Abstract
The Genomic Disulfide Analysis Program (GDAP) provides web access to computationally predicted protein disulfide bonds for over one hundred microbial genomes, including both bacterial and achaeal species. In the GDAP process, sequences of unknown structure are mapped, when possible, to known homologous Protein Data Bank (PDB) structures, after which specific distance criteria are applied to predict disulfide bonds. GDAP also accepts user-supplied protein sequences and subsequently queries the PDB sequence database for the best matches, scans for possible disulfide bonds and returns the results to the client. These predictions are useful for a variety of applications and have previously been used to show a dramatic preference in certain thermophilic archaea and bacteria for disulfide bonds within intracellular proteins. Given the central role these stabilizing, covalent bonds play in such organisms, the predictions available from GDAP provide a rich data source for designing site-directed mutants with more stable thermal profiles. The GDAP web application is a gateway to this information and can be used to understand the role disulfide bonds play in protein stability both in these unusual organisms and in sequences of interest to the individual researcher. The prediction server can be accessed at http://www.doe-mbi.ucla.edu/Services/GDAP.
INTRODUCTION
Disulfide bonds have long been implicated as a key factor in stabilizing proteins in extracellular environments. More recently, evidence has grown to suggest that, in certain thermophilic bacteria and archaea, disulfide bonds play a pervasive role in stabilizing proteins inside the cell as well (1). The discovery of widespread disulfide bonds by both experimental and computation techniques in certain microbial genomes highlights the importance of these covalent interactions as a general strategy for protein stability. Yet, despite the considerable size of the Protein Data Bank (PDB) database, its coverage of any particular microbial genome is generally very low, with many completely sequenced genomes having no known structures in the PDB. In the absence of structures, the direct visualization of disulfide bonds across various organisms is impossible. However, the abundance of complete genomes and the continued pace of sequencing offer an opportunity for systematic computational predictions. In this paper, we report a convenient web application that uses fast sequence-to-structure mapping techniques and specific residue distance criteria to identify potential disulfide bonds.
The abundance of disulfide predictions across microbial genomes is a helpful tool in studying protein structure and stability. Predictions contained on this site can be used to analyze the specific role cysteines play in stabilizing proteins. Specifically, the large number of predictions from many microbial genomes can be examined for general preferences in cysteine placement. This could prove helpful in designing point mutations by site-directed mutagenesis to engineer more thermostable protein structures. The Genomic Disulfide Analysis Program (GDAP) site can also be queried to explore the evolution and conservation of disulfide bonds within the phylogenetic context of the species analyzed. Homologs between species can be compared, yielding clues about protein evolution as it relates to disulfide bond utilization. These and other possible applications for GDAP should further our understanding of the role disulfide bonds play in protein stabilization, structure and evolution.
DISULFIDE PREDICTION PROCESS
The prediction process utilized by the GDAP web application is based on previous disulfide bond prediction techniques (1). Proteins of unknown structure from 196 microbial genome records (available as of May 2003) were downloaded from the National Center for Biotechnology Information (NCBI, ftp://ftp.ncbi.nih.gov/). These records contained protein sequences for genomes, individual chromosomes or plasmids that together represented complete genome sequences for 101 non-redundant microbial species. Each protein was queried against the PDB (2) using a step-wise process to identify homologous structures. The BLAST (3) program was used first to identify a sequence-to-PDB structure match with an E-value < 0.0001 (version dated April 23, 2002). This threshold was chosen so that only reliable sequence-to-structure mappings would be included. A lower threshold would have precluded accurate alignment of the genomic sequence to the known structure's sequence. If a match was not identified with BLAST, the process was repeated with PSI-BLAST (4) (version dated April 23, 2002), again using a match cutoff of E < 0.0001. PSI-BLAST allowed for more sensitive homology detection where BLAST initially failed. Finally, if no match was found in the PDB using either of these two algorithms, the method of Sequence Derived Properties (SDP) was used (5). SDP, a fold recognition algorithm, was chosen to maximize the possibility of selecting a structural homolog despite lower sequence similarity. An SDP-derived match was only accepted if the resulting SDP score was ≥5, which corresponds to an accuracy of 95% (6). If no homolog could be found among the known structures, prediction of disulfide bonding for that protein was not possible.
If a match was found between a genomic protein sequence and a PDB structure's sequence, the two were aligned using a local alignment algorithm. The mlocalS program from the SeqAln package (version 2.0) was used to implement the Smith and Waterman local alignment algorithm (7). This alignment formed the basis for the sequence-to-structure mapping. Cysteines were identified in the query sequence and the coordinates for the corresponding, aligned structure residues were extracted. Every combination of cysteine pairings was examined, with distances being calculated between every pair of cysteine residues. Generally, two cysteines were considered to be potentially disulfide bonded if the distance between their Cα positions was within 8 Å when mapped onto a homologous structure. This criterion has previously been shown to maximize true positives and minimize false positives, predicting true disulfide bonds in a test set with an overall accuracy of ∼80% (1). All such occurrences were included. No attempt has been made to rank multiple predicted disulfides or to exclude potentially mutually exclusive disulfide bond predictions. To minimize false positives, a primary structure criterion was enforced. Cysteine residues were only considered for disulfide bond prediction if they were three or more residues apart in linear sequence. This restriction eliminated predictions of disulfide bonds between residues in the CXXC motif, which is often involved in metal binding or catalysis. It should be understood that disulfide bonds can form between two residues more closely spaced in sequence, but the present structure mapping approach is not able to provide strong statistical inferences of disulfide bonds for such cases.
In addition to making specific disulfide predictions, the GDAP process also actively annotates the query genomic sequences. This information is provided to help in the analysis of disulfide bond-containing proteins. Three annotation filters are applied to each protein: SignalP (8), TMpred (9), and ScanProsite (10). Proteins predicted by SignalP (version 2.0) to contain a signal peptide in the first 70 N-terminal residues are flagged as extracellular. A protein is considered positive for the export signal if more than one test in SignalP's test suite is positive. Transmembrane proteins are identified using the TMpred program (version dated October 30, 1998) with a cutoff score of 1000. The transmembrane prediction is helpful in identifying proteins that contain either extracellular or periplasmic domains, both of which affect the expected presence of disulfide bonds. Finally, proteins that contain known metal binding motifs are flagged. A collection of these motifs was extracted from the PROSITE database (11) and the ScanProsite (version 1.3) program was used to flag sequences matching these motifs. The identification of proteins with suspected and known metal binding motifs is important because clusters of cysteine residues in these proteins might actually serve catalytic or metal binding roles, rather than forming a structural disulfide bond. Together, the annotations allow the user to restrict the disulfide bond predictions to proteins that meet any combination of these three criteria.
ACCESS TO THE WEBSITE
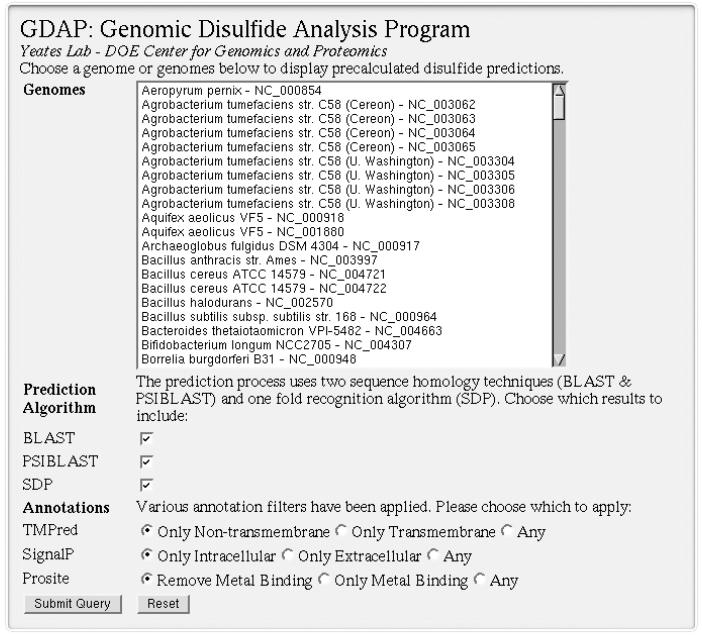
Genome-wide disulfide predictions are accessible through the GDAP website and include over one hundred complete, non-redundant microbial genomes. The primary interface for GDAP is a web form that allows users to select genomes of interest and display all proteins in those genomes that contain predicted disulfide bonds (Figure 1). This includes the ability to aggregate sets of predictions across multiple genomes. To support various types of filtering by the user, annotation criteria can be selected and enforced for each search. Each prediction includes the algorithm used to identify the sequence-to-structure map, its associated significance score expressed as an E-value (if available) and the cysteine residues predicted to be in disulfide bonds. When available, basic protein annotations are also provided for query sequences. The resulting lists of proteins with predicted disulfide bonds can be further explored via web links to NCBI sequence records and PDB structures.
Figure 1.
Users can view disulfide bond predictions for proteins across 196 microbial genome records on the GDAP website. The search interface for these pre-calculated predictions can aggregate results from multiple genome records and can also apply filtering based on several criteria.
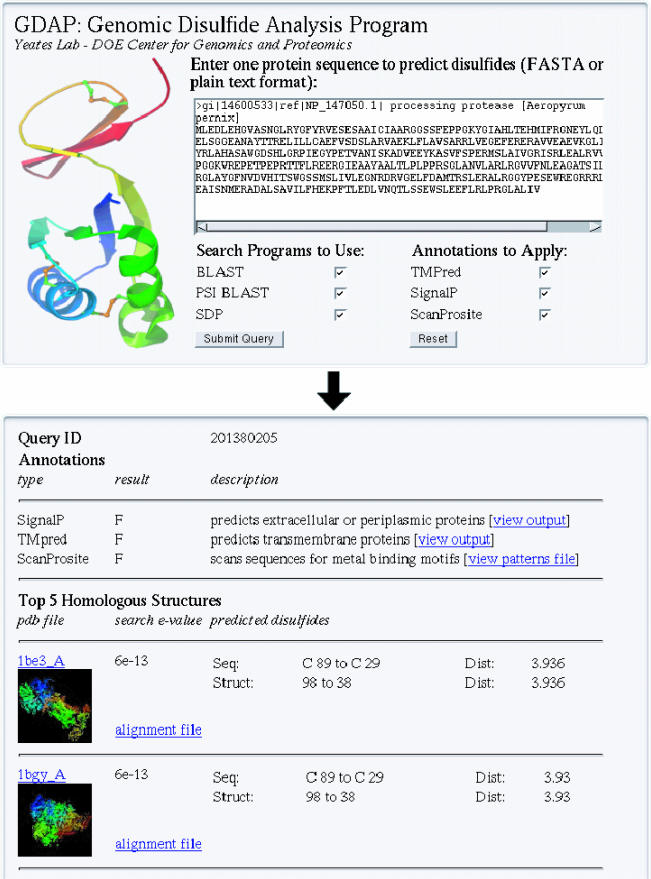
In addition to genome-wide, pre-calculated disulfide bond predictions, individual sequences can be submitted via the GDAP web application and run through the prediction process in real time. Cysteines predicted to be in disulfide bonds are listed along with links to the PDB structures onto which the user-supplied sequence maps (Figure 2). Unlike the pre-calculated disulfide predictions, user-supplied sequences processed with GDAP report the top five significant PDB structure matches from all three homology search techniques (BLAST, PSI-BLAST and SDP). Each match is tested for disulfide bonds and, if available, the cysteine residues involved in each prediction are displayed. Finally, the user-supplied sequence is annotated using TMpred, SignalP and ScanProsite. This additional information, along with any sequence-to-structure mapping that predicts disulfide bonds, provides valuable information about protein structure and function even when the query protein itself may lack annotation or a clear functional role.
Figure 2.
GDAP allows users to upload a protein sequence of interest in FASTA format. Various annotation filters and homology techniques can be chosen by the user. Disulfide bond predictions are made, if possible, on the basis of the top matches to PDB structures.
STATISTICS
The analysis was performed on 196 genome accession files, which represented 101 different microbial species. On average, the homology search protocol identified high-quality matches to PDB structures for ∼33% of each genome. This corresponded to 119 894 protein sequences mapped to homologous PDB structures out of a total of 359 555 queried proteins. This partial coverage reflects the relatively stringent criteria adopted for homology detection and sequence alignment. Among those sequences that could be mapped onto structures, 16 511 probable disulfide-bonded proteins could be identified.
Several of the examined organisms have previously been shown to contain a high percentage of disulfide-bonded cysteines (1). Specifically, disulfide richness has been implicated mainly in certain thermophilic archaea. That trend is evident from the data provided on the GDAP web site. Taken together, the large number of genomes examined and the relative abundance of disulfides in some of these organisms results in a large collection of structurally relevant disulfide bond predictions.
CONCLUSIONS
The GDAP web application provides a convenient and powerful way to identify potential disulfide bonds in sequences of unknown structure. Despite the limitations inherent in modeling disulfides using sequence-to-structure alignments, the breadth of the predictions across so many genomes should prove useful. This dataset will continue to grow as new genomes are sequenced and added to GDAP on a regular basis. Possible future applications include protein fold prediction and a general exploration of protein stabilization strategies employed by various organisms in nature. Beyond the pre-calculated, genome-wide predictions, the ability to upload protein sequences to GDAP extends the predictions to sequences of special interest to the end user. This functionality will be expanded to enable on-demand, genome-wide predictions of disulfide bonds for new or partially sequenced genomes as they become available.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank D. Boutz, M. Beeby and P. Mallick for their contributions to this work. This work was supported by grants from the Department of Energy and the National Institutes of Health. B.O. was supported by a USPHS National Research Service Award GM07185.
REFERENCES
- 1.Mallick P, Boutz,D.R, Eisenberg,D. and Yeates,T.O. (2002) Genomic evidence that the intracellular proteins of archaeal microbes contain disulfide bonds. Proc. Natl Acad. Sci., USA, 15, 9679–9684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 4.Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischer D. and Eisenberg,D. (1996) Protein fold recognition using sequence-derived predictions. Protein Sci., 5, 947–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mallick P., Goodwill,K.E., Fitz-Gibbon,S., Miller,J.H. and Eisenberg,D. (2000) Selecting protein targets for structural genomics of Pyrobaculum aerophilum: validating automated fold assignment methods by using binary hypothesis testing. Proc. Natl Acad. Sci., USA, 97, 2450–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith T.F. and Waterman,M.S. (1981) Identification of common molecular subsequences. J. Mol. Biol., 147, 195–197. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen H., Engelbrecht,J., Brunak,S. and von Heijne,G. (1997) A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int. J. Neural Syst., 5, 581–599. [DOI] [PubMed] [Google Scholar]
- 9.Hofmann K. and Stoffel,W. (1993) TMbase—a database of membrane spanning proteins segments. Biol. Chem., 374, 166. [Google Scholar]
- 10.Gattiker A., Gasteiger,E. and Bairoch,A. (2002) ScanProsite: a reference implementation of a PROSITE scanning tool. Appl. Bioinformatics, 1, 107–108. [PubMed] [Google Scholar]
- 11.Sigrist C.J.A., Cerutti,L., Hulo,N., Gattiker,A., Falquet,L., Pagni,M., Bairoch,A. and Bucher,P. (2002) PROSITE: a documented database using patterns and profiles as motif descriptors. Brief Bioinform., 3, 265–274. [DOI] [PubMed] [Google Scholar]