Abstract
In addition to two well-recognized proteasome subtypes—constitutive proteasomes and immunoproteasomes—mounting evidence also suggests the existence of intermediate proteasome subtypes containing unconventional mixtures of catalytic subunits. While they appear to play unique biological roles, the lack of practical methods for detecting distinct proteasome subtypes has limited functional investigations. Here, we report the development of activity-based probes that crosslink two catalytic subunits within intact proteasome complexes. Identification of the crosslinked subunit pairs provides direct evidence on the catalytic subunit composition of proteasomes. Using these probes, we found that U266 multiple myeloma cells contain intermediate proteasomes comprising both β1i and β2, but not β1 and β2i, consistent with previous findings with other cell types. Our bifunctional probes can be utilized in functional investigations of distinct proteasome subtypes in various biological settings.
Keywords: Proteasome subtype, constitutive proteasome, immunoproteasome, intermediate proteasome, crosslinking agents
Graphical abstract
Introduction
Proteasomes process and degrade numerous cellular proteins, thereby serving as crucial regulators of a wide array of cellular processes including cell cycle progression,[1] apoptosis,[2] and immune responses.[3] These multiprotease complexes are of particular interest due to their validation as effective therapeutic targets by the FDA approval of the proteasome inhibitor drugs bortezomib and carfilzomib for the treatment of multiple myeloma.[4] Studies investigating the utility of proteasome inhibitors in the treatment of additional cancer types as well as autoimmune and inflammatory diseases are ongoing.[5]
Proteasomes structurally comprise a cylindrically-shaped 20S catalytic core particle, which may be capped on one or both ends by regulatory particles.[6] The 20S core particle is formed by 28 subunits arranged in four stacked rings, each containing seven subunits. Three different catalytically active proteasome subunits reside in each of the two inner rings, known as the β-rings.[7] These catalytic subunits are synthesized as inactive precursors with N-terminal propeptides, which are removed during the final steps of 20S proteasome assembly to expose their catalytic threonine residues.[8] Unique combinations of these catalytically active subunits can incorporate into otherwise identical 20S core particles, thereby forming distinct proteasome subtypes. For a number of years, the prevailing model of proteasome regulation acknowledged the predominant existence of two proteasome subtypes: constitutive proteasomes and immunoproteasomes. Constitutive proteasomes contain catalytic subunits β1, β2, and β5 and are expressed in most cell types, whereas immunoproteasomes contain a homologous set of catalytic immunosubunits—β1i, β2i, and β5i—and are constitutively expressed in hematopoietic cells[9] and inducible in other cell types following exposure to cytokines such as interferon-γ.[10] The immunosubunits replace their constitutive counterparts during proteasome assembly, leading to alterations in proteolytic activities suggested to promote the production of MHC class I antigens.[11] More recently, an immunoproteasome variant known as the thymoproteasome was discovered in cortical thymic epithelial cells. The thymoproteasome contains immunosubunits β1i and β2i, together with the thymoproteasome-specific subunit β5t, and was shown to function in the positive selection of T cells.[12]
Additionally, results from an increasing number of studies suggest the existence of intermediate proteasome subtypes comprising non-prototypical mixtures of constitutive proteasome and immunoproteasome catalytic subunits. Since their initial discovery in murine tissues,[13] the presence of intermediate proteasome subtypes has been indicated in a variety of non-diseased tissues and cancer cells of murine and human origin.[14] Proteasome subtypes with distinct catalytic subunit compositions appear to have unique proteolytic activity profiles as well as tissue and subcellular distribution patterns, suggesting that they may have specialized functions.[13–15] Consistent with these observations, results from several investigations of antigenic peptide production suggested that intermediate proteasome subtypes serve to enhance the diversity of peptides presented on MHC class I molecules to CD8+ T cells.[14b, 16] The exclusive production of specific tumor antigens by intermediate proteasome subtypes demonstrated the importance of identifying proteasome subtypes present in cells targeted by immunotherapy.[14b] Furthermore, distinguishing proteasome compositions were associated with pathological conditions such as Crohn’s disease and ulcerative colitis and were suggested as potential biomarkers for these diseases.[17] Distinct proteasome subtypes are also linked to differential sensitivity to specific proteasome inhibitors;[15b, 15c] thus, identifying proteasome subtypes present in diseased cells may ultimately facilitate the selection of therapeutic approaches which target the relevant subtypes with improved efficacy or with minimal toxicity to non-diseased cells.[15c] However, much regarding the unique functions of each subtype or their roles as drug targets remains undiscovered, largely due to limitations in methods currently available for determining the catalytic subunit composition of individual proteasome subtypes within cells and tissues.
Earlier efforts to examine proteasome composition typically involved measuring the abundance of proteasome catalytic subunits in cell extracts or purified 20S proteasomes using various methods such as immunoblotting or 2D-PAGE[8b, 14c, 15b, 15e, 17–18] as well as label-free LC-MS-based or ELISA-based quantification.[14b, 14d, 15b, 15d] In these studies, the relative abundance of each catalytic subunit was then used to deduce the proteasome subtypes present within the cells or tissues being examined. However, examination of overall catalytic subunit abundance is insufficient to conclusively define the catalytic subunit composition of individual proteasome subtypes, especially if multiple subtypes are present.[13]
More rigorous studies of distinct proteasome subtypes involved separation of purified 20S proteasome complexes by anion-exchange or hydrophobic interaction chromatography,[13–14, 14e, 15c, 15f] isoelectric focusing-free flow electrophoresis,[15a] or immunoprecipitation of exogenously expressed tagged forms of catalytic β-subunits,[8b, 18] followed by evaluation of catalytic subunit composition by immunoblotting[8b, 13, 14e, 15a, 18] or 2D-PAGE.[14a, 15c, 15f] For example, Guillaume et al.[14b] developed antibodies recognizing specific catalytic β-subunits in their native states, facilitating the identification of proteasome subtypes by sequential immunoprecipitation steps to deplete proteasomes containing specific catalytic subunits and subsequent immunoblotting analysis. Using this approach, they confirmed the existence of two intermediate proteasome subtypes: one comprised of constitutive subunits β1 and β2 with immunosubunit β5i and the other comprised of constitutive subunit β2 with immunosubunits β1i and β5i.[14b] While these techniques are considered improvements in detecting different proteasome subtypes, they require multiple technically-challenging steps and depend on the availability and affinity of antibodies that can bind to catalytic proteasome subunits in their native states.
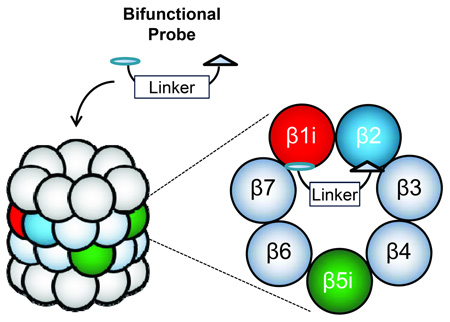
An unmet need therefore exists to develop a more practical method for determining the catalytic subunit composition of distinct proteasome subtypes present within cell or tissue samples. We recently reported a fluorescence resonance energy transfer (FRET)-based approach for assessing the catalytic subunit composition of proteasomes.[19] Expanding upon the designs of reversibly-binding bifunctional proteasome inhibitors previously reported by Moroder and colleagues,[20] and subsequently by Vidal, Reboud-Ravaux and colleagues,[21] we now report the development of an alternative approach in which irreversibly-binding bifunctional activity-based probes are utilized to crosslink pairs of catalytic subunits within individual 20S proteasomes, and thus to provide direct evidence of the catalytic subunit composition of each subtype (Figure 1).
Figure 1.
Crosslinking strategy for proteasome subtype identification. Bifunctional proteasome probes, comprised of two peptide epoxyketone proteasome inhibitors coupled by a linker, crosslink different pairs of proteasome catalytic subunits when they are present within the same 20S core complex. Crosslinking efficiency of a particular subunit pair is determined by the subunit selectivity of the two inhibitors as well as the length of the linker by which they are coupled. Following treatment of cell lysates or purified 20S proteasomes with bifunctional proteasome probes, the crosslinked subunit pairs are identified by immunoblotting analysis.
Several proteasome inhibitors containing two reactive groups were previously developed from reversibly-binding inhibitors in effort to obtain highly potent or subunit-selective proteasome inhibitors.[20–22] Indeed, joining of two peptide aldehyde proteasome inhibitors,[20] or of two tripeptide linear mimics of the noncovalent proteasome inhibitor TMC-95A,[21] by polyethylene glycol (PEG) or polyaminohexanoic acid spacers gave rise to bifunctional inhibitors displaying substantial increases in inhibitory potency of up to 2 orders of magnitude over their respective monomeric parent inhibitors. Unlike these reversible bifunctional inhibitors, the bifunctional probes reported here were synthesized by coupling two irreversible, subunit-selective peptide epoxyketone proteasome inhibitors via hydrocarbon or PEG linkers (Figure 1). The crystal structure-derived distances between catalytic threonine Oγ atoms of different subunit pairs within the mammalian 20S proteasome guided our selection of linkers of the appropriate lengths to allow crosslinking of the desired catalytic subunit pairs.[23] The epoxyketone pharmacophore binds specifically to the catalytic threonine residues of proteasome subunits covalently and irreversibly,[24] facilitating the detection of these crosslinked subunit pairs by immunoblotting analysis and the subsequent identification of distinct proteasome subtypes present within cells.
Results
β1-β2/β2i Crosslinking
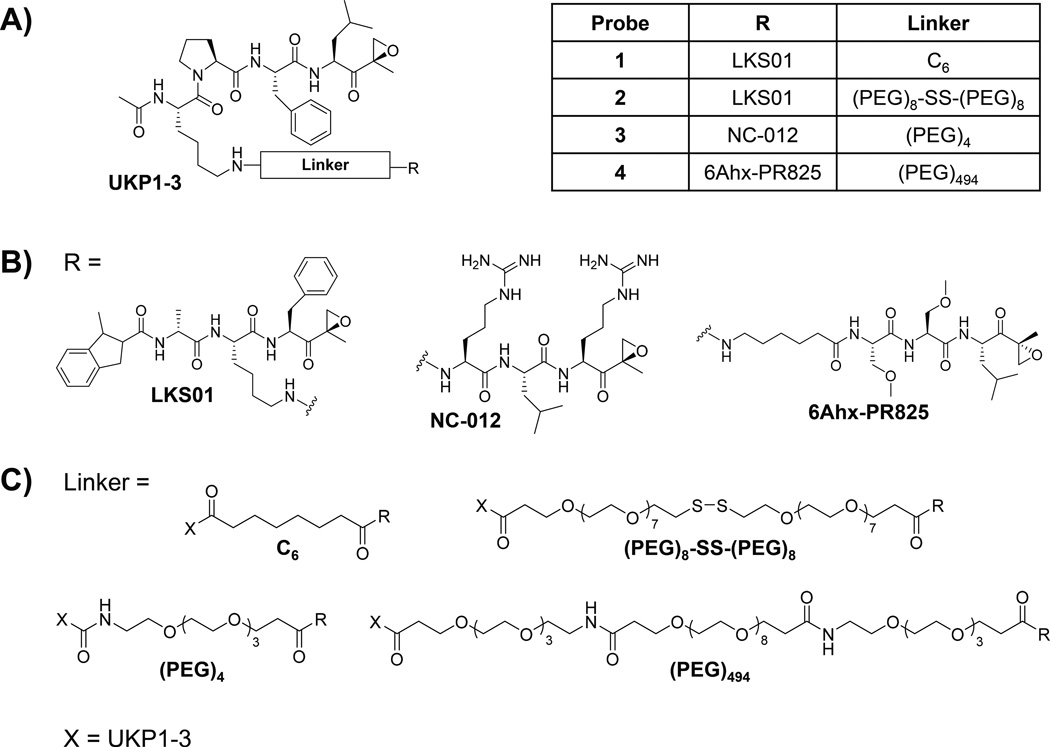
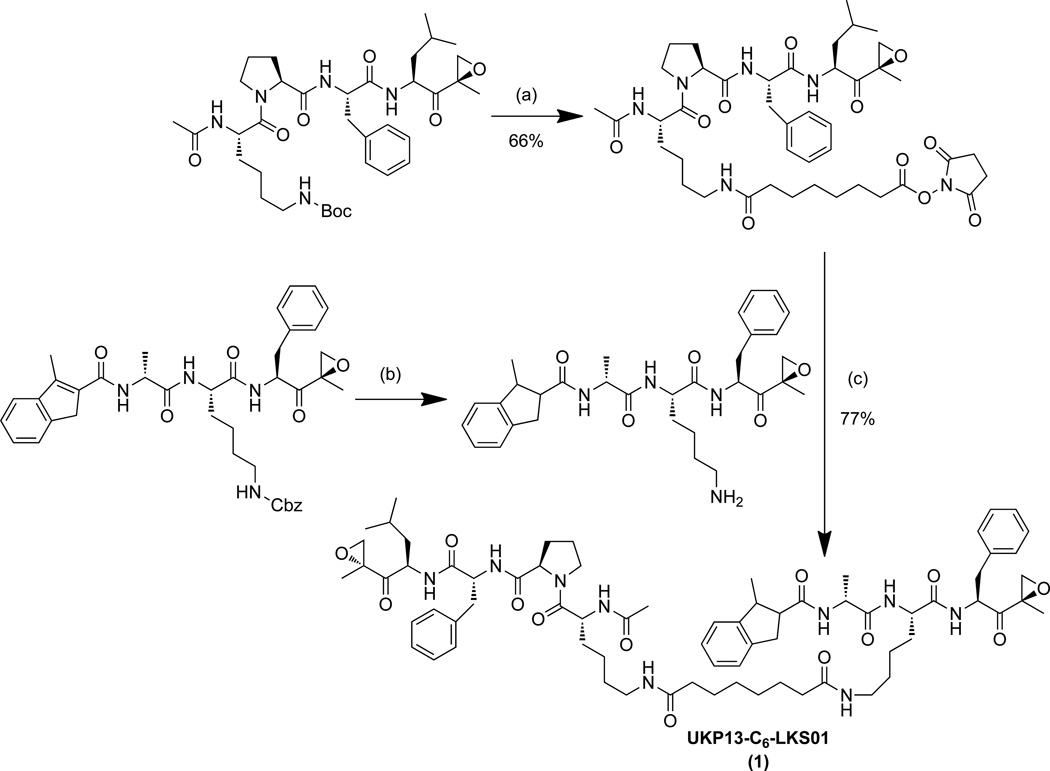
Our laboratory and others have developed several subunit-selective proteasome inhibitors.[19, 25] To determine the feasibility of our crosslinking strategy, we first utilized two of these inhibitors readily available in our laboratory—the β1/β1i-selective inhibitor UKP1-3[19] and the β5/β5i-selective inhibitor LKS01[25c]—to generate the first peptide epoxyketone-based bifunctional proteasome probe. UKP1-3 and LKS01 are derivatives of the β1/β1i-selective inhibitor YU-102[25a, 26] and the β5i-selective inhibitor IPSI,[25b] respectively, in which the P4 glycine side chain of YU-102 and the P2 tryptophan side chain of IPSI were replaced with that of lysine to facilitate their coupling via an amine-reactive linker. Modifications at these positions do not alter the subunit selectivities of these inhibitors.[19, 25c, 25e] Coupling of UKP1-3 and LKS01 via a short hydrocarbon linker with an estimated length of ~11.4 Å produced UKP13-C6-LKS01 (probe 1) (Figure 2, Scheme 1).
Figure 2.
Structures of bifunctional proteasome probes, each comprising the β1/β1i-selective proteasome inhibitor UKP1-3 (A) coupled with another peptide epoxyketone proteasome inhibitor (B) via a hydrocarbon or polyethylene glycol linker (C).
Scheme 1.
Synthesis of UKP13-C6-LKS01 (probe 1). (a) 1) TFA, dichloromethane, 1 hr; 2) DSS (Disuccinimidyl suberate), DIPEA (N,N-diisopropylethylamine), THF, overnight; (b) H2, Pd/C, MeOH/EtOAc, 4 hr; (c) DIPEA, THF, overnight.
We first ensured that probe 1 displays the expected inhibitory profile by conducting proteasome activity assays with subunit-selective fluorogenic substrates[27] in purified human 20S proteasomes and in U266 multiple myeloma cell lysates, which abundantly express all six constitutive proteasome and immunoproteasome catalytic subunits.[28] As expected, this probe was found to preferentially inhibit the β1i and β5i subunits of the purified immunoproteasome (Table 1). Preferential inhibition of β1 and β5 of the purified constitutive proteasome was also observed. In contrast, concentrations of 10 µM or higher were required to achieve 50% inhibition of the β2 and β2i activity (Table 1). Consistent with these findings, treatment of U266 cell lysates with 1 µM of probe 1 resulted in nearly complete inhibition of all β1/β1i and β5/β5i activity, whereas a concentration of 10 µM was required to achieve ~50% inhibition of β2/β2i activity (Supplemental Table 1). Together, these results indicate that the subunit binding preferences of the parent inhibitors were in fact maintained by bifunctional probe 1.
Table 1.
Inhibitory potencies (IC50 values, µM) of bifunctional probes 1–4 towards constitutive proteasome and immunoproteasome catalytic subunits.[a]
| Probe | β1 | β2 | β5 | β1i | β2i | β5i |
|---|---|---|---|---|---|---|
| 1 | 0.655 | 10.64 | 0.895 | 0.402 | >10 | 0.349 |
| 2 | 0.094 | 6.669 | 0.165 | 0.038 | >10 | 0.017 |
| 3 | 1.153 | 0.128 | 3.93 | 0.803 | 0.03 | 4.35 |
| 4 | 0.188 | 15.28 | 0.075 | 0.130 | 28.07 | 0.38 |
Inhibition of hydrolysis of subunit-selective fluorogenic peptide substrates was evaluated following a 1 hr incubation of the purified constitutive proteasome or immunoproteasome with each bifunctional probe.
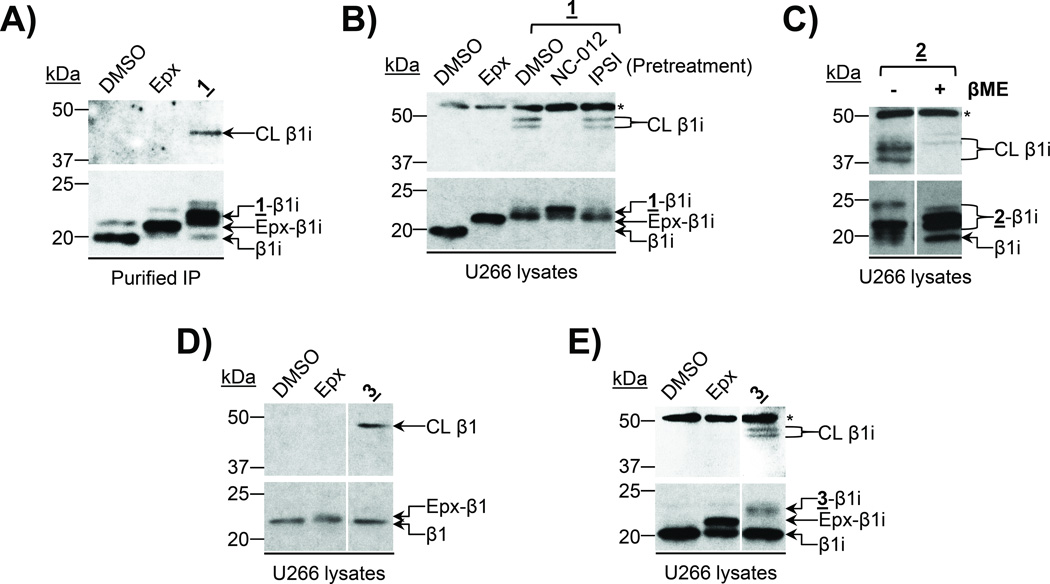
We next assessed the ability of probe 1 to crosslink two proteasome catalytic subunits via immunoblotting with antibodies recognizing proteasome catalytic subunits. In these assays, crosslinked subunits can be visualized by the detection of immunoreactive bands between 40 and 50 kDa (the combined molecular weight of two proteasome catalytic subunits). As shown in Figure 3A, treatment of the purified immunoproteasome with probe 1 produced an anti- β1i-immunoreactive band at ~45 kDa—consistent with the crosslinked β1i subunit. We further validated these findings by treating U266 cell lysates with probe 1 and were able to detect two bands at ~45–50 kDa on a β1i immunoblot (Figure 3B), in contrast to the single band observed in this molecular weight range following treatment of the purified immunoproteasome with this probe. The appearance of two crosslinked β1i bands suggested that β1i was likely crosslinked with two different subunits by probe 1. In order to identify the other subunits crosslinked with β1i, we pretreated U266 lysates with the subunit-selective inhibitors NC-012 (β2/β2i-selective)[25f] or IPSI (β5i-selective)[25b] prior to treatment with probe 1. Pretreatment with NC-012, but not with IPSI, was able to completely block β1i crosslinking, as indicated by the complete depletion of these two crosslinked β1i bands (Figure 3B), demonstrating that the subunits crosslinked with β1i by probe 1 are β2 and β2i. These results indicate that, in addition to proteasome subtypes containing both β1i and β2i, intermediate proteasome subtypes containing the immunosubunit β1i together with the constitutive subunit β2 are present in U266 multiple myeloma cells. It should be noted that probe 1 was found to crosslink β1i with β2/β2i, despite its preferential inhibition of β1/β1i and β5/β5i as revealed by proteasome activity assay results (Supplemental Table 1). This apparent discrepancy between the inhibitory and crosslinking preferences of probe 1 is likely due to the spatial constraint incurred by the C6 hydrocarbon linker, driving crosslinking between adjacent subunits.
Figure 3.
UKP13-C6-LKS01 (probe 1) and UKP13-(PEG)4-NC012 (probe 3) crosslink β1/β1i with β2/β2i. A) The purified 20S immunoproteasome (IP) was treated with DMSO, epoxomicin (Epx), or probe 1 prior to immunoblotting for β1i. B) U266 cell lysates were treated with DMSO or epoxomicin, or pretreated with DMSO or the indicated subunit-selective inhibitors prior to treatment with probe 1 as indicated. β1i was detected by immunoblotting. C) High-molecular-weight immunoblot signals detected following treatment with bifunctional proteasome probes result from crosslinking of proteasome catalytic subunits. U266 cell lysates were treated with probe 2and proteins were subsequently denatured in sample buffer with or without β-mercaptoethanol (βME), which cleaves the internal disulfide bond of the linker. β1i was detected by immunoblotting. D) U266 cell lysates were incubated with DMSO, epoxomicin, or probe 3 prior to immunoblotting for β1. E) U266 cell lysates were incubated with DMSO, epoxomicin, or probe 3 prior to immunoblotting for β1i. Irrelevant lanes were cropped out of blots shown in C–E. CL = crosslinked β-subunit, * = non-specific band.
To further verify that the higher-molecular-weight bands observed on our immunoblots following treatment with probe 1 in fact result from crosslinking of proteasome catalytic subunits, we synthesized the bifunctional proteasome probe UKP13-SS-LKS01 (probe 2) in which the C6 linker of probe 1 was replaced with a PEG linker containing a scissile disulfide bond (Figure 2, Supplemental Information). Cleavage of this disulfide bond by adding a reducing agent, such as β-mercaptoethanol, to probe 2-treated samples should reverse the crosslinks formed by this probe. Assessment of the subunit selectivity of this probe in the purified immunoproteasome revealed selective binding to β5i and β1i (Table 1). Likewise, β1 and β5 were preferred binding targets of this probe in the purified constitutive proteasome. Similar to probe 1the IC50 values of probe 2 for β2 and β2i were significantly higher than those for β1/β1i and β5/β5i (Table 1).
We treated U266 cell lysates with probe 2 and denatured the proteins in the presence or absence of β-mercaptoethanol. In the absence of β-mercaptoethanol, two major bands between 37 and 50 kDa were observed on a β1i immunoblot (Figure 3C), indicating that this probe crosslinks β1i. These bands were no longer detected following disulfide bond cleavage by β-mercaptoethanol (Figure 3C), confirming that they resulted from β1i crosslinking by probe 2. (Of note, although it is possible that this disulfide bond may be cleaved by intracellular glutathione present in cell lysates, the observance of bands consistent with crosslinked β1i in the absence of β-mercaptoethanol indicates that the linker remains intact under these experimental conditions.) These results further confirm that the immunoreactive bands in the 40–50 kDa range observed following treatment with our bifunctional probes result from crosslinking of proteasome catalytic subunits.
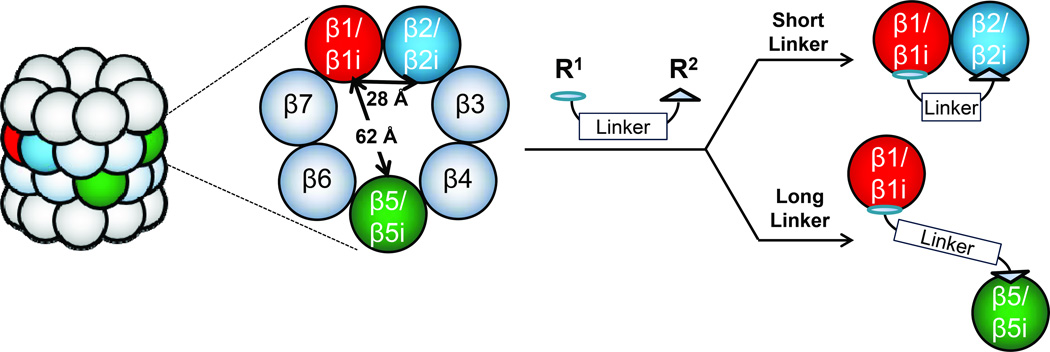
Encouraged by the successful crosslinking of β1i with β2/β2i by probe 1we synthesized UKP13-(PEG)4-NC012 (probe 3), in which, in effort to improve the β1/β1i-β2/β2i crosslinking efficiency, we replaced the LKS01 inhibitor and short C6 hydrocarbon linker of probe 1 with the β2/β2i-selective inhibitor NC-012 [25f] and a more flexible and water-soluble PEG linker, respectively. The N-terminus of NC-012 was derivatized to facilitate linker attachment without compromising the inhibitor’s subunit selectivity [25f]. Given the relatively short (~28 Å) distance between the catalytic Thr1Oγ atoms of β1/β1i and β2/β2i within a β-ring (Figure 1),[23] we selected a short (PEG)4 linker with an estimated length of ~23.2 Å (Figure 2). Using proteasome activity assays, we verified that probe 3 maintained comparable β1/β1i inhibition to that exhibited by probe 1 (Table 1). Conversely, in comparison with probe 1, probe 3 displayed a markedly enhanced preference for β2/β2i inhibition and a reduced preference for inhibition of β5/β5i (Table 1). Data obtained in U266 cell lysates are in agreement with these altered binding preferences (Supplemental Table 1).
Treatment of either the purified constitutive proteasome (Supplemental Figure 1A) or U266 cell lysates (Figure 3D) with probe 3 followed by immunoblotting analysis led to the appearance of an anti-β1-immunoreactive band slightly below 50 kDa, consistent with β1-β2 crosslinking. A competition assay with the subunit-selective inhibitors IPSI (β5i-selective),[25b] Ac-PAL-ek (β1i-selective), and NC-012 (β2/β2i-selective)[25f] in the purified immunoproteasome verified that probe 3 also crosslinks β1i with β2i, but not with β5i (Supplemental Figure 1B). β1i crosslinking was also observed in U266 cell lysates following treatment with probe 3. Immunoblotting analysis revealed two anti- β1i-immunoreactive bands at ~45–50 kDa (Figure 3E), similar to the results obtained using probe 1strongly indicating β1i-β2/β2i crosslinking by probe 3.
Taken together, the results obtained with probes 1 and 3 indicate that U266 cells harbor proteasome subtypes containing both β1i and β2i, as well as those containing both β1i and β2. Interestingly, we did not detect proteasome subtypes containing constitutive subunit β1 with immunosubunit β2i, despite the results indicating that proteasomes containing both β1 and β2 are also present in this cell line (Figure 3D).
β1/β1i-β5/β5i Crosslinking
Next, we sought to develop a bifunctional proteasome probe capable of crosslinking β1/β1i with β5/β5i. To this end, we coupled the β1/β1i-selective inhibitor UKP1-3 with a derivative of the β5-selective inhibitor PR-825[25g, 25h] in which the N-cap of PR-825 was replaced by a 6-aminohexanoyl group to provide UKP13-(PEG)494-6Ahx-PR825 (probe 4) (Figure 2). The N-terminal amine of this PR-825 derivative served as the attachment point for coupling to UKP1-3 via a PEG linker with an estimated length of ~83.9 Å, which we expected to be more than sufficient for crosslinking β1/β1i with β5/β5i, whose catalytic residues are separated by ~62 Å (Figure 1).[23] Activity assays demonstrated that probe 4 binds preferentially to the β5 and β1 subunits of the purified constitutive proteasome, and to the β1i and β5i subunits of the purified immunoproteasome (Table 1). Treatment with 5 µM of probe 4 completely inhibits β1/β1i and β5/β5i activity, as expected (Supplemental Table 1).
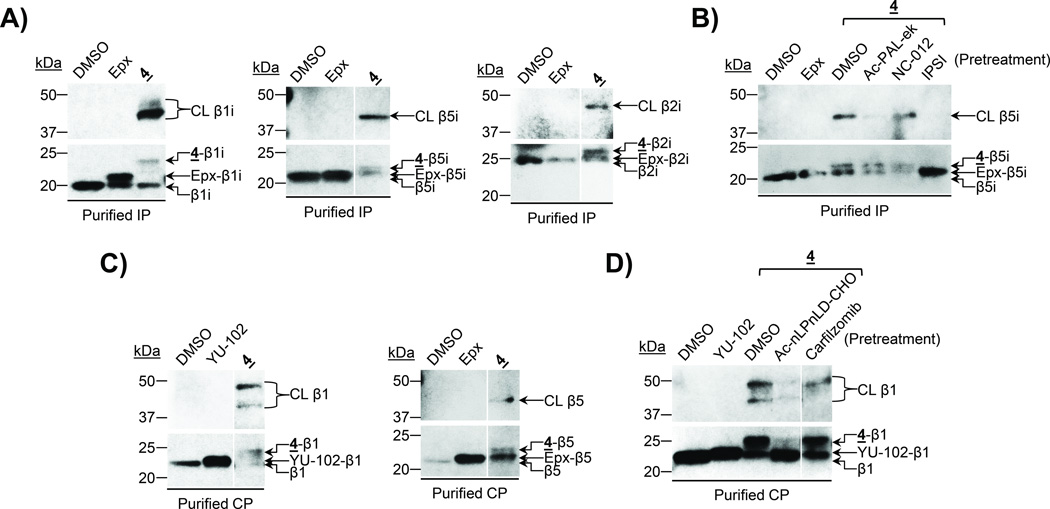
Treatment of the purified immunoproteasome with probe 4 led to the appearance of two immunoreactive bands between ~40–50 kDa on a β1i immunoblot (Figure 4A, left panel). Upon similar analysis using a β5i antibody, a crosslinked β5i band was detected at an apparent molecular weight comparable to that of the major crosslinked β1i band (Figure 4A, middle panel), suggesting that it represents β1i-β5i crosslinking. The minor immunoreactive crosslinked β1i band likely represents β1i-β2i crosslinking, based on the immunoblotting results obtained using a β2i antibody (Figure 4A, right panel). These results indicate that probe 4 preferentially crosslinks β1i with β5i, and crosslinks β1i with β2i to a much lesser extent. Competition assays were conducted to confirm these results. Specifically, pretreatment of the purified immunoproteasome with the β5i-selective inhibitor IPSI[25b] prior to treatment with probe 4 competed away the higher-molecular-weight anti- β5i-immunoreactive band (Figure 4B), confirming that this band indeed represents crosslinked β5i. Pretreatment with the β1i-selective inhibitor Ac-PAL-ek also effectively depleted this band (Figure 4B), confirming that the subunit crosslinked with β5i is β1i.
Figure 4.
UKP13-(PEG)494-6Ahx-PR825 (probe 4) crosslinks multiple pairs of catalytic subunits. A) The purified 20S immunoproteasome (IP) was treated with DMSO, epoxomicin (Epx), or probe 4 prior to immunoblotting for β1i, β5i, or β2i. B) The purified 20S immunoproteasome was treated with DMSO or epoxomicin, or pretreated with DMSO or the indicated subunit-selective inhibitors prior to treatment with probe 4 as indicated. β5i was detected by immunoblotting. C) The purified 20S constitutive proteasome (CP) was treated with DMSO, YU-102, epoxomicin, or probe 4 prior to immunoblotting for β1 or β5. D) The purified 20S constitutive proteasome was treated with DMSO or YU-102, or pretreated with DMSO or the indicated subunit-selective inhibitors prior to treatment with probe 4 as indicated. β1 was detected by immunoblotting. Irrelevant lanes were removed from blots shown in A, Cand D. CL = crosslinked subunit.
To further examine the subunits crosslinked by probe 4we next treated the purified constitutive proteasome with this probe and, upon immunoblotting analysis, observed the appearance of two anti-β1-immunoreactive bands resulting from β1 crosslinking (Figure 4C, left panel). When a similar analysis was performed using a β5 antibody, a crosslinked β5 band was detected at an apparent molecular weight comparable to the lower crosslinked β1 band (Figure 4C, right panel), suggesting β1-β5 crosslinking by probe 4. Alternatively, alignment of the upper crosslinked β1 band observed for the probe 4-treated constitutive proteasome with the band resulting from β1 crosslinking by probe 3 (Supplemental Figure 2A) suggests that this band represents β1-β2 crosslinking. Competition assays were again conducted to verify the identities of the subunits crosslinked by probe 4. Pretreatment of the purified constitutive proteasome with the β1-selective inhibitor Ac-nLPnLD-CHO prior to treatment with probe 4 substantially attenuated both of the higher-molecular-weight anti-β1-immunoreactive bands (Figure 4D and Supplemental Figure 2B), confirming that they both represent crosslinked β1. Conversely, pretreatment with the β5/β5i-selective inhibitor carfilzomib[29] selectively competes away only the lower crosslinked β1 band (Figure 4D), while pretreatment with the β2/β2i-selective inhibitor NC-012[25f] selectively competes away only the upper crosslinked β1 band (Supplemental Figure 2B), confirming that these bands result from β1-β5 and β1-β2 crosslinking, respectively.
Discussion
Here, we report for the first time the development of irreversibly-binding bifunctional activity-based probes which can be used in a crosslinking strategy to determine the catalytic subunit composition of intact proteasome complexes present within cells. These probes, generated by coupling two epoxyketone pharmacophore-based proteasome inhibitors via linkers of varying lengths and chemical compositions, crosslink different pairs of proteasome catalytic subunits. Several bifunctional proteasome-targeting compounds were previously developed from reversible inhibitors in effort to improve inhibitory potency and subunit selectivity over the monomeric inhibitors of which they are comprised.[20–21] In contrast to these reversible bifunctional inhibitors, our probes are designed to irreversibly crosslink specific pairs of catalytic subunits within individual proteasome complexes, permitting identification of the two probe-bound subunits by SDS-PAGE and immunoblotting. Results obtained using our bifunctional probes provide direct evidence of catalytic subunit colocalization within individual 20S proteasome core complexes, thereby allowing proteasomes of distinct compositions to be identified.
While the inhibitory preferences of each bifunctional probe are largely determined by those of their respective monomeric inhibitors, the length and composition of the linker by which these inhibitors are coupled appears to be an important determinant of the crosslinking efficiency of a particular catalytic subunit pair. For example, although probe 1 preferentially inhibits the activities of β1/β1i and β5/β5i, it preferentially crosslinks β1i with β2 and with β2i (Figure 3A and B). Alternatively, crosslinking of β1/β1i with β5/β5i could be achieved by coupling inhibitors targeting these subunits by a significantly longer PEG linker, as observed with probe 4 (Figures 2 and 4). In addition, probe 4 also crosslinks β1/β1i with β2/β2i (Figure 4 and Supplemental Figure 2), demonstrating that the long flexible PEG linker allows crosslinking of multiple catalytic subunit pairs within the 20S proteasome complex.
Compared with previous approaches in which the identities of proteasome subtypes present within a particular cell or tissue type are indirectly deduced from the overall expression levels of individual proteasome catalytic subunits,[8b, 14b–d, 15b, 15d, 15e, 17–18] our crosslinking approach allows proteasomes with distinct compositions to be clearly identified, even when multiple subtypes are present.[13] Additionally, in contrast to approaches requiring separation of subtypes via chromatographic methods, isoelectric focusing-free flow electrophoresis, or immunoprecipitation,[8b, 13–14, 14e, 15a, 15c, 15f, 18] our crosslinking strategy does not require the purification of proteasomes or separation of individual proteasome subtypes; incubation of our bifunctional probes with cell extracts can be immediately followed by protein denaturation and immunoblotting analysis without additional processing steps. Furthermore, in contrast to our previously-reported FRET-based approach,[19] the crosslinking approach reported here does not require the careful selection of subunit-specific concentrations of each probe for accurate interpretation of the experimental results.
Studies of proteasome assembly pathways indicate that catalytic subunits are not incorporated at random; there appear to be regulatory mechanisms in place to restrict the combinations of catalytic subunits that exist together within mature 20S core particles. These studies suggested that incorporation of β2i into proteasome complexes requires the presence of β1i,[8a, 8b, 30] thus precluding the formation of intermediate proteasomes containing β2i without β1i. Furthermore, β5i is required for the proper maturation of proteasomes containing either β1i or β2i. Consequently, intermediate proteasomes comprised of β1i and/or β2i with β5 cannot form, while the incorporation of β5i into proteasomes containing β1i and β2i leads to the formation of mature immunoproteasomes.[8a, 8b, 31]
However, in contrast to the requirement of β1i for β2i’s incorporation, β1i can incorporate into proteasomes without β2i.[8b] Additionally, β5i can efficiently incorporate into proteasomes comprised of either constitutive subunits or immunosubunits.[8a, 8b, 30–31] Therefore, in addition to pure constitutive proteasomes and pure immunoproteasomes, intermediate proteasomes comprised of β1i-β2-β5i and β1-β2-β5i can also form. Although these subtypes were originally identified in transfected cell lines and cells from knockout mice,[8a, 8b, 30–31] their endogenous formation in murine and human cells has been confirmed.[13, 14b] β1i-containing subtypes (β1i-β2-β5i and β1i-β2i-β5i) can be distinguished from one another by their discrete combinations of β1i with β2 or β2i. Likewise, β1-containing subtypes (β1-β2-β5 and β1-β2-β5i) can be distinguished from one another by their discrete combinations of β1 with β5 or β5i. In this regard, our bifunctional probes that crosslink these subunit pairs can be utilized to detect the presence of these distinct proteasome subtypes. Results obtained using our probes in the current study indicate that, in addition to the two conventional proteasome subtypes (pure constitutive proteasomes and immunoproteasomes), U266 cells contain intermediate proteasome subtypes of composition β1i-β2- β5i (Figure 3 B and E). In contrast, we were unable to detect the presence of subtypes containing both β1 and β2i (Figure 3D), consistent with previous reports suggesting that coincorporation of these subunits is not preferred.[8a, 14b, 30]
Conclusion
In summary, it is clear that recent advances in understanding of proteasome biology have led to an increased interest in dissecting the unique functions of each proteasome subtype. This in turn necessitates the development of improved methods to readily identify the subtypes present within complex biological samples. Our crosslinking strategy, which allows the direct analysis of proteasome catalytic subunit composition in cell or tissue lysates without requiring their prior purification or separation, is well suited for this application. Studies employing this new strategy will facilitate evaluations of the functional roles of distinct proteasome subtypes and their relevance as drug targets or disease biomarkers.
Experimental Section
Cell Culture and Whole Cell Lysis
U266 human multiple myeloma cells were purchased from the American Type Culture Collection and cultured under the recommended conditions at 37°C and 5% CO2. The cells were lysed in Passive Lysis Buffer (Promega) according to the manufacturer’s instructions, and the protein concentration of the lysate was determined using the Bio-Rad Protein Assay (Bio-Rad).
Proteasome Activity Assays
Purified human 20S constitutive proteasome or immunoproteasome (Boston Biochem) (50 ng/well) were incubated with DMSO or increasing concentrations of each bifunctional probe for 1 hr at room temperature in 20S proteasome assay buffer (20 mM Tris/Cl, pH 8.0, 0.5 mM EDTA, 0.035% SDS) in 96-well plates (total volume/well = 90 µL). (For the trypsin-like substrate, SDS was omitted from the assay buffer.) 10 µL of each substrate diluted in assay buffer was then added to obtain a final volume of 100 µL/well. The following subunit-selective fluorogenic substrates were used at a final concentration of 100 µM: Ac-nLPnLD-AMC (β1-selective),[27a] Ac-PAL-ek (β1i-selective), Ac-ANW-AMC (β5i-selective).[27b] Ac-RLR-AMC (β2/β2i-selective)[27a] and Ac-WLA-AMC (β5-selective)[27b] were used at a final concentration of 20 µM. Fluorescence produced by the release of AMC was measured once per min over 1hr on a SpectraMax M5 microplate reader (Molecular Devices) using an excitation wavelength of 360 nm and an emission wavelength of 460 nm. Reaction velocities were determined using linear regression in GraphPad Prism, and those of the DMSO-treated controls were set as 100%. IC50 values were determined by nonlinear regression using GraphPad Prism and represent the averages of 3 replicates.
Western Blotting
U266 cell lysates were diluted to a concentration of 1 µg/µL in Passive Lysis Buffer (Promega). Alternatively, purified human 20S constitutive proteasome or immunoproteasome (Boston Biochem) was diluted to a concentration of 5–15 ng/µL in 20S proteasome assay buffer (20 mM Tris/Cl, pH 8.0, 0.5 mM EDTA, 0.035% SDS). Samples were then treated with DMSO, 5 µM epoxomicin (a broadly-acting proteasome inhibitor used as a positive control for covalent modification of β1i, β2, β2i, β5, and β5i), 10 µM YU-102 (a β1/β1i-selective inhibitor[25a, 26] used as a positive control for covalent modification of β1), or 1–10 µM of each bifunctional probe for 4 hrs at room temperature. Where indicated, samples were treated with DMSO, 3 µM Ac-nLPnLD-CHO (β1-selective, unpublished results), 3 µM NC-012 (β2/β2i-selective),[25f] 0.02 µM carfilzomib (β5/β5i-selective),[29] 1 µM Ac-PAL-ek (β1i-selective, unpublished results), or 0.1 µM IPSI (β5i-selective)[25b] for 1 hr at room temperature prior to the addition of bifunctional probes. 2X Laemmli sample buffer (Sigma-Aldrich) was added to each sample, and proteins were denatured at 100°C for 10 min. An equivalent protein amount of each sample was resolved by 14% SDS-PAGE, and proteins were subsequently transferred onto PVDF membranes (Bio-Rad). Membranes were incubated in 5% non-fat dry milk (Bio-Rad) in Tris-buffered saline with 0.05% Tween-20 (TBST) for 1 hr at room temperature. The following primary antibodies were diluted in TBST solutions containing 3% BSA: β1i and β5i (Abcam), β1 (Enzo Life Sciences), β5 (Thermo Scientific). Alternatively, the β2i antibody (Santa Cruz) was diluted in 3% milk in TBST. Membranes were incubated in these primary antibody solutions overnight at 4°C. Anti-rabbit or anti-mouse horseradish peroxidase-conjugated secondary antibodies (GE Healthcare) were diluted 1:20,000 in 3% milk in TBST, and membranes were incubated with these antibodies for 1 hr at room temperature. SuperSignal West Femto Chemiluminescent Substrate (Thermo Scientific) and X-ray film (Thermo Scientific) were utilized for protein visualization.
The synthesis of the bifunctional probes and proteasome activity assays are described in the Supplemental Information section.
Supplementary Material
Acknowledgements
We would like to thank the American Foundation for Pharmaceutical Education (KC) and the National Institutes of Health (Grant R01 CA128903 (KBK) and Grant R15 CA156601 (WL)) for financially supporting this work.
Footnotes
Supporting information for this article is given via a link at the end of the document.
Contributor Information
Wooin Lee, Email: wooin.lee@snu.ac.kr.
Kyung-Bo Kim, Email: kbkim2@uky.edu.
References
- 1.a) Glotzer M, Murray AW, Kirschner MW. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]; b) Shkedy D, Gonen H, Bercovich B, Ciechanover A. FEBS Lett. 1994;348:126–130. doi: 10.1016/0014-5793(94)00582-6. [DOI] [PubMed] [Google Scholar]
- 2.a) Sadoul R, Fernandez PA, Quiquerez AL, Martinou I, Maki M, Schroter M, Becherer JD, Irmler M, Tschopp J, Martinou JC. EMBO J. 1996;15:3845–3852. [PMC free article] [PubMed] [Google Scholar]; b) Grimm LM, Goldberg AL, Poirier GG, Schwartz LM, Osborne BA. EMBO J. 1996;15:3835–3844. [PMC free article] [PubMed] [Google Scholar]
- 3.Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 4.Herndon TM, Deisseroth A, Kaminskas E, Kane RC, Koti KM, Rothmann MD, Habtemariam B, Bullock J, Bray JD, Hawes J, Palmby TR, Jee J, Adams W, Mahayni H, Brown J, Dorantes A, Sridhara R, Farrell AT, Pazdur R. Clin. Cancer. Res. 2013;19:4559–4563. doi: 10.1158/1078-0432.CCR-13-0755. [DOI] [PubMed] [Google Scholar]
- 5.a) Kisselev Alexei F, van der Linden WA, Overkleeft Herman S. Chem. Biol. 19:99–115. doi: 10.1016/j.chembiol.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Driscoll JJ, Woodle ES. Semin. Hematol. 2012;49:277–283. doi: 10.1053/j.seminhematol.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Tanahashi N, Murakami Y, Minami Y, Shimbara N, Hendil KB, Tanaka K. J. Biol. Chem. 2000;275:14336–14345. doi: 10.1074/jbc.275.19.14336. [DOI] [PubMed] [Google Scholar]
- 7.a) Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]; b) Unno M, Mizushima T, Morimoto Y, Tomisugi Y, Tanaka K, Yasuoka N, Tsukihara T. Structure. 2002;10:609–618. doi: 10.1016/s0969-2126(02)00748-7. [DOI] [PubMed] [Google Scholar]
- 8.a) Griffin TA, Nandi D, Cruz M, Fehling HJ, Kaer LV, Monaco JJ, Colbert RA. J. Exp. Med. 1998;187:97–104. doi: 10.1084/jem.187.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) De M, Jayarapu K, Elenich L, Monaco JJ, Colbert RA, Griffin TA. J. Biol. Chem. 2003;278:6153–6159. doi: 10.1074/jbc.M209292200. [DOI] [PubMed] [Google Scholar]; c) Murata S, Yashiroda H, Tanaka K. Nat. Rev. Mol. Cell Biol. 2009;10:104–115. doi: 10.1038/nrm2630. [DOI] [PubMed] [Google Scholar]
- 9.a) Stohwasser R, Standera S, Peters I, Kloetzel P-M, Groettrup M. Eur. J. Immunol. 1997;27:1182–1187. doi: 10.1002/eji.1830270520. [DOI] [PubMed] [Google Scholar]; b) Morel S, Lévy F, Burlet-Schiltz O, Brasseur F, Probst-Kepper M, Peitrequin A-L, Monsarrat B, Van Velthoven R, Cerottini J-C, Boon T, Gairin JE, Van den Eynde BJ. Immunity. 2000;12:107–117. doi: 10.1016/s1074-7613(00)80163-6. [DOI] [PubMed] [Google Scholar]; c) Ossendorp F, Fu N, Camps M, Granucci F, Gobin SJP, van den Elsen PJ, Schuurhuis D, Adema GJ, Lipford GB, Chiba T, Sijts A, Kloetzel P-M, Ricciardi-Castagnoli P, Melief CJM. J. Immunol. 2005;174:7815–7822. doi: 10.4049/jimmunol.174.12.7815. [DOI] [PubMed] [Google Scholar]; d) Basler M, Youhnovski N, van den Broek M, Przybylski M, Groettrup M. J. Immunol. 2004;173:3925–3934. doi: 10.4049/jimmunol.173.6.3925. [DOI] [PubMed] [Google Scholar]; e) Hensley SE, Zanker D, Dolan BP, David A, Hickman HD, Embry AC, Skon CN, Grebe KM, Griffin TA, Chen W, Bennink JR, Yewdell JW. J. Immunol. 2010;184:4115–4122. doi: 10.4049/jimmunol.0903003. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Frisan T, Levitsky V, Polack A, Masucci MG. J. Immunol. 1998;160:3281–3289. [PubMed] [Google Scholar]
- 10.a) Glynne R, Powis SH, Beck S, Kelly A, Kerr L-A, Trowsdale J. Nature. 1991;353:357–360. doi: 10.1038/353357a0. [DOI] [PubMed] [Google Scholar]; b) Kelly A, Powis SH, Glynne R, Radley E, Beck S, Trowsdale J. Nature. 1991;353:667–668. doi: 10.1038/353667a0. [DOI] [PubMed] [Google Scholar]; c) Groettrup M, Kraft R, Kostka S, Standera S, Stohwasser R, Kloetzel P-M. Eur. J. Immunol. 1996;26:863–869. doi: 10.1002/eji.1830260421. [DOI] [PubMed] [Google Scholar]; d) Sijts EJAM, Kloetzel PM. Cell. Mol. Life Sci. 2011;68:1491–1502. doi: 10.1007/s00018-011-0657-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) Gaczynska M, Goldberg AL, Tanaka K, Hendil KB, Rock KL. J. Biol. Chem. 1996;271:17275–17280. doi: 10.1074/jbc.271.29.17275. [DOI] [PubMed] [Google Scholar]; b) Groettrup M, Kirk CJ, Basler M. Nat. Rev. Immunol. 2010;10:73–78. doi: 10.1038/nri2687. [DOI] [PubMed] [Google Scholar]
- 12.Murata S, Sasaki K, Kishimoto T, Niwa S-i, Hayashi H, Takahama Y, Tanaka K. Science. 2007;316:1349–1353. doi: 10.1126/science.1141915. [DOI] [PubMed] [Google Scholar]
- 13.Dahlmann B, Ruppert T, Kuehn L, Merforth S, Kloetzel P-M. J. Mol. Biol. 2000;303:643–653. doi: 10.1006/jmbi.2000.4185. [DOI] [PubMed] [Google Scholar]
- 14.a) Klare N, Seeger M, Janek K, Jungblut PR, Dahlmann B. J. Mol. Biol. 2007;373:1–10. doi: 10.1016/j.jmb.2007.07.038. [DOI] [PubMed] [Google Scholar]; b) Guillaume B, Chapiro J, Stroobant V, Colau D, Van Holle B, Parvizi G, Bousquet-Dubouch M-P, Théate I, Parmentier N, Van den Eynde BJ. Proc. Natl. Acad. Sci. USA. 2010;107:18599–18604. doi: 10.1073/pnas.1009778107. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wang X, Zhao Z, Luo Y, Chen G, Li Z. Proteomics Clin. Appl. 2011;5:484–492. doi: 10.1002/prca.201000149. [DOI] [PubMed] [Google Scholar]; d) Fabre B, Lambour T, Delobel J, Amalric F, Monsarrat B, Burlet-Schiltz O, Bousquet-Dubouch M-P. Mol. Cell. Proteomics. 2013;12:687–699. doi: 10.1074/mcp.M112.023317. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Gohlke S, Kloß A, Tsokos M, Textoris-Taube K, Keller C, Kloetzel PM, Dahlmann B. Ann. Hepatol. 2014;13:429–438. [PubMed] [Google Scholar]
- 15.a) Drews O, Wildgruber R, Zong C, Sukop U, Nissum M, Weber G, Gomes AV, Ping P. Mol. Cell. Proteomics. 2007;6:2021–2031. doi: 10.1074/mcp.M700187-MCP200. [DOI] [PubMed] [Google Scholar]; b) Gomes AV, Young GW, Wang Y, Zong C, Eghbali M, Drews O, Lu H, Stefani E, Ping P. Mol. Cell. Proteomics. 2009;8:302–315. doi: 10.1074/mcp.M800058-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Kloß A, Meiners S, Ludwig A, Dahlmann B. Cardiovasc. Res. 2010;85:367–375. doi: 10.1093/cvr/cvp217. [DOI] [PubMed] [Google Scholar]; d) Pelletier S, Schuurman KG, Berkers CR, Ovaa H, Heck AJR, Raijmakers R. Mol. BioSyst. 2010;6:1450–1453. doi: 10.1039/c004989a. [DOI] [PubMed] [Google Scholar]; e) Zheng J, Dasgupta A, Bizzozero OA. J. Neurochem. 2012;121:486–494. doi: 10.1111/j.1471-4159.2012.07699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Gohlke S, Mishto M, Textoris-Taube K, Keller C, Giannini C, Vasuri F, Capizzi E, D’Errico-Grigioni A, Kloetzel P-M, Dahlmann B. Age. 2014;36:57–72. doi: 10.1007/s11357-013-9543-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.a) Guillaume B, Stroobant V, Bousquet-Dubouch M-P, Colau D, Chapiro J, Parmentier N, Dalet A, Van den Eynde BJ. J. Immunol. 2012;189:3538–3547. doi: 10.4049/jimmunol.1103213. [DOI] [PubMed] [Google Scholar]; b) Vigneron N, Van den Eynde BJ. Curr. Opin. Immunol. 2012;24:84–91. doi: 10.1016/j.coi.2011.12.002. [DOI] [PubMed] [Google Scholar]; c) Zanker D, Waithman J, Yewdell JW, Chen W. J. Immunol. 2013;191:52–59. doi: 10.4049/jimmunol.1300802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Visekruna A, Joeris T, Schmidt N, Lawrenz M, Ritz JP, Buhr HJ, Steinhoff U. Inflamm. Bowel Dis. 2009;15:526–533. doi: 10.1002/ibd.20805. [DOI] [PubMed] [Google Scholar]
- 18.Joeris T, Schmidt N, Ermert D, Krienke P, Visekruna A, Kuckelkorn U, Kaufmann SHE, Steinhoff U. PLoS One. 2012;7:e39827. doi: 10.1371/journal.pone.0039827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park JE, Wu Y, Carmony KC, Miller Z, Sharma LK, Lee D-M, Kim D-Y, Lee W, Kim K-B. Mol. BioSyst. 2014;10:196–200. doi: 10.1039/c3mb70471h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loidl G, Groll M, Musiol H-J, Huber R, Moroder L. Proc. Natl. Acad. Sci. USA. 1999;96:5418–5422. doi: 10.1073/pnas.96.10.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.a) Maréchal X, Pujol A, Richy N, Genin E, Basse N, Reboud-Ravaux M, Vidal J. Eur. J. Med. Chem. 2012;52:322–327. doi: 10.1016/j.ejmech.2012.02.052. [DOI] [PubMed] [Google Scholar]; b) Desvergne A, Genin E, Maréchal X, Gallastegui N, Dufau L, Richy N, Groll M, Vidal J, Reboud-Ravaux M. J. Med. Chem. 2013;56:3367–3378. doi: 10.1021/jm4002007. [DOI] [PubMed] [Google Scholar]
- 22.Loidl G, Groll M, Musiol H-J, Ditzel L, Huber R, Moroder L. Chem. Biol. 1999;6:197–204. doi: 10.1016/S1074-5521(99)80036-2. [DOI] [PubMed] [Google Scholar]
- 23.a) Huber Eva M, Basler M, Schwab R, Heinemeyer W, Kirk Christopher J, Groettrup M, Groll M. Cell. 148:727–738. doi: 10.1016/j.cell.2011.12.030. [DOI] [PubMed] [Google Scholar]; b) Lu YF, Sheng H, Zhang Y, Li ZY. J. Zhejiang Univ., Sci., B. 2013;14:816–828. doi: 10.1631/jzus.B1200299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groll M, Kim KB, Kairies N, Huber R, Crews CM. J. Am. Chem. Soc. 2000;122:1237–1238. [Google Scholar]
- 25.a) Myung J, Kim KB, Lindsten K, Dantuma NP, Crews CM. Mol. Cell. 7:411–420. doi: 10.1016/s1097-2765(01)00188-5. [DOI] [PubMed] [Google Scholar]; b) Parlati F, Lee SJ, Aujay M, Suzuki E, Levitsky K, Lorens JB, Micklem DR, Ruurs P, Sylvain C, Lu Y, Shenk KD, Bennett MK. Blood. 2009;114:3439–3447. doi: 10.1182/blood-2009-05-223677. [DOI] [PubMed] [Google Scholar]; c) Sharma LK, Lee N-R, Jang ER, Lei B, Zhan C-G, Lee W, Kim K-B. ChemBioChem. 2012;13:1899–1903. doi: 10.1002/cbic.201200307. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Carmony KC, Lee D-M, Wu Y, Lee N-R, Wehenkel M, Lee J, Lei B, Zhan C-G, Kim K-B. Biorg. Med. Chem. 2012;20:607–613. doi: 10.1016/j.bmc.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Carmony K, Kim K. Cell Biochem. Biophys. 2013;67:91–101. doi: 10.1007/s12013-013-9626-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Mirabella Anne C, Pletnev Alexandre A, Downey Sondra L, Florea Bogdan I, Shabaneh Tamer B, Britton M, Verdoes M, Filippov Dmitri V, Overkleeft Herman S, Kisselev Alexei F. Chem. Biol. 2011;18:608–618. doi: 10.1016/j.chembiol.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Zhou H-J, Aujay MA, Bennett MK, Dajee M, Demo SD, Fang Y, Ho MN, Jiang J, Kirk CJ, Laidig GJ, Lewis ER, Lu Y, Muchamuel T, Parlati F, Ring E, Shenk KD, Shields J, Shwonek PJ, Stanton T, Sun CM, Sylvain C, Woo TM, Yang J. J. Med. Chem. 2009;52:3028–3038. doi: 10.1021/jm801329v. [DOI] [PubMed] [Google Scholar]; h) Muchamuel T, Basler M, Aujay MA, Suzuki E, Kalim KW, Lauer C, Sylvain C, Ring ER, Shields J, Jing J, Shwonek P, Parlati F, Demo SD, Bennett MK, Kirk CJ, Groettrup M. Nat. Med. 2009;15:781–787. doi: 10.1038/nm.1978. [DOI] [PubMed] [Google Scholar]
- 26.Miller Z, Ao L, Kim KB, Lee W. Curr. Pharm. Des. 2013;19:4140–4151. doi: 10.2174/1381612811319220018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.a) Kisselev AF, Goldberg AL. In: Methods Enzymol. Raymond JD, editor. Vol. 398. Academic Press; 2005. pp. 364–378. [DOI] [PubMed] [Google Scholar]; b) Blackburn C, Gigstad KM, Hales P, Garcia K, Jones M, Bruzzese FJ, Barrett C, Liu JX, Soucy TA, Sappal DS, Bump N, Olhava EJ, Fleming P, Dick LR, Tsu C, Sintchak MD, Blank JL. Biochem. J. 2010;430:461–476. doi: 10.1042/BJ20100383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cenci S, Oliva L, Cerruti F, Milan E, Bianchi G, Raule M, Mezghrani A, Pasqualetto E, Sitia R, Cascio P. J. Leukocyte Biol. 2012;92:921–931. doi: 10.1189/jlb.1011497. [DOI] [PubMed] [Google Scholar]
- 29.Kuhn DJ, Chen Q, Voorhees PM, Strader JS, Shenk KD, Sun CM, Demo SD, Bennett MK, van Leeuwen FWB, Chanan-Khan AA, Orlowski RZ. Blood. 2007;110:3281–3290. doi: 10.1182/blood-2007-01-065888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Groettrup M, Standera S, Stohwasser R, Kloetzel PM. Proc. Natl. Acad. Sci. USA. 1997;94:8970–8975. doi: 10.1073/pnas.94.17.8970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kingsbury DJ, Griffin TA, Colbert RA. J. Biol. Chem. 2000;275:24156–24162. doi: 10.1074/jbc.M001742200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.