:474–481. doi: 10.1158/1541-7786.MCR-12-0679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho A, Dowdy SF. Regulation of G(1) cell-cycle progression by oncogenes and tumor suppressor genes. Curr Opin Genetics Dev. 2002;12:47–52. doi: 10.1016/s0959-437x(01)00263-5. [DOI] [PubMed] [Google Scholar]
- 24.Narasimha AM, Kaulich M, Shapiro GS, Choi YJ, Sicinski P, Dowdy SF. Cyclin D activates the Rb tumor suppressor by mono-phosphorylation. eLife. 2014:e02872. doi: 10.7554/eLife.02872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Florenes VA, Bhattacharya N, Bani MR, Ben-David Y, Kerbel RS, Slingerland JM. TGF-β mediated G1 arrest in a human melanoma cell line lacking p15INK4B: evidence for cooperation between p21Cip1/WAF1 and p27Kip1. Oncogene. 1996;13:2447–2457. [PubMed] [Google Scholar]
- 26.Jiang H, Coleman J, Miskimins R, Miskimins WK. Expression of constitutively active 4EBP-1 enhances p27Kip1 expression and inhibits proliferation of MCF7 breast cancer cells. Cancer Cell Int. 2003;3:2. doi: 10.1186/1475-2867-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 28.Muise-Helmericks RC, Grimes HL, Bellacosa A, Malstrom SE, Tsichlis PN, Rosen N. Cyclin D expression is controlled post-transcriptionally via a phosphatidylinositol 3-kinase/Akt-dependent pathway. J Biol Chem. 1998;273:29864–29872. doi: 10.1074/jbc.273.45.29864. [DOI] [PubMed] [Google Scholar]
- 29.Noh WC, Mondesire WH, Peng J, Jian W, Zhang H, Dong J, Mills GB, Hung MC, Meric-Bernstam F. Determinants of rapamycin sensitivity in breast cancer cells. Clin Cancer Res. 2004;10:1013–1023. doi: 10.1158/1078-0432.ccr-03-0043. [DOI] [PubMed] [Google Scholar]