Abstract
ConPred II (http://bioinfo.si.hirosaki-u.ac.jp/~ConPred2/) is a server for the prediction of transmembrane (TM) topology [i.e. the number of TM segments (TMSs), TMS positions and N-tail location] based on a consensus approach by combining the results of several proposed methods. The ConPred II system is constructed from ConPred_elite and ConPred_all (previously named ConPred), proposed earlier by our group. The prediction accuracy of ConPred_elite is almost 100%, which is achieved by sacrificing the prediction coverage (20–30%). ConPred_all predicts TM topologies for all the input sequences with accuracies improved by up to 11% over individual proposed methods. In the ConPred II system, the TM topology prediction of input TM protein sequences is executed following a two-step process: (i) input sequences are first run through the ConPred_elite program; (ii) sequences for which ConPred_elite does not give the TM topology are delivered to the ConPred_all program for TM topology prediction. Users can get access to the ConPred II system automatically by submitting sequences to the server. The ConPred II server will return the predicted TM topology models and graphical representations of their contents (hydropathy plots, helical wheel diagrams of predicted TMSs and snake-like diagrams).
INTRODUCTION
The functions of transmembrane (TM) proteins are inferable, at least roughly, from knowing the TM topology, i.e. the number of TM segments (TMSs), TMS positions and N-tail location (1,2), and high-quality TM topology data are required for the comprehensive functional identification of TM proteins. For this reason, various TM topology prediction methods have been developed to date, but they are not accurate enough, i.e. at most 50–60% accuracy in terms of the prediction of whole TM topology (3–5).
We have proposed two TM topology prediction methods with improved accuracies based on the consensus approach: ConPred_elite (6) and ConPred_all (previously named as ConPred) (4,7). ConPred_elite achieves prediction reliability of >95% by sacrificing prediction coverage (estimated at 20–30%). ConPred_all improves the prediction accuracy of TM topology by up to 11% over individual proposed methods.
In this paper, we present a consensus prediction server, ConPred II, constructed from ConPred_elite and ConPred_all, for obtaining more reliable TM topology models which should serve for e.g. the comprehensive classification and identification of TM protein functions, and three-dimensional (3D) structural modeling of TM proteins.
ALGORITHMS AND PREDICTION ACCURACIES
The ConPred II system predicts the TM topology of an input TM protein sequence using a two-step procedure. First, the ConPred_elite program is applied to the input sequence. In cases when ConPred_elite does not give a TM topology prediction, the ConPred_all program predicts the TM topology. The details of the ConPred_elite and ConPred_all programs are described below.
Dataset and TM topology prediction methods used
As a training dataset for tuning ConPred II (both ConPred_elite and ConPred_all), we used the TMPDB_alpha_non-redundant dataset (Release 6.3), which is composed of 138 prokaryotic and 93 eukaryotic sequences with experimentally characterized TM topology information (7). The prediction methods used for consensus are KKD (8), TMpred (9), TopPred II (10), DAS (11), TMAP (12), MEMSAT 1.8 (13), SOSUI (14), TMHMM 2.0 (15) and HMMTOP 2.0 (16).
ConPred_elite
ConPrede_elite makes use of five TM topology prediction methods which can predict N-tail location too: TMpred, TMAP, MEMSAT 1.8, TMHMM 2.0 and HMMTOP 2.0. ConPred_elite targets only the sequences to which all five methods assign the same number of TMS(s) (≥1). The largest distance between the center positions of the corresponding predicted TMSs is then calculated. If the distance is within 15 residues for prokaryotic and 11 for eukaryotic sequences, the average of the 5 center positions is calculated. Only when all the TMSs fit this condition the consensus TMS prediction is concluded for the target sequence. And then we determine both ends of the final individual TMSs by extending TMS stretches by as many as 10 residues toward both N- and C-termini from the average center positions. The ConPred_elite prediction is classified into two modes: (i) agree_one mode, when all the five predictions agree on one TM topology model; (ii) split_two mode, when the prediction splits into two models, i.e. all the five predictions agree on the number of TMSs and TMS positions but disagree on N-tail location.
Table 1 shows ConPred_elite's performance for 138 prokaryotic and 93 eukaryotic sequences in the TMPDB_alpha_non-redundant dataset. The prediction accuracy was evaluated on a per-sequence basis. As for TMS positions, when all the center positions of the predicted TMSs coincided within 11 residues with the corresponding TMSs in the actual data, the prediction was regarded as correct. ConPred_elite can predict TM topology almost perfectly, with reliabilities of 0.98 and 0.95 for prokaryotic and eukaryotic sequences, respectively. These high prediction reliabilities are attained by sacrificing prediction coverage (which could be called ‘yield’). The yields for prokaryotic and eukaryotic sequences are 30.4 and 21.5%, respectively, as is shown in Table 1.
Table 1. Prediction performance of ConPred_elite for 138 prokaryotic and 93 eukaryotic sequences in the TMPDB_alpha_non-redundant dataset.
| Super-kingdom | agree_one | split_two | Total | Yield (%) | |||||
|---|---|---|---|---|---|---|---|---|---|
| Correct | Incorrect | Correct | Incorrect | Correct | Incorrect | Rate of correct models included in the predictions | |||
| Majority | Minority | ||||||||
| Prokaryotic | 17 | 1 | 21 | 3 | 0 | 41 | 1 | 0.98 | 30.4 |
| Eukaryotic | 7 | 0 | 10 | 2 | 1 | 19 | 1 | 0.95 | 21.5 |
ConPred_all
ConPred_all comprises four combinations of five prediction methods: (i) KKD, TopPred II, MEMSAT 1.8, SOSUI and TMHMM 2.0 for TMS prediction for prokaryotic sequences; (ii) TopPred II, TMAP, MEMSAT 1.8, TMHMM 2.0 and HMMTOP 2.0 for N-tail location prediction for prokaryotic sequences; (iii) KKD, DAS, MEMSAT 1.8, SOSUI and HMMTOP 2.0 for TMS prediction for eukaryotic sequences; (iv) TMpred, TopPred II, MEMSAT 1.8, TMHMM 2.0 and HMMTOP 2.0 for N-tail location for eukaryotic sequences.
TMS prediction is carried out by iterating the following four steps (from N-terminus to C-terminus): (i) the prediction results of the five individual methods are scanned toward the C-terminus to find the center position of the TMS; (ii) a window of 10 residues for prokaryotic or 11 for eukaryotic sequences is extended toward the C-terminus from the center position; (iii) when at least three TMSs are in the window, the average of the center positions is calculated and the predicted TMS is obtained as a region of 21 residues around the averaged center position; (iv) the TMSs used in the voting are masked, and the scanning of the prediction results is restarted from the residue next to the average-center position. In the consensus N-tail location prediction, a simple ‘majority voting’ system is adopted. The final TM topology prediction is obtained by integrating the two results, i.e. the predicted TMSs and N-tail location.
In Table 2, the prediction accuracies of ConPred_all, together with the nine selected TM topology prediction methods, are shown for 138 prokaryotic and 93 eukaryotic sequences in the TMPDB_alpha_non-redundant dataset. The evaluation criterion with respect to TMS position is the same as in the case of ConPred_elite. It can be clearly seen that ConPred_all has accuracies higher by 5–10% than even the best-performing individual methods.
Table 2. Prediction accuracies of ConPred_all together with the nine selected TM topology prediction methods for prokaryotic sequences (138, consisting of 33 single-spanning and 105 multi-spanning) and eukaryotic sequences (93, consisting of 39 single-spanning and 54 multi-spanning) in the TMPDB_alpha_non-redundant dataset.
| Prediction methods | Prediction accuracies (%) | |||
|---|---|---|---|---|
| No. of TMSs | No. of TMSs and TMS positions | N-tail location | TM topology | |
| Prokaryotic | ||||
| ConPred_all | 79.7a (93.9, 75.2) | 74.6a (93.9, 68.6) | 86.2b (72.7, 90.5) | 68.1e (72.7, 66.7) |
| KKD | 60.1 (90.9, 50.5) | 55.1 (90.9, 43.8) | – | – |
| TMpred | 56.5 (78.8, 49.5) | 50.7 (78.8, 41.9) | 61.6 (57.6, 62.9) | 36.2 (45.5, 33.3) |
| TopPred II | 56.5 (69.7, 52.4) | 47.1 (69.7, 40.0) | 73.9 (63.6, 77.1) | 38.4 (45.5, 36.2) |
| DAS | 41.3 (48.5, 39.0) | 34.8 (48.5, 30.5) | – | – |
| TMAP | 52.9 (87.9, 41.9) | 45.7 (84.8, 33.3) | 57.2 (51.5, 59.0) | 29.0 (42.4, 24.8) |
| MEMSAT 1.8 | 71.0 (90.9, 64.8) | 65.2 (90.9, 57.1) | 84.1 (75.8, 86.7) | 56.5 (66.7, 53.3) |
| SOSUI | 65.2 (90.9, 57.1) | 59.4 (90.9, 49.5) | – | – |
| TMHMM 2.0 | 65.2 (75.8, 61.9) | 60.9 (75.8, 56.2) | 73.9 (69.7, 75.2) | 53.6 (69.7, 48.6) |
| HMMTOP 2.0 | 69.6 (84.8, 64.8) | 63.8 (84.8, 57.1) | 79.7 (66.7, 83.8) | 56.5 (63.6, 54.3) |
| Eukaryotic | ||||
| ConPred_all | 72.0c (92.3, 57.4) | 69.9c (92.3, 53.7) | 78.5d (84.6, 74.1) | 54.8f (79.5, 37.0) |
| KKD | 54.8 (82.1, 35.2) | 49.5 (82.1, 25.9) | – | – |
| TMpred | 59.1 (69.2, 51.9) | 53.8 (69.2, 42.6) | 64.5 (59.0, 68.5) | 35.5 (46.2, 27.8) |
| TopPred II | 51.6 (69.2, 38.9) | 48.4 (69.2, 33.3) | 65.6 (69.2, 63.0) | 36.6 (51.3, 25.9) |
| DAS | 31.2 (38.5, 25.9) | 29.0 (38.5, 22.2) | – | – |
| TMAP | 59.1 (79.5, 44.4) | 52.7 (79.5, 33.3) | 47.3 (53.8, 42.6) | 26.9 (43.6, 14.8) |
| MEMSAT 1.8 | 57.0 (87.2, 35.2) | 54.8 (84.6, 33.3) | 63.4 (71.8, 57.4) | 39.8 (64.1, 22.2) |
| SOSUI | 57.0 (71.8, 46.3) | 53.8 (71.8, 40.7) | – | – |
| TMHMM 2.0 | 59.1 (92.3, 35.2) | 58.1 (92.3, 33.3) | 75.3 (76.9, 74.1) | 46.2 (76.9, 24.1) |
| HMMTOP 2.0 | 68.8 (87.2, 55.6) | 64.5 (87.2, 48.1) | 72.0 (82.1, 64.8) | 51.6 (74.4, 35.2) |
The figures in parentheses are the prediction accuracies for single-spanning and multi-spanning sequences, respectively.
aCombination for prokaryotic: KKD, TopPred II, MEMSAT 1.8, SOSUI and TMHMM 2.0; voting window size = 10 residues.
bCombination for prokaryotic (N-tail location): TopPred II, TMAP, MEMSAT 1.8, TMHMM 2.0 and HMMTOP 2.0.
cCombination for eukaryotic: KKD, DAS, MEMSAT 1.8, SOSUI and HMMTOP 2.0; voting widow size = 11 residues.
dCombination for eukaryotic (N-tail location): TMpred, TopPred II, MEMSAT 1.8, TMHMM 2.0 and HMMTOP 2.0.
eCombination of a and b.
fCombination of c and d.
INPUT, OUTPUT AND BEHAVIOR
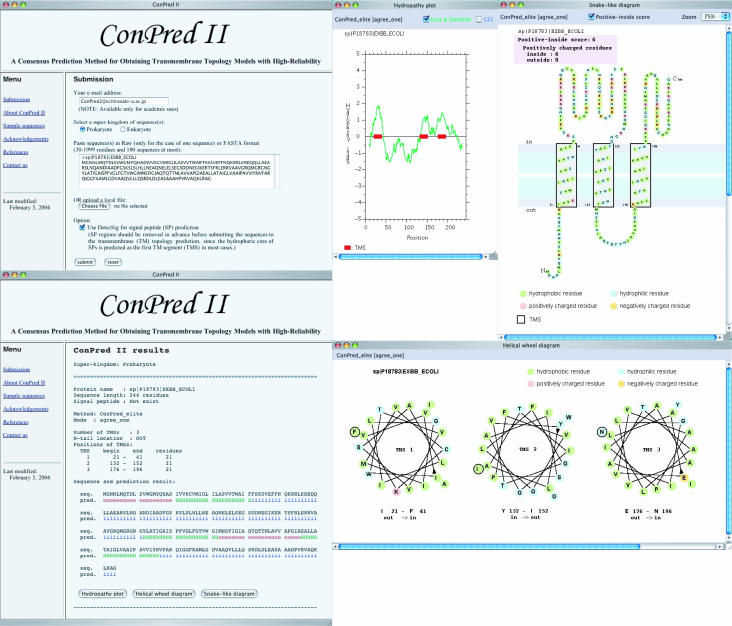
ConPred II runs as a CGI server, written in PHP, and is accessible at http://bioinfo.si.hirosaki-u.ac.jp/~ConPred2/ (Figure 1). Users need first to enter their email addresses in the appropriate text box to prove they are in an academic or governmental organization, and then to select a super-kingdom (prokaryotic or eukaryotic) from the radio buttons. Sequences must be specified in the single-letter amino acid notation; there is no sensitivity between lowercase and uppercase letters. Users have a choice in inputting query sequences between pasting the sequences directly into the input window and uploading the sequences from their local disks. The acceptable formats of input sequences are Raw (only in the case of one sequence) or FASTA. The length of each sequence is limited to a range of 30–1999 residues, since several prediction methods used in ConPred II have a restriction on sequence length.
Figure 1.
A screenshot of the input and output pages of the ConPred II system: top left, an input page to submit protein sequences; bottom left, an output page of prediction results; top middle, a hydropathy plot; top right, a snake-like diagram of the predicted TM topology model; bottom right, helical wheel diagrams of predicted TMSs.
Signal peptide regions should be removed in advance of submitting the sequences for TM topology prediction, since the hydrophobic core of signal peptides is predicted as the first TMS in most cases (17). We provide DetecSig (18) for signal peptide prediction as an input option in the ConPred II system. As for TM proteins that have a signal peptide, the N-tail location is automatically determined as non-cytoplasmic (Nout) without using the N-tail location prediction (19).
When a query sequence is submitted, the individual TM topology prediction programs used in ConPred_elite and ConPred_all start running. First, the ConPred_elite program tries to combine the prediction results of the five methods to obtain a TM topology model. In cases when ConPred_elite gives no prediction result, the ConPred_all program is called for the TM topology prediction.
The default output is an HTML-formatted file which is able to be displayed in any browser. The prediction result (i.e. the number of TMSs, TMS positions and N-tail location) from ConPred II appears on the result page, as shown in Figure 1. It should be noted that the predictions made by individual methods are neither displayed on the result page nor able to be downloaded. As output options for the prediction result, the following three graphical representations (written in Java Applet) are provided (Figure 1): (i) a hydropathy plot, (ii) helical wheel diagrams of the predicted TMSs and (iii) a snake-like diagram of the predicted TM topology model.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the developers of the TM topology prediction methods used in the ConPred II system for providing us with the programs. This research was supported in part by a Grant-in-Aid for Scientific Research on Priority Areas (C) ‘Genome Information Science’ (No. 15014203) and a Grant-in-Aid for Scientific Research (C) (No. 14580665) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
REFERENCES
- 1.Sugiyama Y., Polulyakh,N. and Shimizu,T. (2003) Identification of transmembrane protein functions by binary topology patterns. Protein Eng., 16, 479–488. [DOI] [PubMed] [Google Scholar]
- 2.Inoue Y., Ikeda,M. and Shimizu,T. (2004) Proteome-wide classification and identification of mammalian-type GPCRs by binary topology pattern. Comput. Chem. Biol., 28, 39–49. [DOI] [PubMed] [Google Scholar]
- 3.Möller S., Croning,M.D. and Apweiler,R. (2001) Evaluation of methods for prediction of membrane spanning regions. Bioinformatics, 17, 646–653. [DOI] [PubMed] [Google Scholar]
- 4.Ikeda M., Arai,M., Lao,D.M. and Shimizu,T. (2002) Transmembrane topology prediction methods: a re-assessment and improvement by a consensus method using a dataset of experimentally-characterized transmembrane topologies. In Silico Biol., 2, 19–33. [PubMed] [Google Scholar]
- 5.Chen C.P., Kernytsky,A. and Rost,B. (2002) Transmembrane helix predictions revisited. Protein Sci., 11, 2774–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xia J.-X., Ikeda,M. and Shimizu,T. (2004) ConPred_elite: a highly reliable approach to transmembrane topology prediction. Comput. Biol. Chem., 28, 51–60. [DOI] [PubMed] [Google Scholar]
- 7.Ikeda M., Arai,M., Okuno,T. and Shimizu,T. (2003) TMPDB: a database of experimentally-characterized transmembrane topologies. Nucleic Acids Res., 31, 406–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klein P., Kanehisa,M. and DeLisi,C. (1985) The detection and classification of membrane-spanning proteins. Biochim. Biophys. Acta, 815, 468–476. [DOI] [PubMed] [Google Scholar]
- 9.Hofmann K. and Stoffel,W. (1993) TMbase—a database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler, 347, 166. [Google Scholar]
- 10.Claros M.G. and von Heijne,G. (1994) TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci., 10, 685–686. [DOI] [PubMed] [Google Scholar]
- 11.Cserzö M., Wallin,E., Simon,I., von Heijne,G. and Elofsson,A. (1997) Prediction of transmembrane α-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng., 10, 673–676. [DOI] [PubMed] [Google Scholar]
- 12.Persson B. and Argos,P. (1997) Prediction of membrane protein topology utilizing multiple sequence alignments. J. Protein Chem., 16, 453–457. [DOI] [PubMed] [Google Scholar]
- 13.Jones D.T., Taylor,W.R. and Thornton,J.M. (1994) A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry, 33, 3038–3049. [DOI] [PubMed] [Google Scholar]
- 14.Hirokawa T., Boon-Chieng,S. and Mitaku,S. (1998) SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics, 14, 378–379. [DOI] [PubMed] [Google Scholar]
- 15.Krogh A., Larsson,B., von Heijne,G. and Sonnhammer,E.L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol., 305, 567–580. [DOI] [PubMed] [Google Scholar]
- 16.Tusnády G.E. and Simon,I. (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics, 17, 849–850. [DOI] [PubMed] [Google Scholar]
- 17.Lao D.M., Arai,M., Ikeda,M. and Shimizu,T. (2002) The presence of signal peptide significantly affects transmembrane topology prediction. Bioinformatics, 18, 1562–1566. [DOI] [PubMed] [Google Scholar]
- 18.Lao D.M. and Shimizu,T. (2001) A method for discriminating a signal peptide and a putative 1st transmembrane segment. In Valafar,F. (ed.), Proceedings of the 2001 International Conference on Mathematics and Engineering Techniques in Medicine and Biological Sciences—METMBS '01. CSREA Press, USA, pp. 119–125. [Google Scholar]
- 19.Arai M., Ikeda,M. and Shimizu,T. (2003) Comprehensive analysis of transmembrane topologies in prokaryotic genomes. Gene, 304, 77–86. [DOI] [PubMed] [Google Scholar]