Abstract
Cancer-testis (CT) antigens are a large family of genes that are selectively expressed in human testis germ cells, overexpressed in a variety of tumors and predominantly located on the X chromosome. To date, all known CT antigens are protein-coding genes. Here, we identify miR-888 as the first miRNA with features characteristic of a CT antigen. In a panel of 21 normal human tissues, miR-888 expression was high in testes and minimal or absent in all other examined tissues. In situ hybridization localized miR-888 expression specifically to the early stages of sperm development within the testes. Using The Cancer Genome Atlas database, we discovered that miR-888 was predominately expressed in endometrial tumors, with a significant association to high-grade tumors and increased percent invasion. In a separate panel of endometrial tumor specimens, we validated overexpression of miR-888 by real-time polymerase chain reaction. In addition, miR-888 expression was highest in endometrial carcinosarcoma, a rare and aggressive type of endometrial tumor. Moreover, we identified the progesterone receptor (PR), a potent endometrial tumor suppressor, as a direct target of miR-888. These data define miR-888 as the first miRNA CT antigen and a potential mediator of an aggressive endometrial tumor phenotype through down-regulation of PR.
Introduction
Cancer-testis (CT) antigens are a class of genes that are predominately expressed in the adult testes and are overexpressed in several types of tumors [1]. Within the testes, CT antigen expression localizes to the testicular germ cells termed the spermatogonia [1]. Because of their restricted expression in spermatogonia and the presence of a blood-testis barrier, expression of CT antigens in cancer often induces a tumor-directed immune response [1]. Consequently, CT antigens were historically identified through immunologic techniques such as T-cell epitope cloning and serological expression analysis of cDNA expression libraries [1]. More recently, CT antigens have been identified through analysis of expressed sequence tags for genes exclusively expressed in testes and cancer [1]. These strategies have led to the classification of more than 200 CT antigens (CT antigen database, http://www.cta.lncc.br), with new CT antigens continuing to be discovered [2]. Because their classification mainly relies on tissue expression patterns, the function and immunogenic potential of the majority of CT antigens remain unknown [3].
One of the most intriguing features of CT antigens is their predominant localization to the X chromosome. In fact, almost half of all CT antigens are encoded by the X chromosome [1], and approximately 10% of all protein-coding genes on the X chromosome are CT antigens [4] (Figure 1, bottom). These genes, termed CT-X antigens, recently evolved in eutherian mammals through gene translocation and duplication events to produce large multi-gene families [1]. At least 38 classified CT-X antigens are clustered near the end of the long arm of the X chromosome (Xq27-Xq28; Figure 1, top). Whereas all currently established CT-X antigens are protein-coding genes, this region also contains several reported miRNAs. In particular, genomic duplication events resulted in the evolution of the primate-specific miRNA gene family at Xq27.3 containing miR-888, miR-890, miR-891a, miR-891b, miR-892a, and miR-892b (Figure 1) [5]. The miR-888 gene family has restricted expression in testes, and miR-888 has recently been reported to be overexpressed in cancer [6,7]. However, the function of miR-888 in cancer has not been sufficiently studied [8,9].
Figure 1.
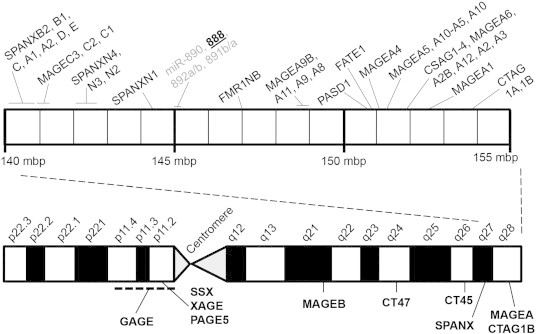
MiR-888 is part of a multicopy gene family on the X chromosome. Using the University of California Santa Cruz Genome Browser and the CT antigen database (http://www.cta.lncc.br), we generated a scaled diagram demonstrating the location of the major CT antigen gene families on the X chromosome (bottom). Within the Xq27-X28 region (approximately 145,000,000 million bp to 155,000,000 million bp), there are 38 classified CT antigens. The miR-888 gene (bold, underlined) is within this region at Xq27.3 and is part of a multicopy miRNA gene family that also includes miR-890, miR-891a/b, and miR-892a/b (gray).
In an analysis of miRNA expression patterns in uterine endometrial cancer (EC), we previously identified miR-888 as highly overexpressed [7]. EC is the fourth most common cancer in women and the most common gynecological malignancy [10]. While patient outcomes have improved for most cancers over the past 10 years, survival for EC patients has alarmingly decreased [10,11]. One of the most potent tumor suppressors in the endometrium is the progesterone receptor (PR), which activates gene expression to induce differentiation, cell cycle arrest, and apoptosis [12–14]. In addition, PR expression is often lost in advanced endometrial tumors [15–17]. Therefore, characterization of the different mechanisms by which PR expression is lost in EC can potentially improve our understanding on how aggressive ECs develop.
Our objective in this study was to determine whether miR-888 is a CT-X antigen and to understand its role in EC. MiR-888 is a primate-specific miRNA that evolved through gene translocation and duplication events on the X chromosome similar to other CT antigen genes. Here, we demonstrate that miR-888 expression is restricted to the testes and localizes to cells in the early stages of spermatogenesis. Using The Cancer Genome Atlas (TCGA) database, we found that miR-888 was most highly expressed in endometrial tumors with a significant association to high-grade tumors and increasing percent invasion. In addition, we describe a novel mechanism of PR inhibition in EC through miR-888. These data suggest that miR-888 functions in endometrial tumors to inhibit PR-mediated anti-proliferative signaling. We also suggest that miRNAs can potentially be classified as CT antigens, with miR-888 as the defining example.
Materials and Methods
Tissue Samples
Endometrial tissues were obtained under informed written consent from patients undergoing hysterectomy at the University of Iowa Hospitals and Clinics. The protocol was approved by the University of Iowa Institutional Review Board (Protocol No. 200209010). A total of 44 endometrial samples was collected, which included 9 samples of benign endometrium (BE), 18 endometrioid adenocarcinomas (EAs), 9 serous adenocarcinomas (SAs), and 8 carcinosarcomas (CSs; Table S1). Rhesus macaque testis samples were generously provided by Dr Jodi McBride of the Oregon National Primate Research Center (Beaverton, OR). In this study, animals were killed by sedation with ketamine followed by deep anesthesia with sodium pentobarbital and then exsanguination as previously reported [18]. Samples were collected under a protocol that was reviewed and approved by the Oregon Health & Science University Institutional Animal Care and Use Committee (Protocol No. IS00002803, “RNA interference therapy in a non-human primate model of Huntington's disease”) as previously reported [18].
Endometrial tissues were obtained under informed written consent from patients undergoing hysterectomy at the University of Iowa Hospitals and Clinics. The protocol was approved by the University of Iowa Institutional Review Board (Protocol No. 200209010). A total of 44 endometrial samples was collected, which included 9 samples of benign endometrium (BE), 18 endometrioid adenocarcinomas (EAs), 9 serous adenocarcinomas (SAs), and 8 carcinosarcomas (CSs; Table S1). Rhesus macaque testis samples were generously provided by Dr Jodi McBride of the Oregon National Primate Research Center (Beaverton, OR). In this study, animals were killed by sedation with ketamine followed by deep anesthesia with sodium pentobarbital and then exsanguination as previously reported [18]. Samples were collected under a protocol that was reviewed and approved by the Oregon Health & Science University Institutional Animal Care and Use Committee (Protocol No. IS00002803, “RNA interference therapy in a non-human primate model of Huntington's disease”) as previously reported [18].
Cell Lines
Six EC cell lines (AN3CA, RL95-2, Hec1A, SK-UT-1B, ECC-1, and KLE), five breast cancer cell lines (MCF7, MDA-MB-231, MDA-MB-453, SKBR3, and T-47D), nine ovarian cancer cell lines (ES-2, Caov-3, Caov-4, OV-90, OVCAR-3, SK-OV-3, SW 626, TOV-112D, and UWB1.298), and three prostate cancer cell lines (DU-145, LNCaP and PC-3) were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and were cultured according to their guidelines. Two EC cell lines, Ishikawa H [19] and the Hec50co subline [20] of the Hec50 cell line [21], were gifts from Dr Erlio Gurpide of New York University and were grown in Dulbecco's modified Eagle's medium supplemented with 10% FBS and 1% penicillin-streptomycin (Life Technologies, Carlsbad, CA) as we previously reported [20]. The Alva-31 cell line [22] was a gift from Dr Thomas Griffith of the University of Iowa and was grown in RPMI 1640 supplemented with 10% FBS and 1% penicillin-streptomycin (Life Technologies). It has previously been reported that the ECC-1 EC cell line originated from the 3-H-12 Ishikawa EC cell subline [23]. Short tandem repeat genotyping was performed on all of our cell lines and confirmed that our Ishikawa cells are from the 3-H-4 subline and that our ECC-1 cells are from the 3-H-12 subline. While these cell lines share a common ancestor, they have distinct genomic profiles and are used in this manuscript to demonstrate data replication within different cellular backgrounds.
RNA Samples
A panel of RNA from 20 different human tissues was obtained from Life Technologies, with each sample containing pooled RNA from three individuals. To this panel, we added our own RNA sample of pooled BE from three patients.
TCGA Data Analysis
Clinical and miR-888 expression data for 450 uterine corpus EC (UCEC) specimens were downloaded from TCGA data portal (National Cancer Institute and National Human Genome Research Institute, accessed 31 May 2013), and clinical characteristics are summarized in Table S2. A full list of individual miR-888 expression values in reads per kilobase per million (RPKM) and clinical information by patient barcode are available in Table S3. MiR-888 expression data were also obtained for a panel of cancers using the University of Iowa Institute for Clinical and Translation Science Compass website (https://research.icts.uiowa.edu/compass/, accessed 28 February 2014). MiR-888 expression data were downloaded for breast invasive carcinoma (BRCA, n = 380), colon adenocarcinoma (COAD, n = 394), lung adenocarcinoma (LUAD, n = 407), ovarian serous cystadenocarcinoma (OV, n = 476), pancreatic adenocarcinoma (PAAD, n = 43), prostate adenocarcinoma (PRAD, n = 187), UCEC (n = 363), and uterine CS (UCS, n = 56), and individual miR-888 expression values in RPKM for each cancer are provided in Table S4.
Clinical and miR-888 expression data for 450 uterine corpus EC (UCEC) specimens were downloaded from TCGA data portal (National Cancer Institute and National Human Genome Research Institute, accessed 31 May 2013), and clinical characteristics are summarized in Table S2. A full list of individual miR-888 expression values in reads per kilobase per million (RPKM) and clinical information by patient barcode are available in Table S3. MiR-888 expression data were also obtained for a panel of cancers using the University of Iowa Institute for Clinical and Translation Science Compass website (https://research.icts.uiowa.edu/compass/, accessed 28 February 2014). MiR-888 expression data were downloaded for breast invasive carcinoma (BRCA, n = 380), colon adenocarcinoma (COAD, n = 394), lung adenocarcinoma (LUAD, n = 407), ovarian serous cystadenocarcinoma (OV, n = 476), pancreatic adenocarcinoma (PAAD, n = 43), prostate adenocarcinoma (PRAD, n = 187), UCEC (n = 363), and uterine CS (UCS, n = 56), and individual miR-888 expression values in RPKM for each cancer are provided in Table S4.
RNA and Protein Extraction from Cells and Tissues
Tissue samples were immersed in RNAlater ICE (Life Technologies) and incubated at − 20°C for at least 16 hours before RNA extraction. The mirVana miRNA Isolation Kit (Life Technologies) was used to isolate total RNA from tissues according to the manufacturer’s instructions. For cultured cells, RNA and protein were isolated using the mirVana PARIS kit (Life Technologies).
Quantitative Reverse Transcription–Polymerase Chain Reaction
Taqman primer/probe sets for hsa-miR-888 and RNU48 (Life Technologies) were used to perform quantitative reverse transcription–polymerase chain reaction (RT-PCR) on 200 ng of RNA using the Taqman Reverse Transcription Kit followed by the 2 × PCR Master Mix with No AmpERASE UNG according to the manufacturer’s instructions (Life Technologies). For PR quantification, primers for PR (5′-ATGTGGCAGATCCCACAGGAGTTT-3′, 5′-ACTGGGTTTGACTTCGTAGCCCTT-3′) and 18S (5′-AACTTTCGATGGTAGTCGCCG-3′, 5′-CCTTGGATGTGGTAGCCGTTT-3′) were designed using the PrimerQuest software from Integrated DNA Technologies (Coralville, IA) and oligonucleotides were ordered from Integrated DNA Technologies. RT was carried out using the SuperScript III First Strand Synthesis System (Life Technologies) and PCR was performed using the 2 × Power SYBR Green PCR Master Mix (Life Technologies) following the manufacturer’s protocol. Quantitative PCR reactions were completed on the Applied Biosystems 7900HT Fast Real-Time PCR System. Fold change was calculated using the Comparative Ct Method (2− ΔΔCt) with RNU48 as the internal control for miR-888 and 18S as the internal control for PR. For the normal tissues (Figure 2A) and cell lines (Figure S1), data are reported as 2− ΔCt × 10,000 because there was no intuitive negative control group to use for fold change comparisons.
Figure 2.
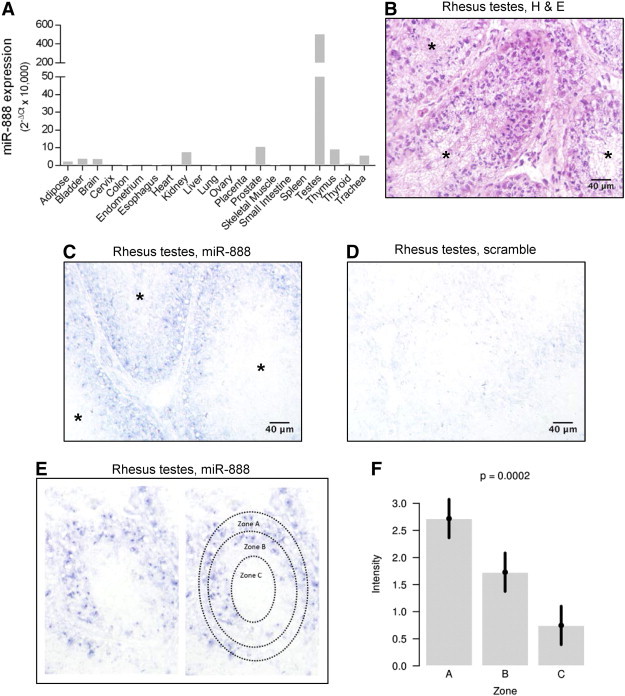
MiR-888 is expressed in the testes during the early stages of spermatogenesis. (A) MiR-888 expression was measured by quantitative RT-PCR in pooled RNA from 21 normal human tissues, and gene expression is reported as 2− ΔCt × 10,000. MiR-888 showed high expression in the testes and a low-level expression in all other tissues examined. In situ hybridization was performed on serial tissue sections of rhesus macaque testes using a probe complementary to miR-888 (C) or a scrambled probe (D) as a negative control. In B, an H&E stain of a serial section of the same testis tissue sample is shown for comparison. In B and C, individual seminiferous tubules are designated with an asterisk (*). (E) MiR-888 staining intensity was scored from 0 to 3 in three self-defined zones of the seminiferous tubule: zone A = spermatogonia, primary spermatocytes, Sertoli cells; zone B = spermatids; zone C = spermatozoa. (F) MiR-888 staining in each zone was analyzed for five images from three different biologic testis samples using a repeated measures ANOVA model. Staining intensity was significantly greater in zone A relative to zones B and C; error bars represent 95% confidence intervals.
Figure S1.
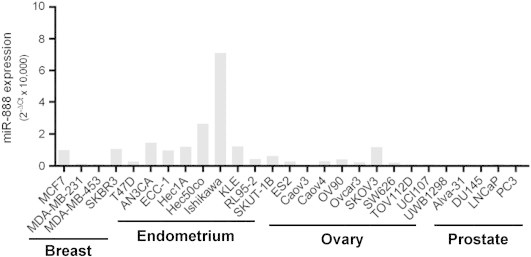
MiR-888 expression in EC cell lines.
Taqman primer/probe sets for hsa-miR-888 and RNU48 (Life Technologies) were used to perform quantitative reverse transcription–polymerase chain reaction (RT-PCR) on 200 ng of RNA using the Taqman Reverse Transcription Kit followed by the 2 × PCR Master Mix with No AmpERASE UNG according to the manufacturer’s instructions (Life Technologies). For PR quantification, primers for PR (5′-ATGTGGCAGATCCCACAGGAGTTT-3′, 5′-ACTGGGTTTGACTTCGTAGCCCTT-3′) and 18S (5′-AACTTTCGATGGTAGTCGCCG-3′, 5′-CCTTGGATGTGGTAGCCGTTT-3′) were designed using the PrimerQuest software from Integrated DNA Technologies (Coralville, IA) and oligonucleotides were ordered from Integrated DNA Technologies. RT was carried out using the SuperScript III First Strand Synthesis System (Life Technologies) and PCR was performed using the 2 × Power SYBR Green PCR Master Mix (Life Technologies) following the manufacturer’s protocol. Quantitative PCR reactions were completed on the Applied Biosystems 7900HT Fast Real-Time PCR System. Fold change was calculated using the Comparative Ct Method (2− ΔΔCt) with RNU48 as the internal control for miR-888 and 18S as the internal control for PR. For the normal tissues (Figure 2A) and cell lines (Figure S1), data are reported as 2− ΔCt × 10,000 because there was no intuitive negative control group to use for fold change comparisons.
miRNA In Situ Hybridization
In situ hybridization was performed using double-digoxigenin (DIG) labeled 2′O-methyl locked nucleic acid (LNA)-ZEN probes (Integrated DNA Technologies) complimentary to miR-888 (5′-DIG-U/ZEN/GACUGACAGCUTUUUGAGU/ZEN/A-DIG-3′) along with a scrambled negative control probe (5′-DIG-C/ZEN/GUAUUAUAGCCGAUUAACG/ZEN/A-DIG-3′), where LNA modifications are underlined. Hematoxylin and eosin (H&E) staining was performed using the Sakura Finetek DRS 601 automatic slide stainer (Sakura Finetek USA Inc, Torrance, CA). All in situ hybridization and H&E stains were performed on adjacent serial sections of the same tissue sample. In situ hybridization was performed following the protocol previously described by McLoughlin et al. with the following modifications [24]. Tissue sections were incubated with ZEN-LNA probes overnight at a hybridization temperature of 55°C and a final concentration of 1 μM. All samples were incubated with an alkaline phosphatase-conjugated antibody to DIG (Roche, San Francisco, CA) at a dilution of 1:1000 overnight at 4°C. All images were taken using the same microscope settings and differential interference contrast (Leica Microsystems, Wetzlar, Germany). Each image background was then edited to white using Adobe Photoshop (San Jose, CA).
miRNA In Situ Hybridization Intensity Scoring
Each seminiferous tubule was divided into three zones: A, B, and C. Zone A represents the outer edge of the tubule in which the Sertoli cells, spermatogonia, and primary spermatocytes are located, zone B represents the region that contained spermatids and zone C represents the lumen where mature sperm are located. For intensity scoring, 0 = no blue staining, 1 = scant blue staining, 2 = moderate blue staining and 3 = abundant blue staining. Scoring was blinded and performed by pathologist Katherine Gibson-Corey, PhD, of the University of Iowa. Each image contained one seminiferous tubule. Scoring was performed for miR-888 staining in three rhesus macaque testis samples with five images from each of three tissue sections.
MiR-888 Transfections
Cells were plated into phenol-red free medium with 10% charcoal-stripped FBS (Life Technologies) to reduce the presence of hormones. ECC-1 or Ishikawa cells were transfected with a plasmid encoding miR-888 (Origene, Rockville, MD) or the empty vector (EV) control (Origene) using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions. At 4 hours post-transfection, cells were treated with 5 nM estradiol (Sigma, St. Louis, MO) to induce PR expression. Cells were collected at 24 hours post-transfection for RNA and protein isolation and analysis.
Western Blot
A rabbit monoclonal antibody against PR isoform A/PR isoform B (PRA/PRB, #3153; Cell Signaling Technology, Danvers, MA) was combined with a rabbit monoclonal antibody against PRB (#3157; Cell Signaling Technology) and both were diluted 1:1000. The mouse monoclonal β-actin antibody (Sigma #A1978) was used as a loading control for all Western blots at a dilution of 1:10,000. The National Institutes of Health ImageJ program was used to perform densitometry quantification and data were normalized to the mock-transfected controls.
Molecular Cloning
An approximately 500-bp region surrounding each of the four miR-888 binding sites in the PR 3′UTR was amplified from ECC-1 genomic DNA using the Platinum PCR SuperMix, High Fidelity (Life Technologies; primer sequences in Table S5). PCR products were purified using the PCR Purification Kit (Qiagen, Valencia, CA) and subcloned into the pGEM-T Easy vector (Promega, Madison, WI). Insert sequences were subsequently cloned into the psiCHECK2 vector and transformed into TOP10 OneShot competent E. coli (Life Technologies).
An approximately 500-bp region surrounding each of the four miR-888 binding sites in the PR 3′UTR was amplified from ECC-1 genomic DNA using the Platinum PCR SuperMix, High Fidelity (Life Technologies; primer sequences in Table S5). PCR products were purified using the PCR Purification Kit (Qiagen, Valencia, CA) and subcloned into the pGEM-T Easy vector (Promega, Madison, WI). Insert sequences were subsequently cloned into the psiCHECK2 vector and transformed into TOP10 OneShot competent E. coli (Life Technologies).
Dual Luciferase Assays
ECC-1 or Ishikawa cells were transfected with miR-888 or the EV in combination with each individual psiCHECK2 luciferase vector. Cells were collected at 24 hours post-transfection, and Firefly and Renilla luciferase activities were measured using the Dual Luciferase Assay System (Promega) according to the manufacturer’s protocol. Renilla luciferase activity was divided by Firefly luciferase activity and normalized to the EV control.
Statistical Analysis
For the in situ hybridization semiquantification, a repeated measures analysis of variance (ANOVA) model was used to analyze the data, because multiple images of each sample were scored. Due to the highly skewed nature of miR-888 expression in TCGA samples, figures are expressed on the square-root scale and observations below the limit of detection are jittered slightly for display purposes. For testing miR-888 expression levels across cancer types from TCGA data, Wilcoxon rank sum tests were used. Penalized regression splines were used for the nonparametric regression model presented in Figure 3B [25]. PCR data involving miR-888 and PR expression, Western blot data, and luciferase assays were all analyzed using ANOVA models. Linear regression was used to assess the significance of the correlation between miR-888 and PR mRNA expression in EC tissue. All miR-888 and luciferase transfections were performed in triplicate on three separate occasions to produce nine individual replicates per experimental condition for statistical analysis.
Figure 3.
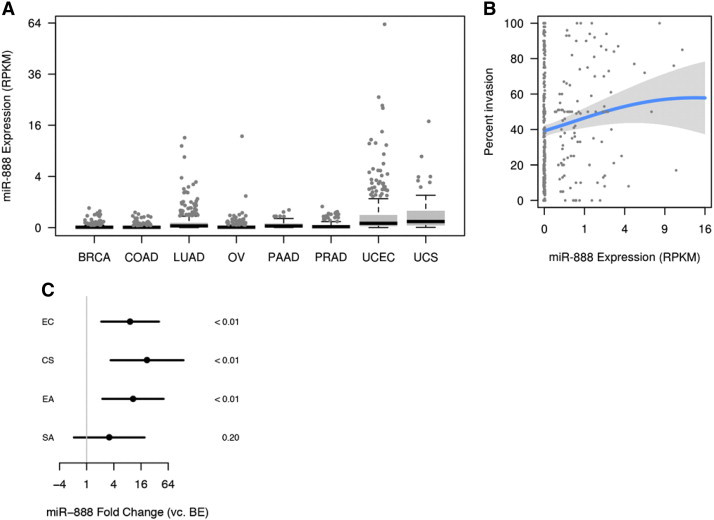
MiR-888 is selectively expressed in aggressive endometrial tumors. (A) MiR-888 deep sequencing expression data in RPKM were downloaded from TCGA for BRCA (n = 380), COAD (n = 394), LUAD (n = 407), OV (n = 476), PAAD (n = 43), PRAD (n = 187), UCEC (n = 363), and UCS (n = 56). MiR-888 expression was significantly greater in UCEC and UCS compared to all other cancers investigated (P < .05, Wilcoxon rank sum test). In A and B, undetectable expression levels are jittered slightly to avoid overplotting at 0. (B) Percent invasion data were available for 396 UCEC tumor specimens and miR-888 RPKM correlated with increasing percent invasion. Penalized regression splines were used to estimate the nonlinear relationship between miR-888 expression and percent invasion (overall association was significant, P = .03). (C) miR-888 expression was measured by quantitative RT-PCR in a group of nine BE and 35 EC primary tumors that included 18 EAs, 9 SAs, and 9 CSs. MiR-888 was 9-fold overexpressed in EC as a whole (P < .01), 11-fold overexpressed in EA (P < .01), 3-fold overexpressed in SA (P = .20), and 22-fold overexpressed in CS (P < .01); bars represent 95% confidence intervals.
Results
MiR-888 is Selectively Expressed in the Testes During the Early Stages of Spermatogenesis
Previously reported deep sequencing data have shown that miR-888 expression is restricted to the testes [6]. To confirm these analyses, we obtained pooled RNA samples from 21 different normal adult human tissues and quantified miR-888 expression through quantitative RT-PCR (Figure 2A). As expected, miR-888 demonstrated exceptionally high expression in the testes and minimal or no detectable expression in all other tissues investigated (Figure 2A). The majority of tissues had very low miR-888 expression [raw cycle threshold (Ct) value of 30–40]. The kidney, prostate, and thymus were the only other tissues that had Ct values less than 30 (Ct ≈ 28–30), but their expression was minimal compared to that observed in the testes (raw Ct = 22). To determine the distribution of miR-888 expression within the testes, we performed miRNA in situ hybridizations on rhesus macaque testis tissue from three individual monkeys. H&E staining was performed on a serial section for tissue structure comparison (Figure 2B). The in situ hybridizations were performed using a probe complimentary to miR-888 (Figure 2C) or a scrambled probe (Figure 2D) as a negative control. miR-888 expression consistently localized to the outer edge of the seminiferous tubule (Figure 2C).
Spermatogenesis occurs within the seminiferous tubules of the testes. As the germ cells mature, they move towards the center of the tubule. To assess miR-888 expression during sperm maturation, the seminiferous tubule was divided into three different zones and scored for miR-888 intensity by a trained pathologist blinded to sample identity (Figure 2E). Zone A encompasses the outer edge of the seminiferous tubule where spermatogonia, primary spermatocytes, and Sertoli cells are located. Zone B represents the region that typically contains the maturing spermatids, and zone C represents the lumen of the testes where mature spermatozoa are located. Using a staining intensity range of 0–3, zone A consistently showed the highest level of staining (Figure 2F). Thus, miR-888 expression predominantly localized to the early stages of spermatogenesis within the seminiferous tubule of the testes.
MiR-888 Is Selectively Overexpressed in Endometrial Tumors
To investigate miR-888 expression in cancer, we used TCGA database and obtained miR-888 expression for a panel of carcinomas that included BRCA (n = 380), COAD (n = 394), LUAD (n = 407), OV (n = 476), PAAD (n = 43), PRAD (n = 187), UCEC (n = 363), and UCS (n = 56; Table S4). MiR-888 expression was significantly elevated in UCEC and UCS compared to all other cancers (P < .05 for all pairwise comparisons involving UCEC and UCS; the two types were not, however, significantly different from each other). Detectable expression of miR-888 was only observed in a minority of specimens from other cancers (Figure 3A). We also investigated miR-888 expression across a panel of cancer cell lines originating from breast, endometrial, ovarian, and prostate tumors and found that miR-888 levels were highest in EC cell lines (Figure S1), albeit low in comparison to testes (Figure 2A) and individual miR-888 expressing endometrial tumors (Figure S2). The cell line data support TCGA results in that miR-888 expression was highest in EC. MiR-888 expression in endometrial tumors from TCGA data was heavily skewed, with 65% of samples having undetectable expression of miR-888 (Figure 3A and Table S2). Such a specific expression pattern is characteristic of CT antigens, which are not endogenously expressed in the tumor tissue of origin [3]. UCEC tumors that had positive expression of miR-888 were substantially more likely to be grade 3 tumors than UCEC tumors with undetectable miR-888 levels (Table 1, P = .001). Furthermore, miR-888 expression in UCEC tumors showed a positive association with percent invasion of the tumor (Figure 3B). Therefore, miR-888 expression in endometrial tumors correlates with a more aggressive tumor phenotype and has an expression pattern in cancer similar to other representative CT antigens.
Figure S2.
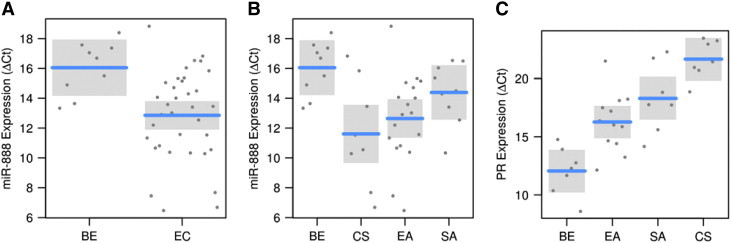
ΔCt values for miR-888 and PR quantification in EC tissues.
Table 1.
MiR-888 Expression by Tumor Grade
| Grade | Percentage of Samples with Nonzero miR-888 Expression |
|---|---|
| G1 | 17% |
| G2 | 24% |
| G3 | 36%⁎ |
P = .001.
To investigate miR-888 expression in cancer, we used TCGA database and obtained miR-888 expression for a panel of carcinomas that included BRCA (n = 380), COAD (n = 394), LUAD (n = 407), OV (n = 476), PAAD (n = 43), PRAD (n = 187), UCEC (n = 363), and UCS (n = 56; Table S4). MiR-888 expression was significantly elevated in UCEC and UCS compared to all other cancers (P < .05 for all pairwise comparisons involving UCEC and UCS; the two types were not, however, significantly different from each other). Detectable expression of miR-888 was only observed in a minority of specimens from other cancers (Figure 3A). We also investigated miR-888 expression across a panel of cancer cell lines originating from breast, endometrial, ovarian, and prostate tumors and found that miR-888 levels were highest in EC cell lines (Figure S1), albeit low in comparison to testes (Figure 2A) and individual miR-888 expressing endometrial tumors (Figure S2). The cell line data support TCGA results in that miR-888 expression was highest in EC. MiR-888 expression in endometrial tumors from TCGA data was heavily skewed, with 65% of samples having undetectable expression of miR-888 (Figure 3A and Table S2). Such a specific expression pattern is characteristic of CT antigens, which are not endogenously expressed in the tumor tissue of origin [3]. UCEC tumors that had positive expression of miR-888 were substantially more likely to be grade 3 tumors than UCEC tumors with undetectable miR-888 levels (Table 1, P = .001). Furthermore, miR-888 expression in UCEC tumors showed a positive association with percent invasion of the tumor (Figure 3B). Therefore, miR-888 expression in endometrial tumors correlates with a more aggressive tumor phenotype and has an expression pattern in cancer similar to other representative CT antigens.
To validate TCGA miR-888 expression data, we obtained RNA from 9 benign endometrial tissues and 35 endometrial tumor specimens that included 18 EAs, 9 SAs, and 8 CSs (Table S1). MiR-888 was nine-fold overexpressed in endometrial tumor specimens compared to BE with a P value < .01 (Figures 3C and S2). Within endometrial tumor subtypes, miR-888 was significantly overexpressed in EA (11-fold, P < .01) and most predominantly overexpressed in CS (22-fold, P < .01). Endometrial CS is a rare but very aggressive form of EC that has a very poor prognosis and limited treatment options [26]. In contrast, EA often presents at an early stage and grade and can be treated by surgical removal alone [27]. Thus, these data suggest that miR-888 is associated with an aggressive tumor phenotype.
To validate TCGA miR-888 expression data, we obtained RNA from 9 benign endometrial tissues and 35 endometrial tumor specimens that included 18 EAs, 9 SAs, and 8 CSs (Table S1). MiR-888 was nine-fold overexpressed in endometrial tumor specimens compared to BE with a P value < .01 (Figures 3C and S2). Within endometrial tumor subtypes, miR-888 was significantly overexpressed in EA (11-fold, P < .01) and most predominantly overexpressed in CS (22-fold, P < .01). Endometrial CS is a rare but very aggressive form of EC that has a very poor prognosis and limited treatment options [26]. In contrast, EA often presents at an early stage and grade and can be treated by surgical removal alone [27]. Thus, these data suggest that miR-888 is associated with an aggressive tumor phenotype.
To determine what cell types expressed miR-888 in endometrial tumors, we performed miRNA in situ hybridization on EA and CS samples (EA38 and CS114; Table S1) that had particularly high expression of miR-888. H&E staining for the EA and CS samples was performed on adjacent tissue sections as a reference (Figure 4, A and B). In EA, miR-888 expression localized to the cancerous glandular epithelium (Figure 4C), with minimal staining in the surrounding stroma and the absence of staining observed for the scrambled negative control probe (Figure 4D). CS is unique in that it contains both carcinomatous and sarcomatous components within a single tumor. It is hypothesized that the cancer was originally an endometrial carcinoma that dedifferentiated to form sarcoma-like cells [28]. MiRNA in situ hybridization of an endometrial CS revealed that miR-888 expression remained in the carcinomatous portion of the tumor (Figure 4, E and F). Interestingly, the carcinomatous portion of the tumor is typically the cell type functionally involved in tumor invasion and metastasis [28]. These data demonstrate that miR-888 expression is restricted to the cancerous glandular epithelium of endometrial tumors and points to the role of miR-888 in cancer cell signaling pathways.
Figure 4.
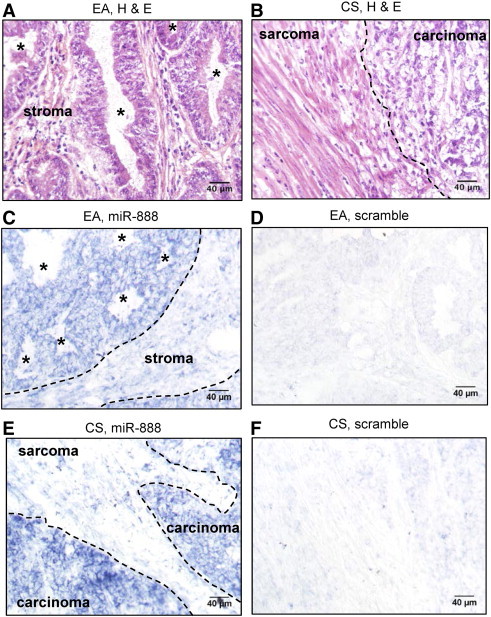
MiR-888 expression localizes to the cancerous epithelium. Serial tissue sections were cut from the EA38 EA tissue (A, C, and D) and the CS114 CS tissue (B, E, and F; see Table S1 for tissue sample details). These tissue samples were chosen for their high expression of miR-888 demonstrated by quantitative RT-PCR. For comparison, H&E-stained sections are shown in A and B. MiRNA in situ hybridization was performed using a probe complimentary to miR-888 (C and E) or a scrambled negative control probe (D and F). For C, the border between the cancerous epithelium and adjacent normal stromal is designated by a dotted line. For B and E, the border between the carcinomatous and sarcomatous elements is designated by a dotted line. In A and C, endometrial glands are designated by an asterisk (*).
To determine what cell types expressed miR-888 in endometrial tumors, we performed miRNA in situ hybridization on EA and CS samples (EA38 and CS114; Table S1) that had particularly high expression of miR-888. H&E staining for the EA and CS samples was performed on adjacent tissue sections as a reference (Figure 4, A and B). In EA, miR-888 expression localized to the cancerous glandular epithelium (Figure 4C), with minimal staining in the surrounding stroma and the absence of staining observed for the scrambled negative control probe (Figure 4D). CS is unique in that it contains both carcinomatous and sarcomatous components within a single tumor. It is hypothesized that the cancer was originally an endometrial carcinoma that dedifferentiated to form sarcoma-like cells [28]. MiRNA in situ hybridization of an endometrial CS revealed that miR-888 expression remained in the carcinomatous portion of the tumor (Figure 4, E and F). Interestingly, the carcinomatous portion of the tumor is typically the cell type functionally involved in tumor invasion and metastasis [28]. These data demonstrate that miR-888 expression is restricted to the cancerous glandular epithelium of endometrial tumors and points to the role of miR-888 in cancer cell signaling pathways.
PR Expression in EC
In the same subset of tumor tissues used for miR-888 quantification, we also quantified PR mRNA expression (7 benign samples of endometrium, 12 EAs, 7 SAs, and 7 CSs; Table S1; samples not used did not have enough remaining RNA). As has been reported previously [16,29], we observed a loss of PR expression in endometrial tumors compared to BE (Figures 5A and S2). PR expression was 20-fold decreased in EA (P < .001) and 100-fold decreased in SA relative to BE (P < .0001). In CS, PR expression was essentially absent (1000-fold decreased, P < .0001; Figure 5A). Linear regression demonstrated a statistically significant negative correlation between miR-888 and PR mRNA expression (R = − 0.579, P value < .001; Figure 5B). In particular, this implies that a five-fold increase in miR-888 concentration is associated with a three-fold decrease in PR concentration (95% confidence interval: 1.7 to 5.3).
Figure 5.
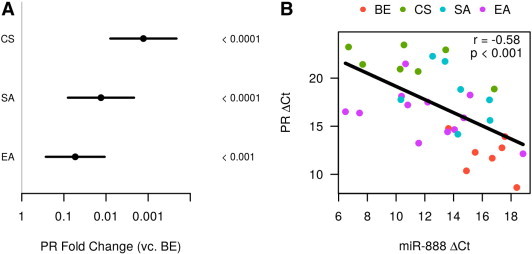
PR mRNA expression negatively correlates with miR-888 expression. (A) PR mRNA was measured by quantitative RT-PCR in 7 BEs, 12 EAs, 7 SAs and 7 CSs. PR mRNA was significantly decreased in all three types of EC; bars represent 95% confidence intervals and P values are displayed to the right. (B) A linear regression was performed using the PR mRNA and miR-888 ΔCt values and a significant negative correlation was observed with a correlation coefficient of R = − 0.58 and P < .001. Cancer subtype is indicated by different colored data points.
In the same subset of tumor tissues used for miR-888 quantification, we also quantified PR mRNA expression (7 benign samples of endometrium, 12 EAs, 7 SAs, and 7 CSs; Table S1; samples not used did not have enough remaining RNA). As has been reported previously [16,29], we observed a loss of PR expression in endometrial tumors compared to BE (Figures 5A and S2). PR expression was 20-fold decreased in EA (P < .001) and 100-fold decreased in SA relative to BE (P < .0001). In CS, PR expression was essentially absent (1000-fold decreased, P < .0001; Figure 5A). Linear regression demonstrated a statistically significant negative correlation between miR-888 and PR mRNA expression (R = − 0.579, P value < .001; Figure 5B). In particular, this implies that a five-fold increase in miR-888 concentration is associated with a three-fold decrease in PR concentration (95% confidence interval: 1.7 to 5.3).
MiR-888 Inhibits the PR in EC
Given the significant association between miR-888 and PR mRNA levels, we next analyzed the 3′UTR of PR for potential binding sites for miR-888 using TargetScanHuman 6.2 (www.targetscan.org). Similar to other steroid hormone receptors, PR has a very long 3′UTR (≈ 10 kilobases) with several AU-rich elements (AREs; Figure 6A, stars), suggesting a very complex post-transcriptional regulatory mechanism. Intriguingly, the PR 3′UTR contained four binding sites for miR-888 in its 3′UTR (Figure 6A, triangles), with the first three sites having 7mer-m8 complementarity and the last site having 8mer complementarity (Figure 6F). To investigate the role of miR-888 regulation of PR, we chose to use the ECC-1 and Ishikawa EC cells because they had a low level of miR-888 expression (Figure S1) and are positive for PR expression [20,30]. The ECC-1 and Ishikawa EC cell lines were transfected with a plasmid containing the miR-888 gene (miR-888) or an EV as a negative control, and miR-888 expression was confirmed by quantitative RT-PCR (Figure S3). Mock transfections were performed as a negative control and were used for data normalization. At 4 hours post-transfection, cells were treated with 5 nM estradiol to induce PR expression. Protein was isolated at 24 hours post-transfection, and Western blot analysis was performed for PR isoforms PRA and PRB. Only PRB was predominantly expressed in ECC-1 and Ishikawa cells, and miR-888 overexpression was capable of reducing PRB at the protein level in both cell lines (Figure 6, B and C). Western blots were quantified by densitometry using ImageJ software, and a significant decrease in PR protein was observed with miR-888 transfection, but not EV transfection, relative to the mock-transfected controls in the ECC-1 (Figure 6D) and Ishikawa (Figure 6E) cell lines.
Figure 6.
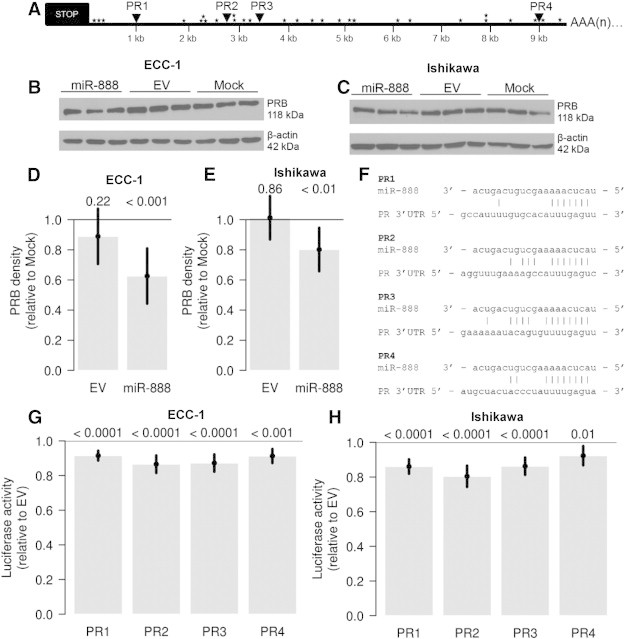
MiR-888 directly targets the PR. (A) PR contains four miR-888 binding sites in its 3′UTR (PR1-PR4, triangles) along with several AREs (stars). Expression of miR-888 was capable of decreasing PR expression at the protein level in both ECC-1 (B) and Ishikawa (C) EC cell lines. Western blots were quantified by densitometry, and PR protein was significantly decreased by miR-888, but not EV transfection, relative to the mock-transfected controls (D: ECC-1, E: Ishikawa). In F, the individual miR-888 binding sites in the PR 3′UTR are aligned with the miR-888 miRNA nucleotide sequence and nucleotide base interactions are denoted with vertical lines. ECC-1 (G) or Ishikawa (H) cells were transfected with luciferase reporters containing each PR 3′UTR miR-888 binding site (PR1-PR4) downstream of Renilla luciferase in combination with miR-888 or the EV control. MiR-888 expression resulted in a significant decrease in luciferase activity compared to the EV control for all four miR-888 binding sites (G: ECC-1, H: Ishikawa). For D, E, G, and H, statistical significance was determined using ANOVA models and error bars represent 95% confidence intervals. P values are listed above each bar graph.
Figure S3.
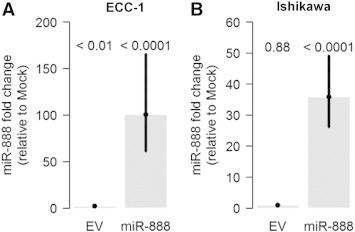
Transfection of ECC-1 and Ishikawa cells with the miR-888 plasmid results in high expression of miR-888.
Given the significant association between miR-888 and PR mRNA levels, we next analyzed the 3′UTR of PR for potential binding sites for miR-888 using TargetScanHuman 6.2 (www.targetscan.org). Similar to other steroid hormone receptors, PR has a very long 3′UTR (≈ 10 kilobases) with several AU-rich elements (AREs; Figure 6A, stars), suggesting a very complex post-transcriptional regulatory mechanism. Intriguingly, the PR 3′UTR contained four binding sites for miR-888 in its 3′UTR (Figure 6A, triangles), with the first three sites having 7mer-m8 complementarity and the last site having 8mer complementarity (Figure 6F). To investigate the role of miR-888 regulation of PR, we chose to use the ECC-1 and Ishikawa EC cells because they had a low level of miR-888 expression (Figure S1) and are positive for PR expression [20,30]. The ECC-1 and Ishikawa EC cell lines were transfected with a plasmid containing the miR-888 gene (miR-888) or an EV as a negative control, and miR-888 expression was confirmed by quantitative RT-PCR (Figure S3). Mock transfections were performed as a negative control and were used for data normalization. At 4 hours post-transfection, cells were treated with 5 nM estradiol to induce PR expression. Protein was isolated at 24 hours post-transfection, and Western blot analysis was performed for PR isoforms PRA and PRB. Only PRB was predominantly expressed in ECC-1 and Ishikawa cells, and miR-888 overexpression was capable of reducing PRB at the protein level in both cell lines (Figure 6, B and C). Western blots were quantified by densitometry using ImageJ software, and a significant decrease in PR protein was observed with miR-888 transfection, but not EV transfection, relative to the mock-transfected controls in the ECC-1 (Figure 6D) and Ishikawa (Figure 6E) cell lines.
To validate PR as a direct target of miR-888 regulation, we cloned 500-bp regions surrounding each PR 3′UTR miR-888 binding site (Figure 6F) into the psiCHECK2 vector downstream of Renilla luciferase. ECC-1 and Ishikawa cells were transfected with the miR-888 plasmid in combination with each PR-psiCHECK2 vector (sites PR1-PR4; Figure 6F), and luciferase activity was measured 24 hours post-transfection. Renilla luciferase activity was normalized to Firefly luciferase activity and data are reported relative to the EV transfection (Figures 6, G and H). Expression of miR-888 caused a significant reduction in luciferase activity of approximately 10% to 20% for all four binding sites in both cell lines (Figures 6, G and H). Therefore, these data support that PR is a direct target of miR-888 regulation in EC cells.
Discussion
Here, we describe for the first time a miRNA, miR-888, that embodies the cardinal characteristics of a CT antigen. This is the first noncoding RNA to meet these criteria. Though CT antigens were historically identified through their ability to elicit an immune response when expressed in tumors, the term CT antigen has evolved to include a broader range of characteristics. There are more than 200 CT antigens currently classified in the CT antigen database (http://www.cta.lncc.br), many of which have yet to be investigated for their immunogenic potential [3]. Further, several CT antigens that were originally described as immunogenic have more recently been shown to elicit only a minimal immune response in vivo[3]. Therefore, even though the term antigen signifies an immunogenic molecule, not all CT antigens display this trait. A more fundamental characteristic of CT antigens is their highly restricted expression pattern in testes and cancer. Therefore, we suggest that noncoding RNAs displaying this expression pattern can be appropriately categorized as CT antigens.
In 2010, Li et al. reported the evolution of a primate-specific X-linked miRNA gene family spanning a 33-kilobase region at Xq27.3 [5]. The miR-888 gene family is located in a 3-kilobase cluster within this region and includes the miRNAs miR-888, miR-890, miR-891a, miR-891b, miR-892a, and miR-892b [5]. Rapid evolution through gene duplication events resulted in the emergence of the miR-888 family members in primates [5]. Ohno’s law predicts that X-linked genes are more highly conserved across placental mammals than autosomal genes due to X chromosome inactivation and gene dosage effects [31]. However, CT-X antigen genes are often exceptions to this rule [4,32]. It has been hypothesized that recessive genes that are selectively beneficial to males become fixed more rapidly on the X chromosome than on the autosomes [33]. This makes the X chromosome a hotspot for CT-X antigens, while the remaining genes on the X-chromosome are highly conserved and follow Ohno’s law [33]. MiR-888 follows the specific evolutionary pattern of CT-X antigens with its recent evolution on the X-chromosome and restricted expression in testis [5]. In addition, miR-888 has predicted targets that are known to be involved in sperm cell production and regulation within the testes [5,6]. We extend this literature by identifying miR-888 expression in cells during the early stages of spermatogenesis within the seminiferous tubules of the testes. This information supports the evolution of the miR-888 gene locus in primates as a CT-X antigen that likely plays a role in testis cell biology.
Landgraf et al. first discovered the miR-888 gene family through small RNA library sequencing and reported highly specific expression of the miRNA family in the epididymis of the testes [6]. Belleannee et al. performed microarrays to investigate miRNA expression across different regions of the epididymis [34]. All members of the miR-888 family, except for miR-888, were found to be differentially expressed throughout the epididymis [34]. Furthermore, miR-888 expression was very low in the epididymis relative to its other family members, suggesting that it might not play a prominent role in this tissue [34]. Our data show that miR-888 expression is highly specific to the testes, with a low level of expression in the kidney, prostate, thymus, and trachea. MiR-888 has previously been detected in regions outside the testes but not in normal tissue under endogenous conditions [35,36]. The miR-888 opposing strand, miR-888*, was detected in human oocytes, yet miR-888* has a different miRNA seed sequence [37]. Therefore, we conclude that miR-888 expression is highly enriched in the human testes. It is also important to note that quantitative RT-PCR is a very sensitive technique and has demonstrated expression of many CT antigens in tissues in addition to testes, albeit at much lower levels [3]. Therefore, miR-888 expression across normal human tissues is consistent with observations of other CT antigens.
Heterogeneous expression of miR-888 across various cancer types was observed wherein miR-888 was only expressed in a small subset of tumors. This expression pattern is characteristic of CT antigens, which are not expressed in the tumor tissue of origin and are only expressed in a portion of tumors [1]. In a panel of eight different types of tumors, miR-888 was most predominantly expressed in EC. This is particularly relevant in that similar to the testes, the endometrium is a tissue governed by extensive hormonal regulation. Association of miR-888 expression with endometrial tumors of high grade and increased invasion implies that miR-888 expression indicates a more advanced form of EC. Furthermore, miR-888 was most highly expressed in the very rare and aggressive endometrial CS, which has often metastasized at the time of diagnosis and has a poor five-year survival rate of only 20% to 35% [26]. Expression of other CT antigens has also been correlated with aggressive tumors [38]. In EC, CT antigens MAGEC1, MAGEA3, and MAGEA4 expression correlated with high-grade tumors [39], and MAGEA4 and NY-ESO were most predominantly expressed in endometrial CSs [40]. Thus, miR-888 expression in endometrial tumors is consistent with the characteristic expression patterns observed for CT antigens.
One of the most important findings reported herein is the identification of PR as a direct target of miR-888. Throughout evolution, 3′UTR length has expanded, with steroid hormone receptors demonstrating the most extensive elongation [41]. The PR 3′UTR underwent recent expansion in primates to approximately 10 kilobases in length, generating new miRNA binding sites for evolutionarily novel miRNAs such as miR-888 [42]. The PR 3′UTR also contains binding sites for many other miRNAs, such as the recently validated miR-96 [43] and miR-126-3p [44], as well as the predicted regulators miR-181 and miR-26a [45]. The PR 3′UTR contains many AREs as well, rendering the mRNA highly unstable [46]. Therefore, the post-transcriptional regulation of PR is multifactorial and complex. The miR-888–dependent decrease in PR protein expression and 3′UTR luciferase reporter activity that we report here was relatively modest. Due to the AU-rich nature of miR-888, this is not surprising and is consistent with previous reports. For example, Garcia et al. report that AU-rich miRNA seeds have lower seed pairing stability and higher target abundance, rendering them less proficient [47]. In addition, a modest reduction in target protein levels by miRNAs has been reported by others previously [48–50]. Baek et al. performed protein arrays after knockout of miR-223 and found that a large number of targets were only reduced by about 30% [51]. The authors hypothesized that many miRNAs function through a fine-tuning mechanism rather than causing a dramatic change in protein translation of target mRNAs [51]. We suggest that miR-888 is but one of several factors using the long PR 3′UTR to regulate mRNA stability and protein translation. These different regulators likely work together through cross-talk mechanisms to cause a more pronounced and cumulative effect on PR protein expression than any one could have alone.
The importance of PR in the testes has recently been investigated. Shah et al. reported that PR expression is low during the early stages of spermatogenesis and highest during late stages IV and V [52]. Here, we show an opposite staining pattern for miR-888 in Rhesus testes, with high expression in the early stages of spermatogenesis and the absence of expression in the later stages. Therefore, we suggest that miR-888 might function to keep PR low during the early stages of spermatogenesis, followed by a loss of miR-888 expression during the late stages of spermatogenesis, permitting PR to induce germ cell differentiation. Consistent with this hypothesis, Abid et al. reported that PR expression was lost in men with infertility due to nonobstructive azoospermia, in which germ cells fail to mature into spermatozoa [53]. In addition, miR-888 appeared in a screen for miRNAs differentially expressed in sperm from infertile males with oligozoospermia and asthenozoospermia [54]. MiR-888 was overexpressed in both types of male infertility, despite the fact that the P values did not reach statistical significance in this relatively small sample size [54]. Nevertheless, the preponderance of the literature indicates that tight regulation of PR signaling is essential for proper germ cell maturation, thus establishing a potential role of miR-888 in male fertility.
Conclusions
We propose that miR-888 should be considered the first miRNA CT antigen. MiR-888 recently evolved in primates on the X chromosome and is expressed specifically during the early stages of spermatogenesis within the testes. MiR-888 is also overexpressed in cancer, specifically in endometrial tumors. Moreover, expression was only observed in a subset of endometrial tumors and was associated with high-grade and enhanced tumor invasion into the myometrium. In EC cells, we confirm PR as a direct target of miR-888 regulation. With PR well described as a tumor suppressor of the endometrium, miR-888 is a potential oncogene in EC. MiR-888 likely works in combination with other miRNAs and factors to post-transcriptionally regulate PR. The role of miR-888 in the testes is not yet fully defined, but regulation of PR by miR-888 in the spermatogenic cells of the testes is an intriguing hypothesis for future research.
The following are the supplementary data related to this article.
Endometrial Tumor Panel
TCGA UCEC Clinical Characteristics
TCGA UCEC Patient Clinical Information
MiR-888 Expression across Multiple Cancers from TCGA
PR Insert PCR Cloning Primers
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.tranon.2015.02.001.
Acknowledgements
James Schappet assisted with TCGA data analyses. Michael Goodheart provided tumor specimens from the University of Iowa Department of Obstetrics and Gynecology Tumor Bank. Katherine Gibson-Corey provided pathologic analysis of testis samples.
Footnotes
This work was supported by NIH R01CA99908 (K.K.L.) and the Department of Obstetrics and Gynecology Research Development Fund (K.K.L.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Disclosure: D.D. and K.W.T. are co-owners of Immortagen, L.L.C. All other authors have declared that no competing interests exist.
This article refers to supplementary materials, which are designated by Tables S1 to S5 and Figures S1 to S3 and are available online at www.transonc.com.
References
- 1.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 2.Scanlan MJ, Stauffer Y, Theiler G, Zahn M, Jongeneel V. 2013. Cancer Immunity CT Gene, Database. [ http://www.cta.lncc.br] [Google Scholar]
- 3.Scanlan MJ, Gure AO, Jungbluth AA, Old LJ, Chen YT. Cancer/testis antigens: an expanding family of targets for cancer immunotherapy. Immunol Rev. 2002;188:22–32. doi: 10.1034/j.1600-065x.2002.18803.x. [DOI] [PubMed] [Google Scholar]
- 4.Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, Platzer M, Howell GR, Burrows C, Bird CP. The DNA sequence of the human X chromosome. Nature. 2005;434:325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, Liu Y, Dong D, Zhang Z. Evolution of an X-linked primate-specific micro RNA cluster. Mol Biol Evol. 2010;27:671–683. doi: 10.1093/molbev/msp284. [msp284; pii] [DOI] [PubMed] [Google Scholar]
- 6.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [S0092-8674(07)00604-6; pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devor EJ, Hovey AM, Goodheart MJ, Ramachandran S, Leslie KK. microRNA expression profiling of endometrial endometrioid adenocarcinomas and serous adenocarcinomas reveals profiles containing shared, unique and differentiating groups of microRNAs. Oncol Rep. 2011;26:995–1002. doi: 10.3892/or.2011.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis H, Lance R, Troyer D, Beydoun H, Hadley M, Orians J, Benzine T, Madric K, Semmes OJ, Drake R. miR-888 is an expressed prostatic secretions-derived microRNA that promotes prostate cell growth and migration. Cell Cycle. 2014;13:227–239. doi: 10.4161/cc.26984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang S, Cai M, Zheng Y, Zhou L, Wang Q, Chen L. miR-888 in MCF-7 side population sphere cells directly targets E-cadherin. J Genet Genomics. 2014;41:35–42. doi: 10.1016/j.jgg.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 10.Cancer Facts and Figures 2014. 2014. http://www.cancer.org/acs/groups/content/@research/documents/document/acspc-041770.pdf
- 11.Howlader N. SEER Cancer Statistics Review, 1975–2010. 2013. http://seer.cancer.gov/statfacts/html/corp.html
- 12.Dai D, Wolf DM, Litman ES, White MJ, Leslie KK. Progesterone inhibits human endometrial cancer cell growth and invasiveness: down-regulation of cellular adhesion molecules through progesterone B receptors. Cancer Res. 2002;62:881–886. [PubMed] [Google Scholar]
- 13.Dai D, Litman ES, Schonteich E, Leslie KK. Progesterone regulation of activating protein-1 transcriptional activity: a possible mechanism of progesterone inhibition of endometrial cancer cell growth. J Steroid Biochem Mol Biol. 2003;87:123–131. doi: 10.1016/j.jsbmb.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Davies S, Dai D, Wolf DM, Leslie KK. Immunomodulatory and transcriptional effects of progesterone through progesterone A and B receptors in Hec50co poorly differentiated endometrial cancer cells. J Soc Gynecol Investig. 2004;11:494–499. doi: 10.1016/j.jsgi.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 15.Yang S, Thiel KW, Leslie KK. Progesterone: the ultimate endometrial tumor suppressor. Trends Endocrinol Metab. 2011;22:145–152. doi: 10.1016/j.tem.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh M, Zaino RJ, Filiaci VJ, Leslie KK. Relationship of estrogen and progesterone receptors to clinical outcome in metastatic endometrial carcinoma: a Gynecologic Oncology Group Study. Gynecol Oncol. 2007;106:325–333. doi: 10.1016/j.ygyno.2007.03.042. [DOI] [PubMed] [Google Scholar]
- 17.Yang S, Jia Y, Liu X, Winters C, Wang X, Zhang Y, Devor EJ, Hovey AM, Reyes HD, Xiao X. Systematic dissection of the mechanisms underlying progesterone receptor downregulation in endometrial cancer. Oncotarget. 2014;5:9783–9797. doi: 10.18632/oncotarget.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McBride JL, Pitzer MR, Boudreau RL, Dufour B, Hobbs T, Ojeda SR, Davidson BL. Preclinical safety of RNAi-mediated HTT suppression in the rhesus macaque as a potential therapy for Huntington's disease. Mol Ther. 2011;19:2152–2162. doi: 10.1038/mt.2011.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishida M. The Ishikawa cells from birth to the present. Hum Cell. 2002;15:104–117. doi: 10.1111/j.1749-0774.2002.tb00105.x. [DOI] [PubMed] [Google Scholar]
- 20.Albitar L, Pickett G, Morgan M, Davies S, Leslie KK. Models representing type I and type II human endometrial cancers: Ishikawa H and Hec50co cells. Gynecol Oncol. 2007;106:52–64. doi: 10.1016/j.ygyno.2007.02.033. [DOI] [PubMed] [Google Scholar]
- 21.Kumar NS, Richer J, Owen G, Litman E, Horwitz KB, Leslie KK. Selective down-regulation of progesterone receptor isoform B in poorly differentiated human endometrial cancer cells: implications for unopposed estrogen action. Cancer Res. 1998;58:1860–1865. [PubMed] [Google Scholar]
- 22.Loop SM, Rozanski TA, Ostenson RC. Human primary prostate tumor cell line, ALVA-31: a new model for studying the hormonal regulation of prostate tumor cell growth. Prostate. 1993;22:93–108. doi: 10.1002/pros.2990220202. [DOI] [PubMed] [Google Scholar]
- 23.Korch C, Spillman MA, Jackson TA, Jacobsen BM, Murphy SK, Lessey BA, Jordan VC, Bradford AP. DNA profiling analysis of endometrial and ovarian cell lines reveals misidentification, redundancy and contamination. Gynecol Oncol. 2012;127:241–248. doi: 10.1016/j.ygyno.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLoughlin HS, Fineberg SK, Ghosh LL, Tecedor L, Davidson BL. Dicer is required for proliferation, viability, migration and differentiation in corticoneurogenesis. Neuroscience. 2012;223:285–295. doi: 10.1016/j.neuroscience.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wood SN. Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. J R Stat Soc Series B Stat Methodol. 2011;73:3–36. [Google Scholar]
- 26.Bansal N, Herzog TJ, Seshan VE, Schiff PB, Burke WM, Cohen CJ, Wright JD. Uterine carcinosarcomas and grade 3 endometrioid cancers: evidence for distinct tumor behavior. Obstet Gynecol. 2008;112:64–70. doi: 10.1097/AOG.0b013e318176157c. [DOI] [PubMed] [Google Scholar]
- 27.Sorosky JI. Endometrial cancer. Obstet Gynecol. 2012;120:383–397. doi: 10.1097/AOG.0b013e3182605bf1. [DOI] [PubMed] [Google Scholar]
- 28.Kernochan LE, Garcia RL. Carcinosarcomas (malignant mixed Müllerian tumor) of the uterus: advances in elucidation of biologic and clinical characteristics. J Natl Compr Canc Netw. 2009;7:550–556. doi: 10.6004/jnccn.2009.0037. [quiz 557] [DOI] [PubMed] [Google Scholar]
- 29.Wei JJ, Paintal A, Keh P. Histologic and immunohistochemical analyses of endometrial carcinomas: experiences from endometrial biopsies in 358 consultation cases. Arch Pathol Lab Med. 2013;137:1574–1583. doi: 10.5858/arpa.2012-0445-OA. [DOI] [PubMed] [Google Scholar]
- 30.Mo B, Vendrov AE, Palomino WA, DuPont BR, Apparao KB, Lessey BA. ECC-1 cells: a well-differentiated steroid-responsive endometrial cell line with characteristics of luminal epithelium. Biol Reprod. 2006;75:387–394. doi: 10.1095/biolreprod.106.051870. [DOI] [PubMed] [Google Scholar]
- 31.Ohno S. vol. 1. Springer-Verlag; Berlin: 1967. Sex Chromosomes and Sex-Linked Genes; pp. 1–73. [Google Scholar]
- 32.Mueller JL, Skaletsky H, Brown LG, Zaghlul S, Rock S, Graves T, Auger K, Warren WC, Wilson RK, Page DC. Independent specialization of the human and mouse X chromosomes for the male germ line. Nat Genet. 2013;45:1083–1087. doi: 10.1038/ng.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ellegren H. Sex-chromosome evolution: recent progress and the influence of male and female heterogamety. Nat Rev Genet. 2011;12:157–166. doi: 10.1038/nrg2948. [DOI] [PubMed] [Google Scholar]
- 34.Belleannee C, Calvo E, Thimon V, Cyr DG, Legare C, Garneau L, Sullivan R. Role of microRNAs in controlling gene expression in different segments of the human epididymis. PLoS One. 2012;7:e34996. doi: 10.1371/journal.pone.0034996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Odriozola A, Riancho JA, de la Vega R, Agudo G, Garcia-Blanco A, de Cos E, Fernandez F, Sanudo C, Zarrabeitia MT. miRNA analysis in vitreous humor to determine the time of death: a proof-of-concept pilot study. Int J Legal Med. 2013;127:573–578. doi: 10.1007/s00414-012-0811-6. [DOI] [PubMed] [Google Scholar]
- 36.Wang J, Xiang G, Mitchelson K, Zhou Y. Microarray profiling of monocytic differentiation reveals miRNA-mRNA intrinsic correlation. J Cell Biochem. 2011;112:2443–2453. doi: 10.1002/jcb.23165. [DOI] [PubMed] [Google Scholar]
- 37.Xu YW, Wang B, Ding CH, Li T, Gu F, Zhou C. Differentially expressed micoRNAs in human oocytes. J Assist Reprod Genet. 2011;28:559–566. doi: 10.1007/s10815-011-9590-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen YT, Ross DS, Chiu R, Zhou XK, Chen YY, Lee P, Hoda SA, Simpson AJ, Old LJ, Caballero O. Multiple cancer/testis antigens are preferentially expressed in hormone-receptor negative and high-grade breast cancers. PLoS One. 2011;6:e17876. doi: 10.1371/journal.pone.0017876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chitale DA, Jungbluth AA, Marshall DS, Leitao MM, Hedvat CV, Kolb D, Spagnoli GC, Iversen K, Soslow RA. Expression of cancer-testis antigens in endometrial carcinomas using a tissue microarray. Mod Pathol. 2005;18:119–126. doi: 10.1038/modpathol.3800232. [DOI] [PubMed] [Google Scholar]
- 40.Resnick MB, Sabo E, Kondratev S, Kerner H, Spagnoli GC, Yakirevich E. Cancer-testis antigen expression in uterine malignancies with an emphasis on carcinosarcomas and papillary serous carcinomas. Int J Cancer. 2002;101:190–195. doi: 10.1002/ijc.10585. [DOI] [PubMed] [Google Scholar]
- 41.Chen CY, Chen ST, Juan HF, Huang HC. Lengthening of 3′UTR increases with morphological complexity in animal evolution. Bioinformatics. 2012;28:3178–3181. doi: 10.1093/bioinformatics/bts623. [DOI] [PubMed] [Google Scholar]
- 42.Liu JL, Liang XH, Su RW, Lei W, Jia B, Feng XH, Li ZX, Yang ZM. Combined analysis of microRNome and 3′-UTRome reveals a species-specific regulation of progesterone receptor expression in the endometrium of rhesus monkey. J Biol Chem. 2012;287:13899–13910. doi: 10.1074/jbc.M111.301275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen X, Hu Z, Wang W, Ba Y, Ma L, Zhang C, Wang C, Ren Z, Zhao Y, Wu S. Identification of ten serum microRNAs from a genome-wide serum microRNA expression profile as novel noninvasive biomarkers for nonsmall cell lung cancer diagnosis. Int J Cancer. 2012;130:1620–1628. doi: 10.1002/ijc.26177. [DOI] [PubMed] [Google Scholar]
- 44.Cui W, Li Q, Feng L, Ding W. MiR-126-3p regulates progesterone receptors and involves development and lactation of mouse mammary gland. Mol Cell Biochem. 2011;355:17–25. doi: 10.1007/s11010-011-0834-1. [DOI] [PubMed] [Google Scholar]
- 45.Maillot G, Lacroix-Triki M, Pierredon S, Gratadou L, Schmidt S, Benes V, Roche H, Dalenc F, Auboeuf D, Millevoi S. Widespread estrogen-dependent repression of micrornas involved in breast tumor cell growth. Cancer Res. 2009;69:8332–8340. doi: 10.1158/0008-5472.CAN-09-2206. [DOI] [PubMed] [Google Scholar]
- 46.Ing NH. Steroid hormones regulate gene expression posttranscriptionally by altering the stabilities of messenger RNAs. Biol Reprod. 2005;72:1290–1296. doi: 10.1095/biolreprod.105.040014. [DOI] [PubMed] [Google Scholar]
- 47.Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karres JS, Hilgers V, Carrera I, Treisman J, Cohen SM. The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell. 2007;131:136–145. doi: 10.1016/j.cell.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 49.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 50.Kim SW, Ramasamy K, Bouamar H, Lin AP, Jiang D, Aguiar RC. MicroRNAs miR-125a and miR-125b constitutively activate the NF-κB pathway by targeting the tumor necrosis factor alpha-induced protein 3 (TNFAIP3, A20) Proc Natl Acad Sci U S A. 2012;109:7865–7870. doi: 10.1073/pnas.1200081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shah C, Modi D, Sachdeva G, Gadkar S, Puri C. Coexistence of intracellular and membrane-bound progesterone receptors in human testis. J Clin Endocrinol Metab. 2005;90:474–483. doi: 10.1210/jc.2004-0793. [DOI] [PubMed] [Google Scholar]
- 53.Abid S, Gokral J, Maitra A, Meherji P, Kadam S, Pires E, Modi D. Altered expression of progesterone receptors in testis of infertile men. Reprod Biomed Online. 2008;17:175–184. doi: 10.1016/s1472-6483(10)60192-7. [DOI] [PubMed] [Google Scholar]
- 54.Abu-Halima M, Hammadeh M, Schmitt J, Leidinger P, Keller A, Meese E, Backes C. Altered microRNA expression profiles of human spermatozoa in patients with different spermatogenic impairments. Fertil Steril. 2013;99:1249–1255.e16. doi: 10.1016/j.fertnstert.2012.11.054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Endometrial Tumor Panel
TCGA UCEC Clinical Characteristics
TCGA UCEC Patient Clinical Information
MiR-888 Expression across Multiple Cancers from TCGA
PR Insert PCR Cloning Primers


