Abstract
Background
Current dosing practices for warfarin are empiric and result in the need for frequent dose changes as the international normalized ratio gets too high or too low. As a result, patients are put at increased risk for thromboembolism, bleeding, and premature discontinuation of anticoagulation therapy. Prior research has identified clinical and genetic factors that can alter warfarin dose requirements, but few randomized clinical trials have examined the utility of using clinical and genetic information to improve anticoagulation control or clinical outcomes among a large, diverse group of patients initiating warfarin.
Methods
The COAG trial is a multicenter, double-blind, randomized trial comparing 2 approaches to guiding warfarin therapy initiation: initiation of warfarin therapy based on algorithms using clinical information plus an individual's genotype using genes known to influence warfarin response (“genotype-guided dosing”) versus only clinical information (“clinical-guided dosing”) (www.clinicaltrials.gov Identifier: NCT00839657).
Results
The COAG trial design is described. The study hypothesis is that, among 1,022 enrolled patients, genotype-guided dosing relative to clinical-guided dosing during the initial dosing period will increase the percentage of time that patients spend in the therapeutic international normalized ratio range in the first 4 weeks of therapy.
Conclusion
The COAG will determine if genetic information provides added benefit above and beyond clinical information alone. (Am Heart J 2013;166:435-441.e2.)
Warfarin sodium is a leading cause of adverse drug events.1,2 Although warfarin is highly efficacious at preventing thromboembolism (TE), it must be dosed properly to avoid life-threatening bleeding from overdosing and decreased efficacy from underdosing. The practice of empiric dosing results in widespread improper dosing, and out-of-range international normalized ratios (INRs) are extremely common early in therapy. Improper levels of anticoagulation result in substantial morbidity and cost.3,4 Even minor bleeding can lead to withdrawal of therapy, thus depriving patients of an effective therapy to prevent TE. Minor bleeding also leads to repeat office visits and sometimes emergency department visits. Even absent complications, patients who have out-of-range INRs must be carefully reassessed within a short period and often require dosage changes, which generate additional clinic visits, blood tests, and potential for miscalculations of dosage requirements.5
Warfarin dose requirements vary widely across patients. Despite current understanding of the influence of clinical and genetic factors on variability in warfarin dose requirements, formal testing of the utility of a genetic-guided dosing strategy among a large, diverse group of patients using warfarin has not been rigorously performed. Three small trials comparing genotype-guided to clinical-guided dosing have recently been published, none of which were definitive.6-8 In contrast, observational studies have suggested benefits to genotype-guided dosing.9,10 Nonetheless, the Centers for Medicare and Medicaid Services does not covers the cost of genotyping for warfarin dosing because “available evidence does not demonstrate that pharmacogenomic testing to predict warfarin responsiveness improves health outcomes.”11 The COAG trial is designed to address the clinical utility of genotype-guided dosing on anticoagulation stability.
Scientific and statistical methods
Objectives and design of the COAG study
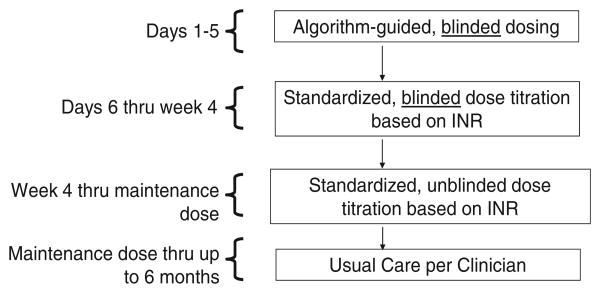
The trial is a multicenter, double-blind, randomized trial comparing genotype-guided dosing with clinical-guided dosing in the first 5 days of therapy. Further dose adjustment will be the same between arms using a standardized dose adjustment protocol. Participants will be followed in the study for 6 months (Figure 1).
Figure 1.
Schematic of entire trial period.
Study population
The study population will be drawn from 18 clinical centers in the United States (see online Appendix A). Participants will be enrolled before initiating warfarin and will include participants with a variety of conditions requiring long-term anticoagulation therapy with warfarin. Eligibility criteria are listed in Table I.
Table I. Inclusion and exclusion criteria for COAG trial.
| Inclusion Criteria |
|
| Exclusion Criteria |
|
Abbreviations: CYP2C9, Cytochrome P450 2C9; VKORC1, vitamin K epoxide reductase complex subunit 1.
Randomization
Randomization will be stratified by participating institutions and by race (African American, estimated 25% of the study population, vs non-African American) because race has been strongly associated with differential benefit of dosing algorithms, particularly with lesser benefit in African Americans.12 The dosing algorithms that will be used in the trial also predict dose differently among African Americans versus non-African Americans. In addition, race is strongly associated with the prevalence of variants in CYP2C9 and VKORC113 (the genes in the dosing algorithm).
Study outcomes
Primary outcome
The primary outcome is the percentage of time participants spend within the therapeutic INR range (PTTR) during the first 4 weeks of therapy. The PTTR will be calculated using linear interpolation,14 which has been shown to be valid and, in the absence of high levels of missing data (eg, ≥20% missing INR values), reproducible.15
The rationale for using PTTR is as follows:
It is one of the most important factors influencing safe and effective anticoagulation: overanticoagulation and underanticoagulation.16 Data from retrospective studies demonstrate that the PTTR predicts adverse events.17
It is often the only factor that can be modified to reduce complications and costs in anticoagulation patients.
It is an acceptable and commonly used measure to judge anticoagulation control.18
Secondary outcomes
Secondary outcomes include major bleeding or TE in the first 4 weeks (the principal, secondary outcome); clinically relevant, nonmajor bleeding;19,20 time to first therapeutic INR and maintenance dose; PTTR <60% or INR ≥4 at least twice during the first 4 weeks; variability in INR; number of warfarin dose changes; PTTR during the first 2 weeks, 3 months, and 6 months of therapy; rate of INRs >4 and INR <2; time to bleeding and TE; cost; and quality of life.
Initial and dose adjustment phase (days 1-5) using algorithms
The criteria for choosing algorithms for the trial were as follows: the algorithm was developed on a derivation dataset and validated separately; the algorithm characteristics are favorable, including the accuracy of prediction of therapeutic maintenance dose; the algorithm is clinically usable in standard clinical environments; and the algorithm has “face validity” in that the predictors and their direction and effect size are consistent with other available studies. The genotype- and clinical-guided dose-initiation algorithms have met these standards.
Genotype dose-initiation algorithm
The algorithm chosen for this study was based on the criteria above and has been published by Gage et al21 (referred to herein as the “genotype dose-initiation algorithm”) and validated by Schelleman et al.12 The algorithm is as follows:
in which CYP2C9*2 and CYP2C9*3 single nucleotide polymorphisms are coded as 0 if absent (no variants), 1 if heterozygous, and 2 if homozygous; VKORC1 is VKORC1 3673G>A (also known as VKORC1 -1639, rs9923231) and is coded 0 (homozygous GG), 1 (heterozygous), or 2 (homozygous AA). Race is coded as 1 if African American and 0 otherwise. Smokes, amiodarone use, and indication for therapy are coded as 1 if yes and 0 if no. Body surface area is calculated as [(weight, kg)0.425 × (height, cm)0.725]/139.2. For the COAG trial, the target INR will be fixed at 2.5. CY2C9 will be set to 0 for the first day.
Genotype dose-revision algorithm
The genotype dose-revision algorithm was derived and validated previously. 22 The algorithm will be applied on days 4 and/or 5 of therapy and is as follows:
in which CYP2C9*2, CYP2C9*3, VKORC1, race, smoke, and amiodarone are coded as in the genotype dose-initiation algorithm above. Diabetes, stroke, and fluvastatin use are coded as 1 if yes and 0 if no. Target INR will be fixed at 2.5. “INR” is the INR measured on the day of dosing. “Dose-i” is the dose given i days before the INR measured.
Clinical dose-initiation algorithm
The algorithm from Gage et al21 is as follows:
Clinical dose-revision algorithm
The clinical dose-revision algorithm22 is as follows:
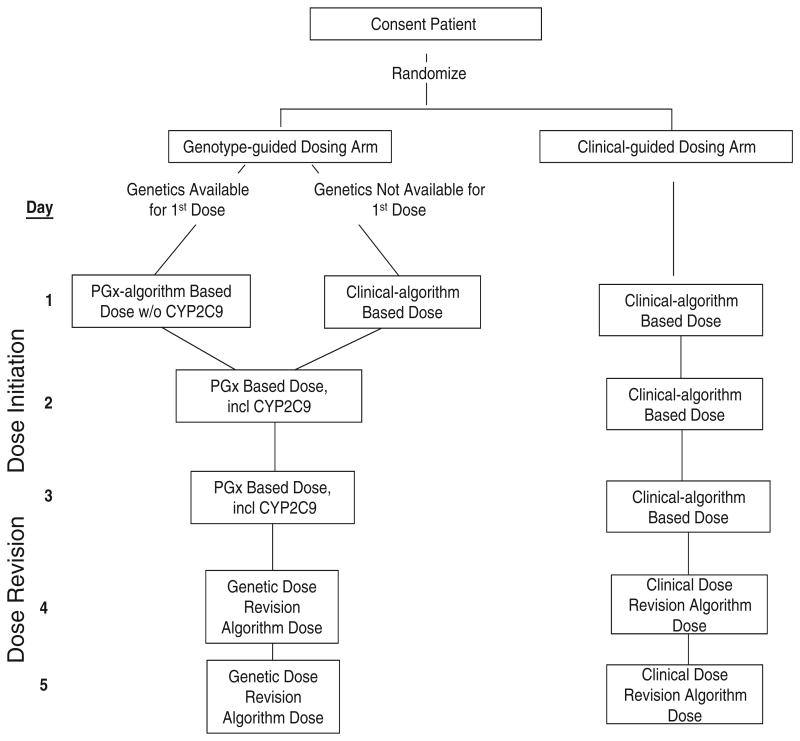
Administration of dosing
Figure 2 diagrams the administration of warfarin dosing during the initial 5-day intervention period. Table II specifies how each day's dose is calculated and adjusted based on INRs. Online Appendix B specifies how dose will be titrated from day 7 through day 28.
Figure 2.
Schematic of first 5 days of protocol showing dose initiation and dose revision in genotype and clinical arms.
Table II. Dosing scheme for both study arms during the intervention period.
| Day | INR | Warfarin dose (adapted from original Crowther algorithm using 5-mg starting dose) | Warfarin dose for trial (both algorithm arms) |
|---|---|---|---|
| 1 | 5 | Initiation algorithm rounded up,* ignoring CYP2C9 variants (if known) for genotype-guided arm (ie, set CYP2C9 variable to 0 in equation); use clinical algorithm for those in clinical-guided arm or those in genotype-guided arm for whom genotype not yet known | |
| 2 | No INR per protocol | Initiation algorithm rounded up, including CYP2C9 variants hereafter* | |
| If INR checked off protocol | <1.5 | 5.0 mg | Initiation algorithm rounded up, including CYP2C9 variants hereafter* |
| 1.5-1.9 | 2.5 | 0.5 * Initiation algorithm, including CYP2C9 variants hereafter† | |
| 2.0-2.5 | 1.0-2.5 | 0.5 * Initiation algorithm, including CYP2C9 variants hereafter† | |
| >2.5 | 0.0 | 0 | |
| 3 | No INR per protocol | Initiation algorithm rounded up* | |
| If INR checked off protocol | <1.5 | 5.0-10.0 mg | Initiation algorithm rounded up* |
| 1.5-1.9 | 2.5-5.0 | 0.75 * Initiation algorithm† | |
| 2.0-2.4 | 0-2.5 | 0.5 * Initiation algorithm† | |
| 2.5-3.0 | 0.0-2.5 | 0.25 * Initiation algorithm† | |
| >3 | 0.0 | 0 | |
| 4# | <1.5 | 10.0 | Dose-revision algorithm rounded up‡ |
| 1.5-1.9 | 5.0-7.5 | ||
| 2.0-2.4 | 0.0-5.0 | Dose-revision algorithm rounded down§ | |
| 2.5-3.0 | 0.0-5.0 | 0.5 * Dose-revision algorithm today, then dose-revision algorithm rounded down‖ | |
| >3.0 | 0.0 | 0 today, then dose-revision algorithm rounded down¶ | |
| 5# | Not available | Dose-revision algorithm (from prior day) | |
| <1.5 | 10.0 | Dose-revision algorithm rounded up‡ | |
| 1.5-1.9 | 7.5-10.0 | ||
| 2.0-2.8 | 0.0-5.0 | Dose-revision algorithm rounded down§ | |
| 2.9-3.0 | 0.0-5.0 | 0.5 * Dose-revision algorithm today, then dose-revision algorithm rounded down‖ | |
| >3.0 | 0 | 0 today, then dose-revision algorithm rounded down¶ |
If predicted dose is ≥3.0 mg, the dose will be rounded up to the nearest 1.0 mg. If predicted dose is <3.0 mg, the dose will be rounded up to the nearest 0.5 mg (eg, 2.1 mg would be rounded to 2.5 mg rather than to an integer value).
If the algorithm dose is adjusted due to INR on days 2 or 3 (eg, 0.5 * Dose from initiation algorithm), the dose will first be calculated using this correction and then rounded to the nearest 0.5 (if calculated dose is <3.0 mg) or 1.0 mg (if calculated dose is ≥3.0 mg): (eg, if on day 2 the INR is 1.7 and the dose-initiation algorithm dose is 4.3, the dose will be 4.3 mg
0.5 = 2.15 mg, and the day 2 dose will be rounded up to 2.0 mg; if on day 2 the INR is 1.7 and the dose-initiation algorithm dose is 7.5, the dose will be 7.5 mg * 0.5 = 3.75 mg,and will be rounded to 4.0 mg). Exact half doses above 3 mg on days 2 and 3 will be rounded up (eg, 3.5 mg will be rounded to 4, 4.5 mg will be rounded to 5, etc).
Weekly dose will be calculated from dose-revision algorithm on days 4 or 5. If the predicted weekly dose is ≥11 mg (eg, >1.5 mg/d), the weekly dose will be rounded up to the nearest 1.0 mg (eg, if weekly dose is 14.4, it will be rounded to 15 mg/week). If the predicted weekly dose is <11 mg (eg, <1.5 mg/d), the weekly dose will be rounded up to the nearest 0.5 mg (eg, if weekly dose is 10.4 mg, it will be rounded to 10.5 mg). This convention of using half doses for weekly doses below 11 mg follows that used in the Couma-Gen trial.
Weekly dose will be calculated from dose-revision algorithm on days 4 or 5. If the predicted weekly dose is ≥11 mg (eg,>1.5 mg/d), the weekly dose will be rounded down to the nearest 1.0 mg (eg, if weekly dose is 14.7, it will be rounded down to 14 mg/wk). If the predicted weekly dose is<11 mg (eg,≤1.5 mg/d), the weekly dose will be rounded down to the nearest 0.5 mg (eg, if weekly dose is 10.6 mg, it will be rounded down to 10.5 mg).
If INR is 2.5 to 3.0 on day 4 or 2.9 to 3.0 on day 5, the dose for that day (day 4 or day 5) will be 0.5 * dose calculated for that day (as described in † above). After that day, the weekly dose will be the weekly dose calculated by the dose-revision algorithm, rounded down as in § above.
If INR is >3.0 on day 4 or day 5, the dose for that day (day 4 or day 5) will be held. After that day, the weekly dose will be the weekly dose calculated by the dose-revision algorithm, rounded down as in § above.
If INR not done on this day, dose from prior day will be used.
The first dose in the genotype-guided arm will not incorporate CYP2C9. This is because initial dosing in poor metabolizers of warfarin (ie, those with CYP2C9 variants) should not be altered based on differences in metabolism, and recent studies suggest that CYP2C9 variants have little influence on INR response early in therapy.23
Based on the available data at the time of planning the COAG trial and recommendations of the American College of Chest Physicians Evidence-Based Clinical Practice Guidelines 8th Edition (2008), we have chosen not to use loading (ie, 2 times predicted) doses in this trial.
Administration of study drug
Rationale for blinding of study arm and warfarin dose for first 4 weeks
The trial will be double blind, with neither treating clinicians nor participants knowing the dosing algorithm used or the actual dose during the first 4 weeks. The outcome of warfarin therapy can be influenced by many factors, including not only proper dosing but also monitoring vigilance, educational efforts by clinicians, patient adherence to therapy, patient adherence to diet, and the use of interacting medications. This would be particularly problematic if the occurrence of these postrandomization factors both differed by study arm and were also related to anticoagulation control, as might be expected. Blinding will mitigate these concerns and ensure that outcomes are assessed similarly in all participants.
Method of blinding
For the first 4 weeks of the trial, warfarin will be provided in blinded form, with the dose overencapsulated based on a previously published method demonstrated not to alter warfarin pharmacokinetics, and used in several prior randomized trials.24 After 4 weeks of therapy (the primary outcome duration), clinicians will be informed of the actual dose that the participant is taking and participants will then receive their warfarin through their usual pharmaceutical outlet.
Genotyping at the central laboratory
Two genotyping platforms have been selected for use in the COAG Trial: the Genmark eSensor XT-8 System and the Autogenomics INFINITI XT Warfarin Assay. Both platforms are Food and Drug Administration approved and have high call and concordance rates, rapid turnaround times, very low failure rates, and the ability to genotype the SNPs needed for the chosen dosing algorithms. To maintain blinding and minimize differential dropouts (eg, because of possible genotyping delays), rapid turnaround genotyping will be performed on all participants, regardless of study arm. All clinical centers have been trained in the use of their chosen platform and have undergone additional quality control measures. To maintain quality control throughout the trial, the central laboratory will also confirm the genotyping results on a weekly basis for all participants enrolled.
Study visits
Participant visits will follow a standard visit schedule that is consistent with usual clinical protocol (ie, additional clinic visits will not be necessary, see online Appendix C).
Statistical considerations
Full details of the statistical approaches to analysis and sample size calculations are provided in previously published articles.25,26 In brief, analysis of the primary outcome will be by intention to treat. We assumed a clinically meaningful minimum detectable difference of 5% to 10% in PTTR between the genotype-guided and clinical-guided dosing arms.26 A 10% improvement in PTTR, for example, has been used, in part, to justify the use of formalized anticoagulation clinics as standard of care.27,28
The evaluation of PTTR at an overall significance level of α = .05 will be performed using an α allocation approach in which 0.04 will be apportioned to test the comparison of the 2 arms in the overall cohort; the significance level for the primary subgroup of interest will be obtained based on the correlation between the 2 tests.25 The subgroup of interest is defined by a difference of 1 mg/d or more in the dose predicted by the genotype dose-initiation algorithm versus that predicted by the clinical dose-initiation algorithm. The difference in predicted initial doses between the 2 dose-initiation algorithms is known at the time of randomization and, therefore,a proper subgroup.29 Additional subgroup analyses will include those defined by allelic variation and race/ethnicity.
Adopting a conservative approach to protect against errors in the estimates of PTTR standard deviation (25%-30%) and distribution of allelic variants in the population as well as to provide adequate power for subgroup and secondary analyses, a sample size of 1,238 was initially chosen, which would have at least 80% power for the full cohort analysis and for the subgroup based on the predicted dose difference.
Data and safety monitoring
An independent Data and Safety Monitoring Board (DSMB) has been established by the National Heart, Lung, and Blood Institute. The DSMB includes experts in the areas of thromboembolic disease, anticoagulation treatment, pharmacogenetics, clinical trials, biostatistics, and bioethics. The trial is monitored periodically for aspects of data quality, adequacy of follow-up, and occurrence of adverse events. The DSMB reviewed the results of a preplanned “internal pilot study” to estimate the SD of PTTR using observed data from the first 310 participants. There are no plans for interim evaluation of efficacy.
Sample size adjustments
A sample size of 1,238 was initially chosen to provide >80% power for the full cohort analysis and for the subgroup based on the predicted dose difference. However, because of a suboptimal recruitment rate, the DSMB approved a decrease in the target sample size to 1,022, which maintained adequate power (at least 80%) for both primary analyses.
Discussion
Warfarin is a difficult drug to manage. There are numerous reasons for this, but one of the major ones is the inability to know which dose an individual patient will require to maintain a steady, therapeutic drug effect. Genetic information may improve warfarin dosing and patient care, but to date, this hypothesis has not been confirmed. Whether it is beneficial to start a patient on a dose of warfarin that is closer to that patient's ultimate maintenance dose is not known. It is possible that starting on a genotype-guided dose will not lead to better anticoagulation control. Therefore, the COAG trial has been designed as an efficacy study to determine the incremental benefit of using genetic information on initial warfarin dosing compared with the best possible nongenetic approach.
There are several questions that the COAG trial is not designed to answer. First, the trial does not have adequate power to determine if genotyping leads to reductions in clinical events (bleeding, TE). Such a trial would require a substantially larger sample size than planned (eg, approximately 11,000 to have 80% power to detect a difference in events from 6% to 4.8% between the study arms). Although PTTR is, therefore, a surrogate for these clinical outcomes, there are several reasons that PTTR is, itself, a valuable outcome. The PTTR is used as a measure of quality of care, and improving anticoagulation could have significant impact on numerous other important outcomes besides bleeding or TE, including patient satisfaction, costs, and quality of life. Second, the COAG trial will not test how genotyping will work in the “real world.” Should genotyping improve anticoagulation control in this carefully performed study, it will be important to further study the impact of genotyping in broad-based practice. Finally, the COAG trial is not evaluating whether genotype-guided warfarin dosing will shift the risk-benefit ratio of using alternative agents, such as dabigatran, rivaroxaban, or apixaban, versus warfarin. The use of these newer agents has its own potential challenges,30-32 and should the COAG trial demonstrate improved results with the use of genetic-based warfarin dosing, further comparisons with newer agents may be warranted.
In summary, the COAG trial is a controlled, randomized, double-blinded trial that will determine if the use of genetic information provides added benefit beyond clinical information. It will provide the rigor needed to test the potential benefits of a personalized medicine approach and will provide insights into the methodological and statistical challenges of performing clinical trials in this field.
Acknowledgments
This work is supported under contract HHSN268200800003C from the National Heart, Lung, and Blood Institute. The authors are solely responsible for the design and conduct of this study, all study analyses, the drafting and editing of the manuscript, and its final contents.
Appendix A
COAG clinical sites.
| University of Texas |
| Mount Sinai School of Medicine |
| University of California, San Francisco |
| Washington University School of Medicine |
| University of Maryland School of Medicine |
| University of Florida |
| Henry Ford Hospital |
| Mayo Clinic College of Medicine |
| Hospital of the University of Pennsylvania |
| Vanderbilt University |
| Intermountain Medical Center |
| Marshfield Clinical Research Foundation |
| Duke University Medical Center |
| Georgia Regents University |
| University of Alabama at Birmingham |
| University of Utah Health Care |
| Tulane University |
| Montefiore Medical Center |
Appendix B
Dose titration scheme for both study arms after the intervention period (days 6 through maintenance dose).
| INR 1.0-1.59 | Inquire about signs/symptoms of clotting* and, if necessary, refer to an appropriate facility for care Immediate extra dose (average of day 4-5 dose if on day 6) Increase weekly dose by 20% Retest in 3-5 d (if INR not yet therapeutic), retest on next protocol specified day (if INR previously therapeutic) |
| INR 1.6-1.79 | Give an extra half dose today (average of days 4-5 for day 6) Increase weekly dose by 10% Retest in 3-5 d (if INR not yet therapeutic), retest on next protocol specified day (if INR previously therapeutic) |
| INR 1.8-1.99 | Increase weekly dose by 5% if participant has received at least 8 warfarin doses Retest in 3-5 d (if INR not yet therapeutic), retest on next protocol specified day (if INR previously therapeutic) |
| INR 2.0-3.0 | No change in dose During first 2 wk of therapy, retest in 3-5 d During weeks 3 and 4, retest in 1 wk After week 4, retest in 1 m |
| INR 3.01-3.39 | First episode Retest in 3 d if INR never therapeutic, in 1 wk if INR previously therapeutic Second, consecutive episode Decrease weekly dose by 5% Retest in 3 d if INR never therapeutic, in 1 wk if INR previously therapeutic If prior INR was between 3.01 and 3.39 and dose has not be reduced within the previous 6 d, decrease weekly dose by 10% |
| INR 3.4-4.99 | Inquire about signs/symptoms bleeding* and, if necessary, refer to an appropriate facility for care Reduce today's dose by a half if INR <4 or omits today's dose if INR ≥4 Decrease weekly dose by 10% Retest in 3 d if INR never therapeutic, in 1 wk if INR previously therapeutic |
| INR ≥5.0 | Inquire about signs/symptoms bleeding,* and if necessary, refer to an appropriate facility for care. Customize care if bleeding. Omit 2 doses Retest in 48 hours When retested, if INR is between 1.8 and 3.39, decrease weekly dose by 15% and retest in 7 d (if INR never therapeutic), 14 d (if INR previously therapeutic) When retested, if INR still >3.39, omit 2 more doses and retest in 48 hours and then repeat as above Note: If INR >9.0 follow special protocol (IHC guidelines) |
If weekly dose is <11 mg/wk, weekly dose will be rounded to the nearest 0.5 mg weekly dose. If weekly dose is ≥11 mg/wk, weekly dose will be rounded to the nearest 1.0 mg weekly dose. Titration algorithm is based on Coumagen trial (Circulation 2007;116:2563–70).
Signs and symptoms of clotting: pain or swelling in the legs, SOB, chest pain, new focal weakness or numbness, slurred speech, vision changes, and others. Signs and symptoms of bleeding: nose bleeds, unusual bruising, dark stools, pink or bloody urine, excessive menstruation, blood in the sputum, and others.
Appendix C
COAG study visit and data collection schedule.
| Month 1 | Month 1 | Month 1 | Month 1 | Month 2* | Month 3 | Month 4* | Month 5* | Month 6 | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|
|
|
|
||||
| Baseline | Week 1 | Week 2 | Week 3 | Week 4 | ||||||||
|
|
|
|
|
|
||||||||
| Day -1/0 | Day 4 | Day 7 | Day A | Day B | ||||||||
|
|
|
|
|
|
||||||||
| Visit 1 | Visit 2 | Visit 3 | Visit 4 | Visit 5 | Visit 6 | Visit 7 | Visit 8 | Visit 9 | Visit 10 | Visit 11 | Visit 12 | |
| Informed consent | X | |||||||||||
| Participant contact information | X | X | X | X | X | X | X | X | X | X | X | X |
| Enrollment information | X | |||||||||||
| Eligibility confirmation | X | |||||||||||
| Randomization | X | |||||||||||
| Genotyping information | X | |||||||||||
| Dose requisition information | X | X | X | X | X | X | X | PRN | PRN | PRN | PRN | PRN |
| Personal history | X | |||||||||||
| Medical history | X | |||||||||||
| Diet information | X | X | X | X | X | X | X | |||||
| EQ-5D Health Questionnaire | X | X | X | X | ||||||||
| Health Status Questionnaire | X | X | X | |||||||||
| INRs | X | X | X | X | X | X | X | X | X | X | X | X |
| Adverse events | X | X | X | X | X | X | X | X | X | X | X | X |
| Concomitant medications | X | X | X | X | X | X | X | X | X | |||
| Medical events | X | X | X | X | X | X | X | X | X | X | X | |
| Hospitalization information | PRN | PRN | PRN | PRN | PRN | PRN | PRN | PRN | PRN | PRN | PRN | |
| Duke Anti-Coagulation Satisfaction Survey | PRN | PRN | PRN | PRN | PRN | |||||||
| Adherence Questionnaire | PRN | PRN | PRN | PRN | ||||||||
These visits may be conducted by telephone.
References
- 1.Gurwitz JH, Field TS, Harrold LR, et al. Incidence and preventability of adverse drug events among older persons in the ambulatory setting. JAMA. 2003;289:1107–16. doi: 10.1001/jama.289.9.1107. [DOI] [PubMed] [Google Scholar]
- 2.Budnitz DS, Lovegrove MC, Shehab N, et al. Emergency hospitalizations for adverse drug events in older Americans. N Engl J Med. 2011;365(21):2002–12. doi: 10.1056/NEJMsa1103053. [DOI] [PubMed] [Google Scholar]
- 3.Levine MN, Raskob G, Landefeld S, et al. Hemorrhagic complications of anticoagulant treatment. Chest. 1998;114:511S–23S. doi: 10.1378/chest.114.5_supplement.511s. [DOI] [PubMed] [Google Scholar]
- 4.Hylek EM, Skates SJ, Sheehan MA, et al. An analysis of the lowest effective intensity of prophylactic anticoagulation for patients with nonrheumatic atrial fibrillation. N Engl J Med. 1996;335:540–6. doi: 10.1056/NEJM199608223350802. [DOI] [PubMed] [Google Scholar]
- 5.Brigden ML, Kay C, Le A, et al. Audit of the frequency and clinical response to excessive oral anticoagulation in an out-patient population. Am J Hematol. 1998;59:22–7. doi: 10.1002/(sici)1096-8652(199809)59:1<22::aid-ajh5>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 6.Anderson JL, Horne BD, Stevens SM, et al. Randomized trial of genotype-guided versus standard warfarin dosing in patients initiating oral anticoagulation. Circulation. 2007;116:2563–70. doi: 10.1161/CIRCULATIONAHA.107.737312. [DOI] [PubMed] [Google Scholar]
- 7.Caraco Y, Blotnick S, Muszkat M. CYP2C9 genotype-guided warfarin prescribing enhances the efficacy and safety of anticoagulation: a prospective randomized controlled study. Clin Pharmacol Ther. 2008;83:460–70. doi: 10.1038/sj.clpt.6100316. [DOI] [PubMed] [Google Scholar]
- 8.Burmester JK, Berg RL, Yale SH, et al. A randomized controlled trial of genotype-based Coumadin initiation. Genet Med. 2011;13(6):509–18. doi: 10.1097/GIM.0b013e31820ad77d. [DOI] [PubMed] [Google Scholar]
- 9.Epstein RS, Moyer TP, Aubert RE, et al. Warfarin genotyping reduces hospitalization rates results from the MM-WES (Medco-Mayo Warfarin Effectiveness study) J Am Coll Cardiol. 2010;55:2804–12. doi: 10.1016/j.jacc.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 10.Anderson JL, Horne BD, Stevens SM, et al. A randomized and clinical effectiveness trial comparing two pharmacogenetic algorithms and standard care for individualizing warfarin dosing (CoumaGen-II) Circulation. 2012;125(16):1997–2005. doi: 10.1161/CIRCULATIONAHA.111.070920. [DOI] [PubMed] [Google Scholar]
- 11.CMS - Centers for Medicare and Medicaid Services. Medicare Coverage Database - Potential NCD Topics. [Accessed 9/9/08]; http://www.cms.hhs.gov/mcd/ncpc_view_document.asp?id=19. 7-30-2008. Ref Type: Electronic Citation.
- 12.Schelleman H, Chen J, Chen Z, et al. Dosing algorithms to predict warfarin maintenance dose in Caucasians and African Americans. Clin Pharmacol Ther. 2008;84(3):332–9. doi: 10.1038/clpt.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi H, Wilkinson GR, Nutescu EA, et al. Different contributions of polymorphisms in VKORC1 and CYP2C9 to intra- and inter-population differences in maintenance dose of warfarin in Japanese, Caucasians and African-Americans. Pharmacogenet Genomics. 2006;16:101–10. doi: 10.1097/01.fpc.0000184955.08453.a8. [DOI] [PubMed] [Google Scholar]
- 14.Rosendaal FR, Cannegieter SC, van der Meer FJ, et al. A method to determine the optimal intensity of oral anticoagulant therapy. Thromb Haemost. 1993;69:236–9. [PubMed] [Google Scholar]
- 15.Hutten BA, Prins MH, Redekop WK, et al. Comparison of three methods to assess therapeutic quality control of treatment with vitamin K antagonists. Thromb Haemost. 1999;82:1260–3. [PubMed] [Google Scholar]
- 16.Landefeld CS, Beyth RJ. Anticoagulant-related bleeding: clinical epidemiology, prediction, and prevention. Am J Med. 1993;95:315–28. doi: 10.1016/0002-9343(93)90285-w. [DOI] [PubMed] [Google Scholar]
- 17.Wan Y, Heneghan C, Perera R, et al. Anticoagulation control and prediction of adverse events in patients with atrial fibrillation. Circ Cardiovasc Qual Outcomes. 2008;1:84–91. doi: 10.1161/CIRCOUTCOMES.108.796185. [DOI] [PubMed] [Google Scholar]
- 18.Heneghan C, Ward A, Perera R, et al. Self-monitoring of oral anticoagulation: systematic review and meta-analysis of individual patient data. Lancet. 2012;379(9813):322–34. doi: 10.1016/S0140-6736(11)61294-4. [DOI] [PubMed] [Google Scholar]
- 19.van Gogh Investigators. Buller HR, Cohen AT, et al. Idraparinux versus standard therapy for venous thromboembolic disease. N Engl J Med. 2007;357:1094–104. doi: 10.1056/NEJMoa064247. [DOI] [PubMed] [Google Scholar]
- 20.ROCKET AF Study Investigators. Rivaroxaban-once daily, oral, direct factor Xa inhibition compared with vitamin K antagonism for prevention of stroke and Embolism Trial in Atrial Fibrillation: rationale and design of the ROCKET AF study. Am Heart J. 2010;159(3):340, e1–7, e1. doi: 10.1016/j.ahj.2009.11.025. [DOI] [PubMed] [Google Scholar]
- 21.Gage BF, Eby D, Johnson JA, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther. 2008;84:326–31. doi: 10.1038/clpt.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lenzini P, Wadelius M, Kimmel S, et al. Integration of genetic, clinical, and INR data to refine warfarin dosing. Clin Pharmacol Ther. 2010;87(5):572–8. doi: 10.1038/clpt.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwarz UI, Ritchie MD, Bradford Y, et al. Genetic determinants of response to warfarin during initial anticoagulation. N Engl J Med. 2008;358:999–1008. doi: 10.1056/NEJMoa0708078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansson S, Ohlsson L, Stenhoff H, et al. No effect of encapsulation on the pharmacokinetics of warfarin. Biopharm Drug Dispos. 2005;26:121–7. doi: 10.1002/bdd.441. [DOI] [PubMed] [Google Scholar]
- 25.Joo J, Geller NL, French B, et al. Prospective alpha allocation in the Clarification of Optimal Anticoagulation through Genetics (COAG) trial. Clin Trials. 2010;7(5):597–604. doi: 10.1177/1740774510381285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.French B, Joo J, Geller NL, et al. Statistical design of personalized medicine interventions: the Clarification of Optimal Anticoagulation through Genetics (COAG), trial. Trials. 2010;11:108. doi: 10.1186/1745-6215-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dolan G, Smith LA, Collins S, et al. Effect of setting, monitoring intensity and patient experience on anticoagulation control: a systematic review and meta-analysis of the literature. Curr Med Res Opin. 2008;24:1459–72. doi: 10.1185/030079908x297349. [DOI] [PubMed] [Google Scholar]
- 28.Ansell J, Hirsh J, Hylek E, et al. Pharmacology and management of the Vitamin K antagonists. Chest. 2008;133:160S–98S. doi: 10.1378/chest.08-0670. [DOI] [PubMed] [Google Scholar]
- 29.Simon R. Validation of pharmacogenomic biomarker classifiers for treatment selection. Cancer Biomark. 2006;2:89–96. doi: 10.3233/cbm-2006-23-402. [DOI] [PubMed] [Google Scholar]
- 30.Cotton BA, McCarthy JJ, Holcomb JB. Acutely injured patients on dabigatran. N Engl J Med. 2011;365(21):2039–40. doi: 10.1056/NEJMc1111095. [DOI] [PubMed] [Google Scholar]
- 31.Lakkireddy D, Reddy YM, Biase LD, et al. Feasibility and safety of dabigatran versus warfarin for periprocedural anticoagulation in patients undergoing radiofrequency ablation for atrial fibrillation. J Am Coll Cardiol. 2012;59(13):1168–74. doi: 10.1016/j.jacc.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 32.Schulman S, Crowther MA. How I anticoagulate in 2012, new and old anticoagulant agents, and when and how to switch. Blood. 2012;119(13):3016–23. doi: 10.1182/blood-2011-10-378950. [DOI] [PubMed] [Google Scholar]