Abstract
Continuum solvation models, such as Poisson–Boltzmann and Generalized Born methods, have become increasingly popular tools for investigating the influence of electrostatics on biomolecular structure, energetics and dynamics. However, the use of such methods requires accurate and complete structural data as well as force field parameters such as atomic charges and radii. Unfortunately, the limiting step in continuum electrostatics calculations is often the addition of missing atomic coordinates to molecular structures from the Protein Data Bank and the assignment of parameters to biomolecular structures. To address this problem, we have developed the PDB2PQR web service (http://agave.wustl.edu/pdb2pqr/). This server automates many of the common tasks of preparing structures for continuum electrostatics calculations, including adding a limited number of missing heavy atoms to biomolecular structures, estimating titration states and protonating biomolecules in a manner consistent with favorable hydrogen bonding, assigning charge and radius parameters from a variety of force fields, and finally generating ‘PQR’ output compatible with several popular computational biology packages. This service is intended to facilitate the setup and execution of electrostatics calculations for both experts and non-experts and thereby broaden the accessibility to the biological community of continuum electrostatics analyses of biomolecular systems.
INTRODUCTION
Due to the ubiquitous nature of electrostatics in biomolecular systems, a variety of computational methods have been developed for calculating these interactions [see refs (1–6) and references therein]. Popular computational electrostatics methods for biomolecular systems can be loosely grouped into two categories: ‘explicit solvent’ methods, which treat solvent molecules in full molecular detail, and ‘implicit solvent’ methods, which include solvent-solute interactions in averaged or continuum fashion. Implicit solvent methods have gained increasing popularity for evaluating the electrostatic properties of biomolecules as they typically require significantly less computational effort than explicit solvent models (1,2,4–7).
The basic ingredients of an implicit solvent electrostatics calculation are environmental parameters such as temperature, solvent dielectric and ionic strength; biomolecular atomic coordinates; and parameters for atomic charges and radii. While the environmental parameters are relatively straightforward to specify, the remaining two ingredients can often be difficult to supply. In particular, most biomolecular structures in the Protein Data Bank (PDB) (8) do not contain hydrogen atoms, and many are also missing a fraction of the heavy atom coordinates. The addition of hydrogens and the reconstruction of these missing coordinates is not a trivial process; electrostatic properties obtained from the ‘repaired’ structures can often be very sensitive to the manner in which missing atoms are added and protonation states are assigned (9,10). Furthermore, inconsistent atomic nomenclature and other force field idiosyncrasies can often make the assignment of atomic charges and radii a cumbersome task.
This paper describes the development of the freely available PDB2PQR service (http://agave.wustl.edu/pdb2pqr/), which was designed to facilitate the setup and execution of continuum electrostatics calculations, particularly by non-experts. As its name implies, this service was designed to convert PDB-format (11) structural information into ‘PQR’-format parameterized files. A PQR file is a popular and compact way to include atomic parameters in a PDB-like format by replacing the occupancy column of a PDB file (‘P’) with the atomic charge (‘Q’) and the temperature factor column with the radius (‘R’). The PQR format is therefore able to be parsed by most visualization programs and contains additional information that can be read by continuum electrostatics software, including APBS (12) and MEAD (13), as well as other computational biology programs, particularly AutoDock (14) and AMBER (15). Finally, there are a number of tools available (12,15) for converting from PQR format to other formats required by continuum electrostatics software such as Delphi (16) and UHBD (17).
METHODS
The PDB2PQR web service is driven by a modular, Python-based collection of routines which provides considerable flexibility to the software and permits non-interactive, high-throughput usage. The service is available via the web at http://agave.wustl.edu/pdb2pqr/ (with an NBCR-supported mirror at http://nbcr.sdsc.edu/pdb2pqr/); the Python software is available by contacting the authors.
Rebuilding missing heavy atoms
The first step in the PDB2PQR pipeline involves identification of potential problems with the initial biomolecular structure file. Specifically, the initial structure file is processed and missing heavy (non-hydrogen) atoms are identified. Next, the PDB2PQR service will determine if it is possible to rebuild the missing atoms and will exit if the structure appears too, incomplete to reconstruct (e.g. >10% of heavy atoms missing from the entire structure too few atoms in the sidechain to reconstruct from topology). If PDB2PQR ascertains that heavy atom reconstruction is feasible, atoms are rebuilt using standard amino acid topologies in conjunction with existing atomic coordinates to determine new positions for the missing heavy atoms.
Additionally, users are presented with an option to ‘debump’ the reconstructed atoms and thereby ensure that they are not being placed within the Van der Waals radii of other nearby atoms. This procedure is carried out by varying the sidechain χ angles until the steric conflict is resolved. Since debumping of newly added atoms can be somewhat time-consuming, users are presented with an option to disable this feature.
Addition of hydrogens
Hydrogen atoms are added to the biomolecular structure after reconstruction of all heavy atoms. Hydrogens are positioned to optimize the global hydrogen-bonding network in the structure. The procedure is similar in purpose to the work of Hooft et al. (18) and Nielsen et al. (10) but uses a newer algorithm and implementation. First, the phases of HIS, ASN and GLN sidechain χ angles are sampled via Monte Carlo for optimum hydrogen-bonding conformation. Second, water hydrogens are placed and undergo rigid body Monte Carlo optimization for maximum water–water and water–protein hydrogen bonding. In addition to optimizing proton placement, these routines also assign protonation states to HIS, ASP and GLU based on optimum hydrogen bonding, local energetics, and model pKa values. By default, newly added hydrogen atoms are checked for steric conflicts via the debumping procedure outlined above. To facilitate faster preparation of PQR structures, both the hydrogen bond optimization and the debumping routines can be disabled at the option of the user.
Parameter assignment
After addition of hydrogen atoms, the PDB2PQR suite assigns atomic charges and radii based on the chosen force field. Currently, PDB2PQR provides parameters from CHARMM22 (19), AMBER99 (20) or PARSE (21) force fields. This step involves translating the atom and residue names found in the force field to those of the input structure file and assigning the appropriate parameters. Several popular variations on naming schemes are attempted for the translation; the service exits with an error message if none of the translation attempts is successful. Currently, parameters are not assigned to non-water HETATM entries as these groups are not consistently present in the available force field files. A list of all unparameterized atoms is both displayed in the PDB2PQR web output and saved as comments in the final PQR file. Additionally, any residues with non-integral charges after parameterization are identified and listed both in the web output and as remarks in the PQR file.
APBS input file generation
Users are also presented with the option to automatically generate an input file to the APBS Poisson–Boltzmann solver software (12). This input file is constructed to perform a solvation energy calculation on the newly generated PQR file with grid spacings, lengths, and so on pre-calculated to give accurate energetic results using typical parameter values (2,22).
CONCLUSIONS
We have described the free PDB2PQR web server, a service which helps users prepare molecular structures for continuum electrostatics calculations by adding missing atoms, optimizing hydrogen bonding and assigning atomic charge and radius parameters. Many of these operations are not unique to continuum electrostatics and should be of use for a wider range of computational biology work, including drug design and docking as well as molecular dynamics simulations. Therefore, we anticipate that the PDB2PQR service will be a helpful addition to the portfolio of tools available to the structural and computational biology communities.
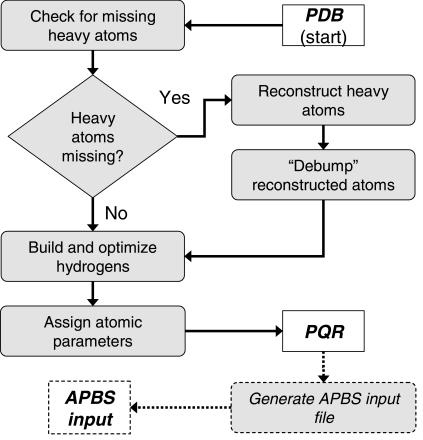
Figure 1.
Flowchart demonstrating the sequence of operations performed by the pipeline. The process begins with an input PDB file and ends with a parameterized PQR file and, optionally, an APBS input file.
Acknowledgments
ACKNOWLEDGEMENTS
N.A.B. and T.J.D. were supported by a grant from NPACI and NIH grant GM069702. N.A.B. is an Alfred P. Sloan Research Fellow. J.E.N. acknowledges support from the Danish Natural Science Council. J.A.M. is supported by NIH, NSF, CTBP, NBCR, W.M. Keck Foundation and Accelrys Inc. The development of the PDB2PQR pipeline was supported by the National Biomedical Computation Resource (NIH P41 RR08605).
REFERENCES
- 1.Baker N.A. and McCammon,J.A. (2002) Electrostatic interactions. In Bourne,P. and Weissig,H. (eds.), Structural Bioinformatics. John Wiley & Sons, Inc., New York. [Google Scholar]
- 2.Gilson M. (2000) Introduction to continuum electrostatics. In Beard,D.A. (ed.), Biophysics Textbook Online. Biophysical Society, Bethesda, MD, Vol. Computational Biology. [Google Scholar]
- 3.Darden T.A. (2001) Treatment of long-range forces and potential. In Becker,O.M., MacKerell,A.D.J., Roux,B. and Watanabe,M. (eds), Computational Biochemistry and Biophysics. Marcel Dekker, Inc., New York, pp. 91–114. [Google Scholar]
- 4.Roux B. (2001) Implicit solvent models. In Becker,O.M., MacKerell,A.D.J., Roux,B. and Watanabe,M. (eds), Computational Biochemistry and Biophysics. Marcel Dekker, New York, pp. 133–152. [Google Scholar]
- 5.Davis M.E. and McCammon,J.A. (1990) Electrostatics in biomolecular structure and dynamics. Chem. Rev., 94, 7684–7692. [Google Scholar]
- 6.Honig B. and Nicholls,A. (1995) Classical electrostatics in biology and chemistry. Science, 268, 1144–1149. [DOI] [PubMed] [Google Scholar]
- 7.Roux B. and Simonson,T. (1999) Implicit solvent models. Biophys. Chem., 78, 1–20. [DOI] [PubMed] [Google Scholar]
- 8.Bourne P.E., Addess,K.J., Bluhm,W.F., Chen,L., Deshpande,N., Feng,Z., Fleri,W., Green,R., Merino-Ott,J.C., Townsend-Merino,W. et al. (2004) The distribution and query systems of the RCSB Protein Data Bank. Nucleic Acids Res., 32, D223–D225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nielsen J.E., Andersen,K.V., Honig,B., Hooft,R.W.W., Klebe,G., Vriend,G. and Wade,R.C. (1999) Improving macromolecular electrostatics calculations. Protein Eng., 12, 657–662. [DOI] [PubMed] [Google Scholar]
- 10.Nielsen J.E. and Vriend,G. (2001) Optimizing the hydrogen-bond network in Poisson–Boltzmann equation-based pK(a) calculations. Proteins, 43, 403–412. [DOI] [PubMed] [Google Scholar]
- 11.(1996) Protein Data Bank Contents Guide: Atomic Coordinate Entry Format Description. 2.1 ed. Research Collaboratory for Structural Bioinformatics. [Google Scholar]
- 12.Baker N.A., Sept,D., Joseph,S., Holst,M.J. and McCammon,J.A. (2001) Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl Acad. Sci., USA, 98, 10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bashford D. (1997) An object-oriented programming suite for electrostatic effects in biological molecules. In Ishikawa,Y., Oldehoeft,R.R., Reynders,J.V.W. and Tholburn,M. (eds), Scientific Computing in Object-Oriented Parallel Environments. Springer, Berlin, Vol. 1343, pp. 233–240. [Google Scholar]
- 14.Morris G.M., Goodsell,D.S., Halliday,R.S., Huey,R., Hart,W.E., Belew,R.K. and Olson,A.J. (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem., 19, 1639–1662. [Google Scholar]
- 15.Pearlmann D.A., Case,D.A., Caldwell,J.W., Ross,W.S., Cheatham,T.E., 3rd, DeBolt,S., Ferguson,D., Seibel,G. and Kollman,P. (1995) AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics, and free energy calculations to simulate the structural and energetic properties of molecules. Comp. Phys. Commun., 91, 1–41. [Google Scholar]
- 16.Rocchia W., Sridharan,S., Nicholls,A., Alexov,E., Chiabrera,A. and Honig,B. (2002) Rapid grid-based construction of the molecular surface and the use of induced surface charge to calculate reaction field energies: applications to the molecular systems and geometric objects. J. Comput. Chem., 23, 128–137. [DOI] [PubMed] [Google Scholar]
- 17.Madura J.D., Briggs,J.M., Wade,R.C., Davis,M.E., Luty,B.A., Ilin,A., Antosiewicz,J., Gilson,M.K., Bagheri,B., Scott,L.R. et al. (1995) Electrostatics and diffusion of molecules in solution—simulations with the University of Houston Brownian Dynamics program. Comput. Phys. Commun., 91, 57–95. [Google Scholar]
- 18.Hooft R.W., Sander,C. and Vriend,G. (1996) Positioning hydrogen atoms by optimizing hydrogen-bond networks in protein structures. Proteins, 26, 363–376. [DOI] [PubMed] [Google Scholar]
- 19.MacKerell A.D.J., Bashford,D., Bellot,M., Dunbrack,R.L., Jr, Evanseck,J.D., Field,M.J., Fischer,S., Gao,J., Guo,H., Ha,S. et al. (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B, 102, 3586–3616. [DOI] [PubMed] [Google Scholar]
- 20.Wang J.M., Cieplak,P. and Kollman,P.A. (2000) How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem., 21, 1049–1074. [Google Scholar]
- 21.Sitkoff D., Sharp,K.A. and Honig,B. (1994) Accurate calculation of hydration free energies using macroscopic solvent models. J. Phys. Chem., 98, 1978–1988. [Google Scholar]
- 22.Baker N.A. (2004) Poisson–Boltzmann methods for biomolecular electrostatics. Methods Enzymol., in press. [DOI] [PubMed] [Google Scholar]