Abstract
Steroid receptors (SRs) bind specific DNA regulatory sequences, thereby activating and repressing gene expression. We previously showed that transcriptional coregulator Hic-5 facilitates glucocorticoid regulation of some genes but blocks glucocorticoid regulation of others. Here, in a genome-wide analysis, Hic-5 depletion dramatically increased the global number of sites occupied by glucocorticoid receptor (GR) α (the major GR isoform), and many binding sites blocked by Hic-5 were associated with genes for which Hic-5 also blocked glucocorticoid-regulated expression. Hic-5 had similar effects on GRγ (a splice variant of GRα) and estrogen receptor α (ERα), facilitating hormonal regulation of some genes and blocking hormonal regulation of others. As with GRα, Hic-5 blocking of hormonal gene regulation mediated by GRγ and ERα was associated with blocking of GRγ and ERα occupancy at nearby sites. Hic-5 supported hormonal regulation of many more genes for GRα than for GRγ or ERα and thus exhibited selective coregulator functions for different SRs. In contrast, the number of Hic-5–blocked genes was similar for all 3 SRs. In addition to classic coregulator activity, Hic-5 influences the genomic occupancy of multiple SRs and thereby blocks some aspects of hormonal regulation. Thus, Hic-5, because of its tissue-specific expression, could contribute to tissue-specific genomic occupancy and gene regulation by SRs.
Steroid receptors (SRs) are ligand-activated transcription factors that bind to specific DNA sites and regulate the expression of several hundred genes in a cell type–specific manner. Like other DNA-binding transcription factors, SRs recruit coregulator proteins that assist them in achieving their effects on chromatin and assembly of active transcription complexes (1). Together, SRs and coregulators regulate transcription in a cell type–, gene-, and chromatin context–specific manner, as evidenced by the fact that different combinations of coregulators are required for regulation of different target genes of one SR in a single cell type (2–6). The mechanism of coregulator specificity of gene regulation is largely unknown but presumably involves several factors inherent to the gene itself: the specific DNA sequence to which the SR binds, which alters SR conformation and thus the complement of coregulators that it can bind and recruit (7, 8); the nearby binding of other transcription factors, which also recruit coregulators and may affect SR binding and conformation; and last, the local chromatin structure of the gene (9), which may dictate the types of coregulator activities required for successful activation or repression. Most of the reported functions of coregulator proteins are downstream of SR binding to DNA, such as chromatin remodeling (10), assembly of an active transcription complex (11), and RNA polymerase II recruitment (12). In contrast, we recently reported the regulation of genomic occupancy of the glucocorticoid receptor (GR) by coregulator Hic-5, indicating that coregulators can influence transcription factor binding to genomic sites by acting before or in concert with transcription factor binding in chromatin (4).
Hic-5 (TGFβ1I1) belongs to the paxillin family of proteins and has four LIM domains at the C terminus and LD motifs at the N terminus (13). Hic-5 has no known enzymatic activity and is believed to function as an adaptor protein involved in assembling higher order protein complexes (14). Through its C-terminal LIM domains, Hic-5 has been shown to bind the τ2 activation domain in the hinge region of GR (14, 15). Hic-5 has distinct functions in the cytosolic and nuclear compartments (16). In the cytosol, it influences signaling from the focal adhesion complexes (17), whereas in the nuclear compartment it mediates and regulates the activities of the SRs androgen receptor, progesterone receptor, and GR (14, 15, 18) and other transcription factors such as vitamin D receptor, SMADs, and peroxisome proliferator–activated receptor γ (PPARγ) (19–21). Using U2OS osteosarcoma cells expressing the major GR isoform, GRα, we previously characterized genome-wide Hic-5 coregulator gene regulation in glucocorticoid signaling and found that Hic-5 differentially affects regulation of multiple subsets of glucocorticoid-regulated genes, using different molecular mechanisms of action (4). On some genes, Hic-5 functioned as a classic coregulator, acting after GRα binding to assist GRα in activating or repressing transcription, in at least some cases by regulating occupancy of the Mediator complex and RNA polymerase II recruitment. Interestingly, Hic-5 also selectively blocked hormonal regulation of a distinct subset of potential GRα target genes. These genes remained transcriptionally unresponsive to hormone when Hic-5 was present but became robustly hormone regulated after Hic-5 depletion. Upon mechanistic examination of 3 of these Hic-5–blocked genes, we found that Hic-5 precluded GRα occupancy and chromatin remodeling of GR binding sites near the blocked genes.
In this study, we examined whether the various modes of Hic-5 action observed with GRα would apply to 2 other SRs: GRγ, a naturally occurring isoform generated by alternative splicing that differs from GRα by a single amino acid insertion within the DNA-binding domain (22), and estrogen receptor α (ERα). GRα and GRγ have similar hormone binding affinities, but they bind to overlapping but different sets of genomic sites and thus regulate overlapping but different sets of target genes when activated by glucocorticoids (23). We examined the selectivity of the Hic-5 coregulator function for the 3 different SRs, all expressed in U2OS cells, and whether Hic-5 blocks hormonal regulation of some potential GRγ and ERα target genes as it did for GRα. In addition, because we previously found that the mechanism used by Hic-5 to block hormonal regulation of 3 selected GRα target genes involved preventing GRα binding to potential GR binding sites associated with the blocked genes, we extended this analysis to selected GRγ and ERα target genes and also performed a genome-wide mapping of GRα binding events in U2OS cells with or without depletion of Hic-5. Our results demonstrate that, although Hic-5 functions as a coregulator for all 3 SRs, the type of regulation observed and the number of target genes affected are specific to each SR. Furthermore, Hic-5 serves a common critical function for multiple SRs in restricting their occupancy on specific subsets of potential target genes. The broad range of tissues in which GRα, GRγ, and ERα are found (24, 25), superimposed on the highly tissue-specific expression of Hic-5 (26), thus suggests a potential role for Hic-5 in influencing SR binding site selection in different tissues and cell types.
Materials and Methods
Cell culture and reagents
U2OS osteosarcoma cells stably expressing wild-type GRα or GRγ were maintained as described previously (4). U2OS cells stably expressing ERα under control of a doxycycline (Dox)-regulated promoter were obtained from Thomas Spelsberg and cultured as described previously (27). Estradiol (E2), dexamethasone (Dex), and Dox were purchased from Sigma-Aldrich. Charcoal-stripped fetal bovine serum (FBS) was purchased from Omega Scientific. Antibodies used in the immunoblot analysis were against Hic-5, GR, ERα (Santa Cruz Biotechnology), hemagglutinin (HA) (Roche), and Flag M2 (Sigma-Aldrich). For hormone-stimulated experiments in U2OS(ERα), steroid-depleted transfection medium was used (5% charcoal-stripped FBS in DMEM/F-12).
Gene expression microarrays
RNA samples from U2OS(GRγ) cells transfected with nonspecific small interfering (si) RNA or siHic-5 and treated with Dex were collected as described previously (4). The siRNA constructs were the same as those used previously (4). U2OS(ERα) cells were transfected with siRNA, grown for 48 hours in steroid-depleted medium, and then exposed to the vehicle ethanol or 10 nM E2 for 4 hours. ERα expression was induced by treating cells with 100 ng/mL Dox 24 hours before E2 treatment. Total RNA of biological quadruplets collected on different days was extracted (RNeasy; QIAGEN), and samples were submitted to the Southern California Genotyping Consortium at the University of California, Los Angeles, where they were placed randomly on Illumina HT12v4 BeadChips and were processed following standard Illumina procedures. Parallel data for GRα were described previously (4) and subjected to new analyses in this study. The Gene Ontology tool was used for functional annotation of different gene subsets (28)
RT-quantitative PCR (qPCR)
RNA samples were converted to cDNA (iScript strand cDNA synthesis kit; Bio-Rad) according to manufacturer's instructions. Expression of target genes with specific primers (Supplemental Table 1) was measured using SYBR Green (Roche) on the LightCycler 480 system (Roche).
Chromatin immunoprecipitation (ChIP) coupled with high-throughput sequencing (ChIP-seq)
The ChIP experiments were performed as described previously (4). In brief, U2OS(GRα) cells (29) were seeded onto 15-cm plates, transfected with siNS or siHic-5, and grown for 72 hours in DMEM (low glucose) supplemented with 5% FBS. Cells were treated with 100 nM Dex or equivalent amounts of ethanol for 1 hour before cross-linking with 1% (v/v) formaldehyde and harvesting for immunoprecipitation. ChIP-seq samples were immunoprecipitated with anti-GR antibody (Santa Cruz Biotechnology). Single-end 50-bp ChIP-seq data were generated using the Illumina HiSeq system at the Next-Generation Sequencing Core of the USC Norris Comprehensive Cancer Center. Two biological replicates were performed for each condition on separate days along with 2 input controls; sequencing of the replicates produced 33 + 26 million reads for siNS, 42 + 28 million reads for siHic-5, 31 million reads for siNS input, and 38 million reads for siHic-5 input. Samples were mapped using bwa (30) against the hg19 with mitochondria, and peaks were called using the SPP+IDR framework developed from ENCODE project (https://sites.google.com/site/anshulkundaje/projects/idr); peaks falling within blacklisted regions were removed (https://sites.google.com/site/anshulkundaje/projects/blacklists). Differential peaks were identified by merging all peaks and using a method based on MAnorm (31) but modified to allow for replicates (https://github.com/ying-w/chipseq-compare/tree/master/MAnorm) (Wu, D.-Y., D. Bittencourt, M. R. Stallcup, and K. D. Siegmund, submitted for publication). The MAnorm technique used is an extension of a previously published method (31) to allow for the utilization of replicates. In brief, we use a peak calling method developed for ENCODE (SPP+IDR) that takes advantage of ChIP-seq replicates to account for variability between experiments. This method will call peaks on pooled replicates, individual replicates, and random subsets of replicates to determine the consistency between and within replicates. We then use a modified version of MAnorm that will identify differential binding after normalizing to shared binding sites between ChIP-seq conditions.
The specific primers used in qPCR analysis of ChIP are listed in Supplemental Table 1.
Accession numbers
The Gene Expression Omnibus (GEO) entry for Illumina Bead array data for GRγ is GSE65401 and for ERα is GSE65378. The GEO entry for the GRα ChIP-seq data is GSE65847.
Results
Hic-5 shows selectivity in its coregulator function for different SRs
We previously showed that Hic-5 mediates and modulates transcriptional regulation by GRα, affecting glucocorticoid-regulated expression of multiple subsets of GRα target genes via diverse mechanisms of action (4). To elucidate the Hic-5 coregulator function for other SRs, we examined and compared with GRα the genome-wide effects of Hic-5 depletion on 2 other SRs: GRγ, a naturally occurring isoform of GR that has a single amino acid insertion in the DNA-binding domain (DBD) (compared with GRα) and ERα. Because Hic-5 failed to enhance activation of transient reporter genes by ERα (15), we wanted to test whether Hic-5 functions as a coregulator for endogenous ERα target genes. To allow for a well-controlled comparison with GRα, experiments were performed with clonal U2OS cells stably expressing GRγ or ERα. The expression levels of GRγ and GRα were approximately equal, and ERα expression was approximately 3-fold higher than in MCF-7 cells (data not shown) and was induced by the addition of Dox (Supplemental Figure 1A). The U2OS cell system offers substantial advantages for investigating the mechanisms of Hic-5 action: the genes regulated by glucocorticoid and estrogen are already known (4, 27); the full set of GR binding regions and a few ER binding regions have been defined in this cell line (32); and specific activation domains of GR required for the global set of glucocorticoid-regulated genes are known, allowing for eventual integration of information about activation domains and coregulator requirements (33). Hic-5 protein expression was efficiently depleted by the siRNA in both U2OS(GRγ) and U2OS(ERα) cells (Supplemental Figure 1, B and C) with ≥80% knockdown at the mRNA level (Supplemental Figure 1, D and E) at 4 and 24 hours of E2 treatment in U2OS(ERα) cells or Dex treatment in U2OS(GRγ) cells. Illumina BeadChip arrays were used to measure genome-wide transcript levels in cells transfected with either nonspecific or Hic-5–specific siRNA and subsequently treated with hormone (Supplemental Figure 1, F and G).
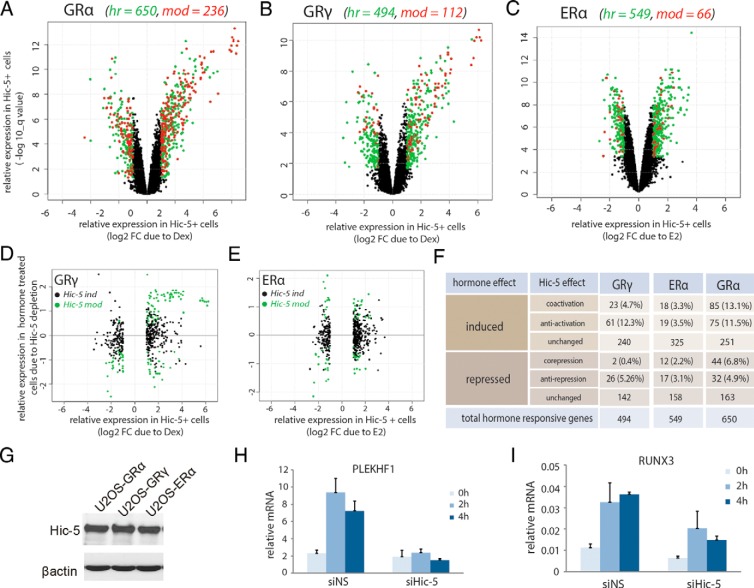
To compare the effect of Hic-5 on hormonal regulation of genes by GRα, GRγ, or ERα, we generated volcano plots, which show log2 fold change caused by 4-hour hormone treatment of the indicated U2OS cell line (containing Hic-5) for expression of all genes represented on the microarray on the x-axis vs the significance of the hormone effect on the y-axis (Figure 1, A–C). Each dot represents a specific gene, with green and red dots representing genes significantly regulated by hormone at 4 hours (2-fold change cutoff and false discovery rate–adjusted P ≤ .02); black dots represent genes that did not meet those statistical criteria and thus were defined as not hormone regulated at 4 hours of hormone treatment. Superimposed on these graphs is color-coded information indicating whether the expression level of the hormone-regulated genes (green and red dots) after 4 hours of hormone treatment was significantly different in Hic-5–depleted cells vs Hic-5–positive cells; red dots represent hormone-regulated genes for which the mRNA level at 4 hours of hormone treatment was significantly altered by Hic-5 depletion (no fold change cutoff and false discovery rate–adjusted P ≤ .02), and green dots represent hormone-regulated genes for which the mRNA level at 4 hours of hormone treatment was not significantly affected by Hic-5 depletion. As in our previous study (4), we refer to the gene sets represented by the red dots as the Hic-5–modulated (mod) genes, and we refer to the gene sets represented by the green dots as Hic-5–independent (ind) genes. Of the total number of hormone-regulated genes for GRα (n = 650), GRγ (n = 494), and ERα (n = 549) (Supplemental Dataset 1, sheet 1 and Supplemental Dataset 2, sheet 1), there were fewer genes in the mod class in U2OS(GRγ) cells (n = 112, 23%) (Supplemental Dataset 1, sheet 2) and U2OS(ERα) cells (n = 66, 12%) (Supplemental Dataset 2, sheet 2) than in U2OS(GRα) cells (n = 236, 36%) (Figure 1F). These results demonstrate selectivity in Hic-5 coregulator function for the 3 different SRs. However, despite our previous finding that Hic-5 failed to enhance transient reporter gene activation by ERα (15), Hic-5 clearly serves as a coregulator for a subset of endogenous target genes of ERα.
Figure 1.
Hic-5 selectively modulates hormonal regulation of GR and ER target genes. A–C, Volcano plots of the Dex-regulated genes in U2OS cells expressing GRα (A) and GRγ (B) and estradiol-regulated genes in U2OS(ERα) cells (C). Log2 fold changes for 4-hour hormone treatment compared with ethanol treatment in Hic-5–positive cells on the x-axis are plotted against the significance of that comparison (log10 q value) on the y-axis. Black, genes that are not significantly regulated by hormone in Hic-5–positive cells; red and green, genes that are significantly regulated by hormone (2-fold change cutoff and false discovery rate–adjusted P ≤ .02). Superimposed on this information, red and green colors indicate comparisons between 4-hour hormone-induced levels of expression in Hic-5–positive vs Hic-5–depleted cells; green, hormone-regulated ind (Hic-5–independent) genes, which are not significantly affected by Hic-5 depletion; red, hormone-regulated mod (Hic-5–modulated) genes, for which the hormone-induced level of expression is significantly affected by Hic-5 depletion; hr, hormone regulated. D and E, scatterplots showing the effect of 4-hour hormone treatment in Hic-5–positive cells (x-axis, log2 fold changes) vs the effect of Hic-5 depletion on the transcript levels in 4-hour hormone-treated cells for genes in the hormone–regulated subset for GRγ (D) and ERα (E) (y-axis, log2 fold changes). Black dots, ind genes; green dots, mod genes. F, Numbers of mod genes in each quadrant of D and E. Hormone-regulated genes are divided into hormone-induced and hormone-repressed sets and are further subdivided according to the effect that Hic-5 protein has on hormone regulation. Numbers in parentheses are percentages of the total number of hormone-regulated genes listed at the bottom of each column. G, Immunoblot analysis showing Hic-5 expression in the 3 U2OS clonal cell lines, with β-actin levels as an internal loading control. H and I, RT-qPCR validation of a mod gene candidate for GRγ (H) and for ERα (I). The duration of hormone treatment is coded by color in the figure. mRNA expression levels were normalized to glyceraldehyde-3-phosphate dehydrogenase mRNA copy numbers for each sample and represent means ± SEM for 3 biological replicates.
Scatterplots for the mod gene sets for GRγ and ERα show the effect of hormone in Hic-5–positive cells on the x-axis vs the corresponding positive or negative effects of Hic-5 depletion in the 4-hour hormone-treated cells on the y-axis (Figure 1, D and E). A comparison with the previously analyzed mod genes of GRα (4) showed that Hic-5 had both positive and negative effects on hormone induction and repression of gene expression for all 3 SRs (Figure 1, D and E; green dots, mod genes). We further subdivided these coregulator effects on hormone regulation into 4 subclasses: Hic-5–coactivated, supporting gene activation by hormone; Hic-5–corepressed, supporting gene repression by hormone; Hic-5–antiactivated, opposing gene activation by hormone; and Hic–5 antirepressed, opposing gene repression by hormone (Figure 1F). Hic-5–independent genes (ind) represent the gene subset for which Hic-5 had no effect on hormone induction or repression (Figure 1, D and E, black dots and F, unchanged and Supplemental Dataset 1, sheet 3 and Supplemental Dataset 2, sheet 3). The number of genes coactivated by Hic-5 for GRα (n = 85) was several-fold larger than for GRγ (23 genes) or ERα (18 genes), showing that Hic-5 had a broader coregulator effect for GRα than for GRγ and ERα. Similarly, Hic-5 had a much broader corepressor effect for GRα (44 genes) than for GRγ (2 genes) and ERα (12 genes). In contrast, Hic-5 was involved in antiactivation and antirepression for similar numbers of genes with GRα and GRγ (Figure 1F). The differences in Hic-5 coregulation observed for the different SRs were not due to differences in Hic-5 levels, because all 3 cell lines expressed similar levels of Hic-5 (Figure 1G). For validation by RT-qPCR of candidate coactivated genes for GRγ and ERα, we showed that induction of PLEKHF1 by Dex in U2OS(GRγ) cells and induction of RUNX3 by E2 in U2OS(ERα) cells were compromised or eliminated by Hic-5 depletion (Figure 1, H and I).
We used Gene Ontology analysis to identify functional pathways that were enriched among the combined mod and block genes for ERα and for GRγ. Several functional categories including regulation of cell proliferation were enriched in the Hic-5 regulated ERα target genes (Supplemental Dataset 3), but the Gene Ontology analysis of the Hic-5–regulated GRγ target genes did not reveal any significantly enriched gene categories (data not shown).
We also compared isoform-specific regulation of target genes by GRα and GRγ. Of all the Dex-regulated genes in the 2 cell lines (650 for GRα and 494 for GRγ), 361 genes are regulated by both receptors (Supplemental Figure 2A), 289 genes were GRα specific, and 133 genes were GRγ specific (Supplemental Dataset 1, sheet 5). Isoform-specific regulation by GRα and GRγ was seen both for genes activated by Dex as well as genes repressed by Dex (Supplemental Figure 2C). Candidate genes with isoform-specific and common regulation by Dex were validated by RT-qPCR (Supplemental Figure 2B): S100P as an example of a GRα-specific gene, JPH2 as a GRγ-specific gene, and SPINK5L3 as a gene regulated by both GRα and GRγ in response to Dex. Hic-5 depletion resulted in significant changes in Dex-induced mRNA levels (mod genes) for 94 of the 289 GRα-specific genes (Supplemental Figure 2D). However, only 20 of the 133 GRγ-specific genes were classified as mod genes (Supplemental Figure 2E), indicating a higher level of cooperation of Hic-5 with GRα than with GRγ. Hic-5 functioned as a coregulator (supporting gene activation or repression by Dex) for most GRα-specific mod genes; in contrast, antiactivation (induction by hormone and repression by coregulator) and antirepression (repression by hormone and activation by coregulator) were the primary functions of Hic-5 for the GRγ-specific mod genes (Supplemental Figure 2F).
Hic-5 interaction with GRα is stronger than with ERα
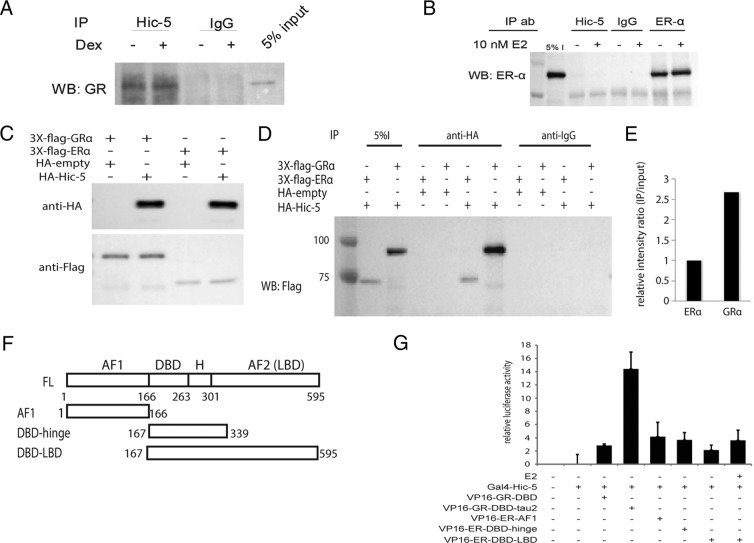
Hic-5 interacts directly with the GR τ2 domain through its C-terminal LIM domains (15). We previously observed a strong hormone-independent interaction of Hic-5 with GRα by coimmunoprecipitation (4) and observed a similar endogenous coimmunprecipitation with GRγ (Figure 2A). However, we failed to detect an interaction between endogenous Hic-5 and ERα under similar conditions (Figure 2B), even though Hic-5 is efficiently immunoprecipitated in these cells (Supplemental Figure 3A). When full-length Flag-tagged GRα and ERα and HA-tagged Hic-5 were overexpressed in Cos-7 cells (Figure 2C), we were able to detect Hic-5 interactions with both GRα and ERα (Figure 2D). However, quantitative analysis of the coimmunoprecipitated GRα and ERα bands, compared with that of the corresponding input bands, indicated that Hic-5 interaction with GRα was approximately 3 times stronger than with ERα (Figure 2, D and E). In addition, we compared the association of these proteins by a mammalian 2-hybrid assay. The Hic-5 interaction with GR was previously mapped to the 30-amino acid τ2 domain in the hinge region of GR (15). Using a GRα fragment that contains the DBD or DBD-τ2 region fused to the VP16 activation domain (AD), we see strong reporter gene activity only in the presence of GRα-τ2 (Figure 2, F and G). However, none of the ER fragments fused to the VP16 AD such as ERα-AF1, ERα-DBD-hinge, and ERα-DBD-ligand-binding domain (LBD) were able to activate the reporter gene through a Hic-5-ERα interaction (Figure 2, F and G). All fragments used in the assay were expressed at relatively similar levels (Supplemental Figure 3B). Whereas there is substantial homology between the DBD domains of GRα and ERα, there is very little homology outside the DBD, including the hinge region (Supplemental Figure 3C). Together, these results indicate that ERα has a weaker interaction with Hic-5 than GRα, and based on our failure to observe interactions of ERα and Hic-5 by endogenous coimmunoprecipitation or 2-hybrid assays, we conclude that the interaction with ERα may be indirect. This difference could be responsible for the narrower coregulation effects observed with ERα in the genome-wide expression studies.
Figure 2.
Differential Hic-5 interaction with various SRs. A and B, Coimmunoprecipitation analysis of GRγ and endogenous Hic-5 in U2OS(GRγ) cells (A) and ERα and Hic-5 in U2OS(ERα) cells (B). Extracts from cells treated with ethanol or 100 nM Dex for 1 hour for U2OS(GRγ) cells or with ethanol or 10 nM E2 for U2OS(ERα) cells were subjected to immunoprecipitation (IP) using a Hic-5–specific antibody (Ab) or nonspecific IgG control. GRγ and ERα were detected by immunoblot analysis. C, Immunoblots showing expression of HA-tagged Hic-5 and 3X Flag tagged GRα or ERα in Cos7 cells in the presence of 100 nM Dex or 10 nM E2. D, Coimmunoprecipitation of Hic-5 from extracts shown in C using HA antibody and Western blot (WB) detection of GRα or ERα using Flag antibody. E, Western blot quantification was determined relative to input using ChemiDoc MP (Bio-Rad) to measure chemiluminescence from the immunoblot. F, Diagram of various ERα domains that were cloned into pVP16 AD plasmids and used in a mammalian 2-hybrid assay. FL, full length. G, Mammalian 2-hybrid assay in CV1 cells transfected with a luciferase reporter plasmid controlled by GAL4 response elements and plasmids encoding GAL4-DBD fused to full-length Hic-5 and pVP16-AD fused with GRα-DBD, GRα-DBD-τ2, ERα-AF1, ERα-DBD-hinge or ERα-DBD-LBD. Relative luciferase activity is measured from 4 biological replicates and shown as average ± SD.
Hic-5 blocks hormonal regulation of some potential SR target genes by selective restriction of SR genomic occupancy
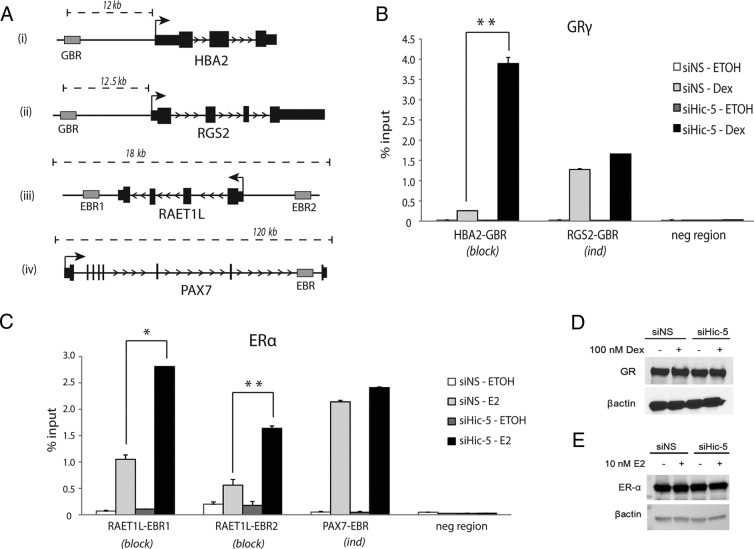
We previously reported that Hic-5 blocks glucocorticoid regulation of 3 potential target genes by restricting the genomic occupancy of GRα and preventing chromatin remodeling at the GR binding sites associated with those genes (4). To assess whether Hic-5 can block hormonal regulation of genes regulated by GRγ and ERα, we overlapped gene sets derived from 3 different comparisons for each cell line: comparison 1, genes regulated by hormone (2-fold change cut-off and false discovery rate-adjusted P ≤ .02) in cells containing Hic-5; comparison 2, genes regulated by hormone (2-fold change cutoff and false discovery rate–adjusted P ≤ .02) in cells depleted of Hic-5; and comparison 3, genes with significantly different mRNA levels (no fold change cutoff and false discovery rate–adjusted P ≤ .02) in hormone-treated cells containing or depleted of Hic-5 (Figure 3A). In this analysis, the mod genes discussed above are found in the overlap of comparisons 1 and 3 (areas shaded gray), whereas the genes that are blocked from hormonal regulation by Hic-5 (block genes) are found in comparison 2 but not in comparison 1 (areas shaded black) (Figure 3B). We find that the numbers of block genes for ERα (n = 201) (Supplemental Dataset 2, sheet 4) and GRγ (n = 190) (Supplemental Dataset 1, sheet 4) were similar to that observed previously (4) for GRα (n = 212) (Figure 3B). RT-qPCR analysis validated candidates of block and ind genes for GRγ and ERα. HBA2 responded to Dex in U2OS(GRγ) cells only after Hic-5 depletion, whereas RGS2 is an ind gene that responded to Dex similarly when Hic-5 was present or absent (Figure 3C). RAET1L behaved as a block gene in U2OS(ERα) cells, whereas PAX7 was induced by E2 in a Hic-5–independent manner (Figure 3D). The block genes were mostly unique to each of the 3 receptors, with about a 25% overlap between the block gene sets for GRα and GRγ and only about 5% of overlap between ERα and either of the 2 GR isoforms (Supplemental Figure 2G).
Figure 3.
Hic-5 blocks hormone regulation of selected target genes of GRγ and ERα as well as those of GRα. A, Venn diagrams show overlap of gene sets generated by 3 different comparisons of gene expression microarray data for each of the 3 SRs: comparison 1, genes that were significantly regulated by 4 hours of hormone treatment in the appropriate U2OS cell line transfected with control siRNA (2-fold change cutoff, false discovery rate–adjusted P ≤ .02); comparison 2, genes that were significantly regulated by 4 hours of hormone treatment in the appropriate U2OS cell line transfected with siRNA against Hic-5 (2-fold change cutoff, false discovery rate–adjusted P ≤ .02); comparison 3, genes with significantly different mRNA levels in hormone-treated cells that were transfected with Hic-5 siRNA compared with control siRNA (no fold change cutoff, false discovery rate–adjusted P ≤ .02). B, As described in A, Venn diagrams were created from the overlapping statistically significant effects of Dex and Hic-5 depletion to visualize the number of genes affected by depletion of Hic-5 in different gene classes in U2OS cells expressing GRγ, ERα, or GRα. Shaded areas show block genes (black) and mod genes (gray). C and D, RT-qPCR validation of mRNA levels for block genes (left panels) and ind genes (right panels) for GRγ (C) and ERα (D). The length of hormone treatment is indicated by shading of the data bars. Data shown are averages ± SEM for 3 biological replicates.
To assess the mechanism by which Hic-5 blocks hormonal regulation of genes by GRγ and ERα, we determined the hormone-induced occupancy of these SRs on sites associated with block and ind genes in cells containing or depleted of Hic-5. GR binding regions (GBRs) near the HBA2 (block) and RGS2 (ind) genes were identified by ChIP-seq in the U2OS(GRα) cells using a GR-specific antibody (Figure 4A). Using ChIP in U2OS(GRγ) cells followed by qPCR, we observed Dex-dependent GRγ occupancy on the GBR of the HBA2 gene, which was weak in cells containing Hic-5 but was enhanced approximately 10-fold when Hic-5 was depleted (Figure 4B). In contrast, Dex-induced GRγ occupancy was unaffected by Hic-5 depletion on the GBR of the ind gene RGS2 (Figure 4B). Similarly, Hic-5 depletion enhanced E2-induced ERα occupancy on 2 ERα binding regions (EBRs) near RAET1L (block) by about 3-fold but had no effect on ERα binding at the EBR near PAX7 (ind) (Figure 4C). The PAX7 EBR was identified in a ChIP-microarray study (34). RAET1L EBR1 was identified by scanning nearby regions by ChIP-qPCR and RAET1L EBR2 contains a canonical estrogen response element motif identified by TRANSFAC (35). Hic-5 depletion did not affect the levels of GRγ in U2OS(GRγ) cells or of ERα in U2OS(ERα) cells (Figure 4, D and E). Thus, the effect of Hic-5 on the occupancy levels of ERα and GRγ on nearby hormone-regulated genes is well correlated with the effect of Hic-5 on the hormonal regulation of those genes. Our results indicate that the mechanism of action by which Hic-5 prevents hormone-induced transcription of block genes is common for GRα, GRγ, and ERα and involves inhibiting occupancy of the SR on receptor binding sites associated with the block genes. Furthermore, the mechanism by which Hic-5 regulates block genes is distinct from its classic coregulator function with the mod genes of GRα, in which Hic-5 assists the SRs in recruiting the Mediator complex and RNA polymerase II (4).
Figure 4.
Hic-5 restricts genomic occupancy of GRγ and ERα on block gene enhancers. A, Diagrams showing gene loci and the GBR or EBR. B, GRγ occupancy at the sites shown in A was determined by quantitative ChIP in U2OS(GRγ) cells transfected with siNS or siHic-5 and treated with ethanol (ETOH) or 100 nM Dex for 1 hour. C, ERα occupancy at the sites shown in A was determined by quantitative ChIP in U2OS(ERα) cells transfected with siNS or siHic-5 and treated with ethanol or 10 nM E2 for 1 hour. U2OS(ERα) cells were treated with 100 ng/mL Dox for 24 hours before E2 treatment to induce ERα expression. Results were normalized to the PCR signal obtained with the same primers using input chromatin as the template. Error bars represent means ± SD from 3 technical replicates in one experiment, and results shown are representative of at least 3 independent experiments. *, P ≤ .05; **, P ≤ .01, calculated using a paired t test for results from 3 independent experiments. D and E, Immunoblot analysis showing levels of GRγ (D) and ERα (E) in cells expressing or depleted of Hic-5, with β-actin levels as an internal loading control.
Hic-5 depletion increases GRα binding events globally
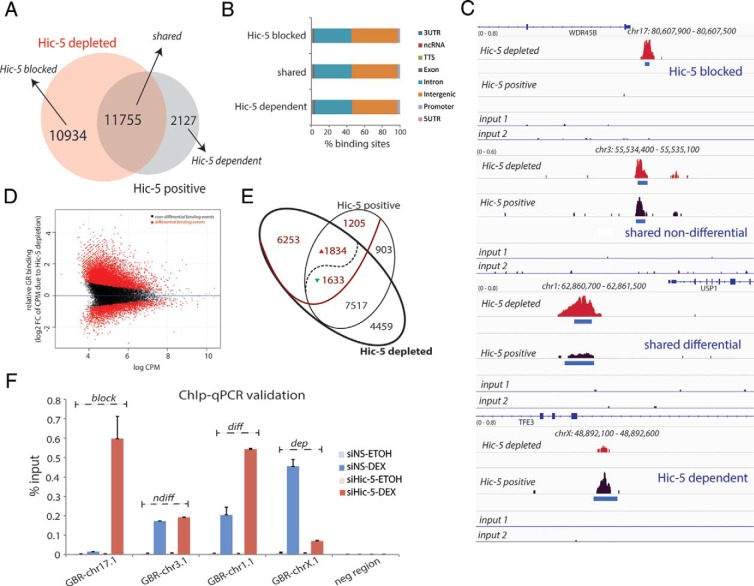
Because Hic-5 prevented chromatin occupancy of GRα, GRγ, and ERα on the few candidate block genes that were tested by quantitative ChIP, we evaluated whether this effect occurs genome-wide by performing ChIP-seq analyses for GRα in U2OS(GRα) cells containing or depleted of Hic-5 and treated with Dex for 1 hour. Hic-5 depletion resulted in an overall gain of GR binding events with many new GR peaks appearing only after Hic-5 depletion (Hic-5–blocked peaks, n = 10 934) (Figure 5A). Most of the GR peaks detected in the Hic-5–positive cells were shared in cells in which Hic-5 was depleted (n = 11 755), and only 2127 peaks were dependent on Hic-5 and were lost upon Hic-5 depletion (Figure 5A). Most of the peaks that were found in the 3 subsets were present in intergenic and intronic regions (Figure 5B). Examples from the ChIP-seq data of GRα binding at Hic-5–blocked, shared, and Hic-5–dependent GRα binding sites are shown (Figure 5C). We also assessed global GR binding events using MAnorm to compare statistically and quantitatively the ChIP-seq datasets from Hic-5–positive and Hic-5–depleted cells (Figure 5D). Each dot represents a GR binding site; the average binding intensity for the combined reads in Hic-5–positive and Hic-5–depleted cells is plotted along the horizontal axis and the log2 fold change of binding caused by Hic-5 depletion on the vertical axis with red dots signifying significant differential binding. There are peaks that are not significantly affected by Hic-5 depletion (black), peaks that are significantly enhanced upon Hic-5 depletion (red dots above 0 on the y-axis), and sites with significantly less binding upon Hic-5 depletion (red dots below 0 on the y-axis). We then combine the information on differential and nondifferential binding sites (red and black dots, respectively) with the Venn diagram shown in Figure 5A to indicate the distribution of differential and nondifferential binding events among shared peaks, peaks that are uniquely called in Hic-5–depleted cells, and peaks that are uniquely called in Hic-5–positive cells (Figure 5E).
Figure 5.
Hic-5 depletion results in gain of GRα occupancy genome-wide. A, Venn diagram showing the overlap of GRα binding events observed in ChIP-seq analysis of Hic-5–positive and Hic-5–depleted U2OS(GRα) cells treated with Dex for 1 hour. B, Distribution of GRα peaks across the human genome, plotted as a percentage of total binding events. C, Examples of GRα peaks observed by ChIP-seq in Hic-5–positive and Hic-5–depleted cells at specific gene loci representing Hic-5–blocked, Hic-5–dependent, and shared classes of GRα binding events, shown in the Integrative Genomics Viewer. Genomic positions of the peaks are shown. D, MAnorm plot showing a quantitative statistical comparison of genome-wide GRα binding in cells expressing or depleted of Hic-5. Log2 of the average binding intensity (sequence tag count in counts per million [CPM]) per binding event in the combined reads for Hic-5–positive and Hic-5–depleted cells on the x-axis is plotted against log2 fold change of binding at each genomic site on the y-axis, which is shown as the ratio of sequence tag counts per binding event in Hic-5–depleted vs Hic-5–positive cells. Each dot represents a GR binding site. Black dots, GR binding events that are not significantly different in the 2 conditions. Red dots above and below 0 on y-axis indicate differential binding and represent peaks with, respectively, significantly increased or decreased (false discovery rate–adapted P < .05) GRα occupancy in Hic-5–depleted cells compared with that in Hic-5–positive cells. E, Venn diagram based on the MAnorm analysis in D. The bold ellipse and nonbold ellipse contain GRα binding events in Hic-5–depleted and Hic-5–positive cells, respectively. GRα binding peaks that were observed under both conditions (shared peaks) are found in the overlapping area of the 2 ellipses, whereas peaks that are unique to one or the other condition are outside of the overlapping area. Red values indicate read count values that were significantly different between Hic-5–positive and Hic-5–depleted cells, and black values indicate GRα binding peaks with read counts that were not significantly different between the 2 conditions. Within the shared peaks, the red and green arrowheads indicate peaks that were significantly larger or smaller, respectively, in Hic-5–depleted cells. F, ChIP followed by qPCR was used to validate the examples of Hic-5–blocked (block), shared differential (diff) and nondifferential (ndiff), and Hic-5–dependent (dep) peaks shown in C.
This comparison involved merging peaks, which results in small differences in the total number of peaks in Figure 5E compared with those in Figure 5A. Overall, 8087 unique or shared GRα binding events (6253 + 1834 in Figure 5E) that occurred after Hic-5 depletion had significantly higher sequence read counts compared with the binding at the same sites that occurred in the presence of Hic-5 (represented in Figure 5D as red dots above 0 on the y-axis), whereas 2838 GRα binding events (1205 + 1633 in Figure 5E) had significantly higher binding when Hic-5 was present (Figure 5D, red dots below 0 on the y-axis). Of all the GRα binding events, 10 925 events (sum of the red numbers in Figure 5E) were significantly different in Hic-5–positive and Hic-depleted cells, with 6253 events (57%) called as peaks only in Hic-5–depleted cells and 1205 events (11%) called as peaks only in Hic-5–positive cells (Figure 5E). From the MAnorm analysis, 10 984 GRα binding events were shared in the 2 datasets (intersection of the 2 ellipses in Figure 5E), with 1834 peaks showing stronger binding in Hic-5–depleted cells, 1633 peaks showing stronger binding in Hic-5–positive cells, and 7517 peaks that were shared but were not significantly different. The 903 unique Hic-5–dependent peaks and the 4459 Hic-5–blocked peaks that were not significantly different in Hic-5–positive and Hic-5–depleted cells presumably represent small peaks that were near the threshold of the peak-calling algorithm. Examples of shared differential and shared nondifferential peaks, along with examples of unique Hic-5–dependent and unique Hic-5–blocked peaks from the ChIP-seq data are shown (Figure 5C), and validation of candidates in each of the four groups by quantitative ChIP is also shown (Figure 5F).
Association of Hic-5–blocked genes in U2OS(GRα) cells with Hic-5–blocked peaks
We correlated the overall changes in genomic occupancy of ligand-activated GRα with the Dex-regulated status of genes. Our previous results indicated that there were 212 block genes in U2OS(GRα) cells treated with Dex for 4 hours (4). To expand the number of GRα block genes for statistical purposes, we compared the 24-hour Dex treatment datasets for Hic-5–positive and Hic-5–depleted cells from the same study and found 584 block genes, ie, genes that were regulated by Dex only after depletion of Hic-5 (2-fold cutoff and false discovery rate–adjusted P ≤ .02) (Supplemental Dataset 4). Quantitative comparison of fold change in expression due to Dex for the block genes in U2OS(GRα) cells containing or lacking Hic-5 indicates that Hic-5 depletion caused very dramatic increases in regulation by Dex for a large fraction of the block genes (Figure 6A, compare pink vs blue bars). This conclusion applies to Dex-induced and Dex-repressed block genes. Next we analyzed whether genomic occupancy of GRα was affected by Hic-5 depletion near block genes. Overall, 40% to 50% of the GRα binding events found within a 100-kb window around the transcription start site (TSS) (±50 kb from the TSS) of all 584 blocked genes were Hic-5–blocked peaks, ie, only observed in Hic-5–depleted cells (Figure 6B, top row). Most of the remaining GRα binding events were shared in Hic-5–positive and Hic-5–depleted cells, whereas Hic-5–dependent peaks made up a relatively low percentage of the total (Figure 6B, middle and bottom rows). Quantitative mRNA and ChIP analyses of 2 candidate block genes (1 activated by Dex and 1 repressed by Dex) validated the effects of Hic-5 on the regulation of these genes by Dex and GRα occupancy on nearby GBRs (Figure 6, C, D, and E). GRAMD4 was induced by Dex at 2, 4, and 24 hours only in Hic-5–depleted cells (Figure 6D); we found 3 GRα binding events near GRAMD4 that were dramatically and significantly increased by Hic-5 depletion, both in the ChIP-seq data (Figure 6C, top panel) and in the quantitative ChIP analysis (Figure 6E). Similarly, GAS1 was repressed by Dex only in Hic-5–depleted cells (Figure 6D); basal expression was elevated by Hic-5 depletion and repressed by Dex. A significant GRα binding event approximately 30 kb from the TSS was identified in the ChIP-seq data analysis when Hic-5 was depleted but not in Hic-5–positive cells (Figure 6C, bottom panel), and Dex-induced GRα occupancy was also dramatically enhanced at this site in quantitative ChIP analysis (Figure 6E). In contrast, Dex-induced GRα occupancy on 2 ind genes, PNLIP, which is induced by Dex, and CCRN4L, which is repressed by Dex (Supplemental Figure 4B), was affected little if at all by Hic-5 depletion, either in ChIP-seq data (Supplemental Figure 4A) or in quantitative ChIP analysis (Supplemental Figure 4C). Our findings indicate that block genes are regulated by Hic-5 at the level of GR occupancy, which is dramatically enhanced by the depletion of Hic-5.
Figure 6.
Regulation of GRα occupancy by Hic-5 on GBRs near block genes. A, Extent to which Hic-5 restricts Dex induction and repression of block genes. U2OS(GRα) cells were transfected with nonspecific siRNA or siHic-5 and treated with ethanol (ETOH) or Dex for 24 hours, and global mRNA levels were determined by microarray analysis (see Supplemental Dataset 4); the Hic-5–blocked gene set was identified as described for Figure 3A. The log2 fold change in the mRNA levels of all Hic-5–blocked genes caused by 24-hour Dex treatment (from microarray data) is shown (y-axis) with results for each gene in Hic-5–depleted cells shown in pink and results for the same gene in Hic-5–positive cells shown in blue. B, Hic-5–blocked, shared, and Hic-5–dependent GRα peaks found near Hic-5–blocked genes. Hic-5–blocked gene sets determined from gene expression microarray analyses of cells treated with Dex for 4 or 24 hours were divided into genes that were induced or repressed by Dex, and the number of GRα binding events observed within ± 50 kb of the TSSs are divided into 3 categories: Hic-5–blocked peaks, shared peaks, and Hic-5–dependent peaks. C, GR binding sites at the GRAMD4 and GAS1 loci are shown in the Integrative Genomics Viewer. The red and black peaks indicate GR binding events in Hic-5–depleted and Hic-5–positive conditions, respectively. Blue bars under the peaks were identified as significant by the peak-calling algorithm. Input tracks for both conditions are shown as controls. The genomic coordinates of GRAMD4 peaks are as follows: GBR1, chr22:47,000,100–47,001200; GBR2, chr22:47,027,100–47028200; GBR3, chr22:47,030,800–47031900, and GAS1-GBR, chr9:89,598,400–89599300. D, RT-qPCR validation of the Dex-regulated expression of Hic-5–blocked genes shown in C. mRNA expression levels were normalized to glyceraldehyde-3-phosphate dehydrogenase mRNA levels for each sample and represent means ± SEM for 3 biological replicates. E, Validation of GR occupancy by ChIP-qPCR. GRα occupancy was analyzed at the sites shown in C by quantitative ChIP (normalized to input chromatin) in U2OS(GRα) cells transfected with siNS or siHic-5 and then treated with ethanol or 100 nM Dex for 1 hour. For GRAMD4, GBRs 1, 2, and 3 are shown left to right in C. Error bars represent means and ranges of variation for 2 technical replicates from one experiment, and results shown are representative of 3 independent experiments.
Discussion
The ability of Hic-5 to support SRs in activating and repressing expression of a subset of SR target genes is typical for all coregulators studied in detail to date. However, a number of coregulator actions by Hic-5 have not previously been described or are not typical of other coregulators. Hic-5 can either support or oppose steroid hormone activation or repression of SR target genes (Figure 1), and Hic-5 depletion has very dramatic effects on hormonal regulation of many SR target genes, serving as an all-or-none switch in many cases (Figure 6A). Furthermore, Hic-5 is the first coregulator shown to prevent expression of many SR target genes, the block gene class (4). In addition, we show here that although Hic-5 serves as a coregulator for ERα and 2 different isoforms of GR (GRα and GRγ), it does so in an SR-specific manner. The coregulator function of Hic-5 for endogenous target genes of ERα is a novel finding, because we previously found that Hic-5 failed to enhance the activation of transient reporter genes by ERα (15). However, Hic-5 still acted selectively on the 3 different SRs. The fraction of GRα target genes for which hormone-regulated expression levels are affected by Hic-5 (the mod gene class) is much larger than the similarly defined fractions for ERα and GRγ (Figure 1F). Furthermore, whereas the functions of Hic-5 on the mod gene class of GRα include coactivation, corepression, antiactivation, and antirepression, Hic-5 functions primarily as an antiactivator and antirepressor for GRγ (Figure 1F).The fact that Hic-5 has a much stronger physical interaction with GRα than with ERα (Figure 2) may help to explain why there are so many more GRα mod genes than ERα mod genes. However, GRα and GRγ interacted similarly with Hic-5 in coimmunoprecipitation assays, and thus the molecular explanation for the difference in the sizes of the GRα and GRγ mod gene classes must lie elsewhere. Hic-5 was also recently shown in prostate stromal cells to function as a coregulator for about half of the target genes of the androgen receptor, which also binds to Hic-5 through the receptor hinge region (16).
Although the Hic-5 mod gene class was much larger for GRα than for GRγ and ERα, the Hic-5 block gene class was similar in size for all 3 SRs (Figure 3). This not only serves as a control against the differences in the sizes of the mod gene class for the 3 SRs, but it also demonstrates that blocking of hormonal regulation of a subset of SR target genes is a common property of Hic-5 that applies to its functional interactions with multiple SRs. In a separate study, we demonstrated that 4 other coregulators for GRα exhibited both a classic coregulator function and the ability to block Dex regulation of a subset of GRα target genes (36). However, the number of block genes, compared with that of mod genes, for these 4 coregulators was much smaller than the number of Hic-5 block genes reported here, and the magnitude of the coregulator effect was not as dramatic for the other 4 coregulators as the all-or-none effect we often observed in this study with Hic-5.
By using quantitative ChIP analysis of several block genes, we found here that the mechanism by which Hic-5 prevents hormonal regulation of the block gene class for all 3 SRs involved inhibition of SR binding to potential SR binding sites associated with block genes (Figures 4–6). The efficient binding of Hic-5 to GRα and GRγ (Figure 2) provides a potential mechanism by which Hic-5 can influence GR binding to genomic sites. However, because ERα binds much more weakly and probably indirectly to Hic-5, the inhibition of ERα occupancy on specific EBRs may be due to a different mechanism. For example, Hic-5 could interact with other coregulators that interact with ERα and thereby inhibit genomic occupancy by ERα. The mechanism of blocking SR binding to genomic sites is unknown but could involve Hic-5 interactions with chromatin remodeling factors to establish a local chromatin conformation at Hic-5–blocked SR binding regions, which precludes efficient occupancy of the site by the hormone-activated SR. Alternatively, Hic-5 could prevent hormone-activated SRs from recruiting chromatin remodelers to specific SR binding regions and thereby inhibit hormone-dependent chromatin remodeling that may be required for the establishment of efficient SR occupancy of the site.
It is important to remember that Hic-5 inhibits GR and ER occupancy of only a subset of their complete set of binding sites in U2OS cells (Figures 4–6). The reason that Hic-5 prevents SR binding to some sites but has no effect on SR binding to other sites remains to be determined. However, we propose that the different effects of Hic-5 on distinct SR target genes involves the inherent properties of each SR binding region. Such inherent properties would probably include the DNA sequence to which the SR is bound, other transcriptional regulators binding near the SR binding site, and the local chromatin environment, all of which vary with the genomic locus. These differences would presumably dictate requirements for different sets of coregulators.
Finally, we demonstrated by ChIP-seq with GRα that Hic-5 has large global effects on the genomic binding of GRα. Although a relatively small number of genomic GRα binding sites are lost upon Hic-5 depletion, a much larger number of GRα binding sites are created by Hic-5 depletion, and some GBRs that exist in the presence of Hic-5 exhibit increased GRα binding after Hic-5 depletion (Figure 5). In fact, Hic-5 depletion almost doubled the number of GRα binding sites in the cells. Thus, Hic-5 regulates genomic occupancy by SRs in a gene-specific manner, by precluding binding of SRs to a large number of potential GBRs. In contrast, there are many sites where the binding of GRα is unaffected by Hic-5 depletion, and these sites serve as important controls to demonstrate the validity of the altered GRα binding at other sites. Furthermore, the site-specific effects of Hic-5 further indicate that different SR target genes have different molecular mechanisms of regulation by Hic-5 and in general.
Both GRα and GRγ are expressed widely in mammalian tissues and cell types, with GRγ mRNA levels accounting for a relatively constant 6% to 8% of total GR mRNA in the tissues examined (24). Similarly, ERα is very widely expressed in human tissues (25), including primary osteoblasts for which U2OS cells serve as a model (37) and both epithelial and stromal cell types in endometrium (18). Hic-5 is expressed in a highly tissue-specific manner (26) and in some organs is preferentially associated with stromal rather than epithelial cells, including prostate where its expression influences androgen receptor and vitamin D receptor action (16, 19). Furthermore, altered Hic-5 expression has been associated with pathological conditions such as Alzheimer disease (38) and endometriosis (18) for which its expression overlaps with ERα. Thus, although it has not been widely examined, Hic-5 expression is quite likely to overlap with GRα, GRγ, and ERα in many tissues. Because Gene Ontology analysis indicated that ERα target genes regulated by Hic-5 are involved in controlling cell proliferation (Supplemental Dataset 3), it will be interesting to test the physiological effects of Hic-5 in connection with estrogen-regulated cell proliferation.
The results presented here strongly suggest that Hic-5 may be an important factor in determining the cell type-specific occupancy of potential binding sites by SRs (39). In addition, it has been previously reported that activation of other signaling pathways and functional interactions with other transcription factors can influence SR binding site selection (40, 41). Similarly, Hic-5 levels or activity in a given cell type could be regulated by protein-protein interactions and posttranslational modifications, resulting in alterations in the specific set of genes regulated by a steroid hormone. In addition, because Hic-5 can serve as a coregulator for various other transcription factors (19–21), it will be interesting in the future to examine whether Hic-5 similarly regulates genomic binding site selection by other transcription factors.
Acknowledgments
We thank Kiran Sriram for technical assistance and Dr Thomas Spelsberg (Mayo Clinic, Rochester MN) for providing U2OS(ERα) cells.
This work was supported by the National Institutes of Health (Grants DK043093 and DK055274 to M.R.S.). The Next Generation Sequencing Core and Bioreagent Core of the USC Norris Comprehensive Cancer Center, which were used in this study, were supported by National Institutes of Health Cancer Center Support (Grant P30CA014089).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- AD
- activation domain
- ChIP-seq
- chromatin immunoprecipitation coupled with high-throughput sequencing
- DBD
- DNA-binding domain
- Dex
- dexamethasone
- Dox
- doxycycline
- E2
- estradiol
- ER
- estrogen receptor
- ERB
- estrogen receptor α binding region
- FBS
- fetal bovine serum
- GR
- glucocorticoid receptor
- GRB
- glucocorticoid receptor binding region
- HA
- hemagglutinin
- Hic-5
- hydrogen peroxide-inducible clone 5 protein
- LBD
- ligand-binding domain
- PPARγ
- peroxisome proliferator–activated receptor γ
- qPCR
- quantitative PCR
- si
- small interfering
- SR
- steroid receptor
- TSS
- transcription start site.
References
- 1. Millard CJ, Watson PJ, Fairall L, Schwabe JW. An evolving understanding of nuclear receptor coregulator proteins. J Mol Endocrinol. 2013;51:T23–T36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Won Jeong K, Chodankar R, Purcell DJ, Bittencourt D, Stallcup MR. Gene-specific patterns of coregulator requirements by estrogen receptor-α in breast cancer cells. Mol Endocrinol. 2012;26:955–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Engel KB, Yamamoto KR. The glucocorticoid receptor and the coregulator Brm selectively modulate each other's occupancy and activity in a gene-specific manner. Mol Cell Biol. 2011;31:3267–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chodankar R, Wu DY, Schiller BJ, Yamamoto KR, Stallcup MR. Hic-5 is a transcription coregulator that acts before and/or after glucocorticoid receptor genome occupancy in a gene-selective manner. Proc Natl Acad Sci USA. 2014;111:4007–4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ou CY, Chen TC, Lee JV, Wang JC, Stallcup MR. Coregulator cell cycle and apoptosis regulator 1 (CCAR1) positively regulates adipocyte differentiation through the glucocorticoid signaling pathway. J Biol Chem. 2014;289:17078–17086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wu DY, Ou CY, Chodankar R, Siegmund KD, Stallcup MR. Distinct, genome-wide, gene-specific selectivity patterns of four glucocorticoid receptor coregulators. Nucl Recept Signal. 2014;12:e002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lefstin JA, Yamamoto KR. Allosteric effects of DNA on transcriptional regulators. Nature. 1998;392:885–888. [DOI] [PubMed] [Google Scholar]
- 8. Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. [DOI] [PubMed] [Google Scholar]
- 10. Burd CJ, Ward JM, Crusselle-Davis VJ, et al. Analysis of chromatin dynamics during glucocorticoid receptor activation. Mol Cell Biol. 2012;32:1805–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim JH, Yang CK, Heo K, Roeder RG, An W, Stallcup MR. CCAR1, a key regulator of mediator complex recruitment to nuclear receptor transcription complexes. Mol Cell. 2008;31:510–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Malik S, Roeder RG. The metazoan mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11:761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shibanuma M, Mashimo J, Kuroki T, Nose K. Characterization of the TGF β1-inducible hic-5 gene that encodes a putative novel zinc finger protein and its possible involvement in cellular senescence. J Biol Chem. 1994;269:26767–26774. [PubMed] [Google Scholar]
- 14. Heitzer MD, DeFranco DB. Mechanism of action of Hic-5/androgen receptor activator 55, a LIM domain-containing nuclear receptor coactivator. Mol Endocrinol. 2006;20:56–64. [DOI] [PubMed] [Google Scholar]
- 15. Yang L, Guerrero J, Hong H, DeFranco DB, Stallcup MR. Interaction of the τ2 transcriptional activation domain of glucocorticoid receptor with a novel steroid receptor coactivator, Hic-5, which localizes to both focal adhesions and the nuclear matrix. Mol Biol Cell. 2000;11:2007–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leach DA, Need EF, Trotta AP, Grubisha MJ, DeFranco DB, Buchanan G. Hic-5 influences genomic and non-genomic actions of the androgen receptor in prostate myofibroblasts. Mol Cell Endocrinol. 2014;384:185–199. [DOI] [PubMed] [Google Scholar]
- 17. Fujita H, Kamiguchi K, Cho D, Shibanuma M, Morimoto C, Tachibana K. Interaction of Hic-5, a senescence-related protein, with focal adhesion kinase. J Biol Chem. 1998;273:26516–26521. [DOI] [PubMed] [Google Scholar]
- 18. Aghajanova L, Velarde MC, Giudice LC. The progesterone receptor coactivator Hic-5 is involved in the pathophysiology of endometriosis. Endocrinology. 2009;150:3863–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Solomon JD, Heitzer MD, Liu TT, et al. VDR Activity is differentially affected by Hic-5 in prostate cancer and stromal cells. Mol Cancer Res. 2014;12:1166–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Drori S, Girnun GD, Tou L, et al. Hic-5 regulates an epithelial program mediated by PPARγ. Genes Dev. 2005;19:362–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Inui S, Shono F, Noguchi F, Nakajima T, Hosokawa K, Itami S. In vitro and in vivo evidence of pathogenic roles of Hic-5/ARA55 in keloids through Smad pathway and profibrotic transcription. J Dermatol Sci. 2010;58:152–154. [DOI] [PubMed] [Google Scholar]
- 22. Rivers C, Levy A, Hancock J, Lightman S, Norman M. Insertion of an amino acid in the DNA-binding domain of the glucocorticoid receptor as a result of alternative splicing. J Clin Endocrinol Metab. 1999;84:4283–4286. [DOI] [PubMed] [Google Scholar]
- 23. Thomas-Chollier M, Watson LC, Cooper SB, et al. A naturally occurring insertion of a single amino acid rewires transcriptional regulation by glucocorticoid receptor isoforms. Proc Natl Acad Sci USA. 2013;110:17826–17831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rivers C, Flynn A, Qian X, et al. Characterization of conserved tandem donor sites and intronic motifs required for alternative splicing in corticosteroid receptor genes. Endocrinology. 2009;150:4958–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Deroo BJ, Korach KS. Estrogen receptors and human disease. J Clin Invest. 2006;116:561–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yuminamochi T, Yatomi Y, Osada M, et al. Expression of the LIM proteins paxillin and Hic-5 in human tissues. J Histochem Cytochem. 2003;51:513–521. [DOI] [PubMed] [Google Scholar]
- 27. Monroe DG, Getz BJ, Johnsen SA, Riggs BL, Khosla S, Spelsberg TC. Estrogen receptor isoform-specific regulation of endogenous gene expression in human osteoblastic cell lines expressing either ERα or ERβ. J Cell Biochem. 2003;90:315–326. [DOI] [PubMed] [Google Scholar]
- 28. Dennis G, Sherman BT, Hosack DA, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 29. Rogatsky I, Trowbridge JM, Garabedian MJ. Glucocorticoid receptor-mediated cell cycle arrest is achieved through distinct cell-specific transcriptional regulatory mechanisms. Mol Cell Biol. 1997;17:3181–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shao Z, Zhang Y, Yuan GC, Orkin SH, Waxman DJ. MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome Biol. 2012;13:R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schiller BJ, Chodankar R, Watson LC, Stallcup MR, Yamamoto KR. Glucocorticoid receptor binds half sites as a monomer and regulates specific target genes. Genome Biol. 2014;15:418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rogatsky I, Wang JC, Derynck MK, et al. Target-specific utilization of transcriptional regulatory surfaces by the glucocorticoid receptor. Proc Natl Acad Sci USA. 2003;100:13845–13850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krum SA, Miranda-Carboni GA, Lupien M, et al. Unique ERα cistromes control cell type-specific gene regulation. Mol Endocrinol. 2008;22:2393–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Matys V, Fricke E, Geffers R, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu DY, Ou CY, Chodankar R, Siegmund KD, Stallcup MR. Distinct genome-wide, gene-specific selectivity patterns of four glucocorticoid receptor coregulators. Nucl Recept Signal. 2014;12:e002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Galea GL, Meakin LB, Sugiyama T, et al. Estrogen receptor α mediates proliferation of osteoblastic cells stimulated by estrogen and mechanical strain, but their acute down-regulation of the Wnt antagonist Sost is mediated by estrogen receptor β. J Biol Chem. 2013;288:9035–9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Caltagarone J, Hamilton RL, Murdoch G, Jing Z, DeFranco DB, Bowser R. Paxillin and hydrogen peroxide-inducible clone 5 expression and distribution in control and Alzheimer disease hippocampi. J Neuropathol Exp Neurol. 2010;69:356–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. John S, Sabo PJ, Thurman RE, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lupien M, Meyer CA, Bailey ST, et al. Growth factor stimulation induces a distinct ERα cistrome underlying breast cancer endocrine resistance. Genes Dev. 2010;24:2219–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ross-Innes CS, Stark R, Teschendorff AE, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]