Abstract
Detecting similar protein surfaces provides an important route for discovering unrecognized or novel functional relationship between proteins. The web server pvSOAR (pocket and void Surfaces Of Amino acid Residues) provides an online resource to identify similar protein surface regions. pvSOAR can take a structure either uploaded by a user or obtained from the Protein Data Bank, and identifies similar surface patterns based on geometrically defined pockets and voids. It provides several search modes to compare protein surfaces by similarity in local sequence, local shape and local orientation. Statistically significant search results are reported for visualization and interactive exploration. pvSOAR can be used to predict biological functions of proteins with known three-dimensional structures but unknown biological roles. It can also be used to study functional relationship between proteins and for exploration of the evolutionary origins of structural elements important for protein function. We present an example using pvSOAR to explore the biological roles of a protein whose structure was solved by the structural genomics project. The pvSOAR web server is available at http://pvsoar.bioengr.uic.edu/.
INTRODUCTION
Protein surface regions such as binding pockets provide specialized environments for biological activity (e.g. active sites, catalytic residues and ligand or substrate binding). The underlying three-dimensional shape and physicochemical texture in these regions facilitate functional interactions. Analysis of protein surfaces can provide insight into the biochemical function and evolutionary relationships of proteins. Identifying similar surfaces between proteins can be useful for understanding protein function and annotating proteins with unknown biological roles.
pvSOAR SERVER
The pvSOAR (pocket and void Surfaces Of Amino acid Residues) web server provides an online resource to identify similar surface regions in three-dimensional protein structures. pvSOAR is based on the methodology described in Binkowski et al. (1) for comparing local sequences, local shapes and local orientations between residues located on a geometrically defined pocket or void. Pockets and voids on protein structures are computed exhaustively for proteins in the Protein Data Bank (2), as in the CASTp web server (http://cast.engr.uic.edu) (3). Surface patterns of pocket and void regions are defined by the concatenated sequence fragment of residues forming the walls of the pocket and by the three-dimensional substructure of those residues. There are currently over 2 000 000 surface patterns for proteins in the Protein Data Bank, which are organized into a queryable format. The data set of surface patterns is updated to stay current with weekly PDB releases.
A pvSOAR search is based on comparison of a query pattern of surface sequence and substructure with a database of patterns from known protein structures. The statistical significance of similarity in the form of E-value and P-value between sequence patterns and between substructures, as measured by the coordinate root mean square distance (cRMSD) (4), is provided for discerning biologically important results. In addition, a newly developed metric for substructure comparison, called orientation root mean square distance (oRMSD), is also provided, along with the corresponding statistical significance (1). In oRMSD measurement, spatial coordinates of residues from a pocket are first projected onto a unit sphere placed at the center of mass; the RMSD between the two sets of transformed residues is then measured. In addition, other structural information is also presented to aid the interpretation of results. This includes pocket surface area and volume, identification numbers of the protein in resources such as SCOP (5), CATH (6), and Enzyme Commission number (7) when applicable.
USING pvSOAR
The pvSOAR web server provides an intuitive graphical user interface. It offers a selection of search methods to query against several specialized protein surface databases (Table 1). Statistically significant search results are returned to the user, at which time he/she can select and visualize interactively the surface patterns under comparison.
Table 1. Databases of protein surface patterns.
| Database | No. of surfaces | Time (sec.) |
|---|---|---|
| CASTp | 2 000 000+ | 43 |
| PDBSelect (90%) | 241 119 | 12 |
| PDBSelect (25%) | 55 840 | 2 |
The full CASTp database contains all computed pocket and void surfaces in the Protein Data Bank. The PDBSelect databases contain the subset of surface patterns found on protein structures contained in the corresponding PDBSelect list at different levels of sequence identity (8). The column ‘Time’ refers to the number of seconds it takes to perform the search using pocket 19 of protein myoglobin from Physeter catodon (PDB id = 109 m) as the database query (actual times will vary depending on network traffic).
Selecting a protein structure for querying
A pvSOAR search begins with the selection of a protein structure by the user for investigation. Users can either upload their own structure in PDB format to the pvSOAR server or type in the four-letter PDB identification code. All surface patterns of pockets and voids are computed on the fly for uploaded structures, or are accessed from pre-computed data for structures in the Protein Data Bank. Structures uploaded will remain accessible for the entire day on the pvSOAR server via log-in using a unique structure identification number which is emailed to the user. Several search options allow the user to compare the query protein with several databases of protein surface patterns. The available surface databases are shown in Table 1. We describe the search options below.
Querying a surface pattern against a database
This search option compares a single surface pattern with the selected surface database. After selecting a query protein structure, the user will then identify the surface region to be compared. Since pvSOAR uses the same pocket identification numbers as the CASTp web server, users can directly enter the pocket identification number identified from CASTp analysis. Alternatively, they can enter a residue number to be mapped to a specific pocket or void. Surfaces containing this residue will be selected, by default, to be compared with all surface patterns contained in the selected database. To facilitate navigation, we recommend that users concurrently open up another window through the CASTp server to identify pockets and voids of interests.
After the completion of a database search, the results are listed in a table sorted by their statistical significance, as measured by their cRMSD P-value. This table conveniently displays the hit surface, the E-value of the aligned surface sequence pattern, the number of aligned residues, cRMSD and oRMSD values. The list can alternatively be sorted by the E-value of the aligned sequence pattern or by the P-value of the oRMSD value, by clicking on the corresponding table header. Additional structural information such as pocket surface area and volume, SCOP and CATH identification number, Enzyme Commission number and information contained in the SITE, HET, and LIG records of the PDB file are also provided when available.
For visual inspection of the alignment of surface sequence patterns and superimposed substructures, users can select a hit from the list and will be taken to an interactive visualization environment for exploration of the matched surfaces. This requires installation of MDL's Chime plug-in (Windows) or a Java-enabled browser to run the JMol applet (Windows, Unix, Linux, OS-X). Visualization consists of the alignment of surface sequence patterns, individual surface substructures and superimposed surface substructures. Selecting the ‘Back’ button on the browser will return the user to the table listing all the hits, at which time he/she can select another hit for exploration.
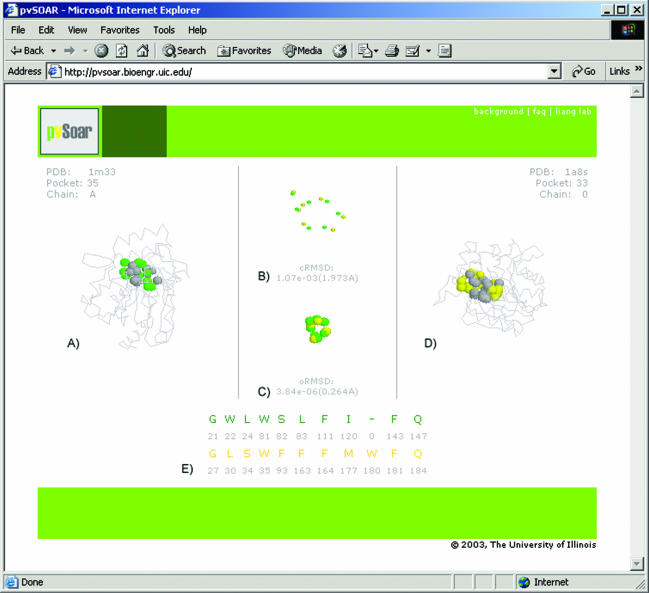
Figure 1 shows the environment for interactive visualization of matched surface patterns. A pocket (CASTp pocket id = 35) from protein BioH in Escherichia coli (PDB id = 1m33) generated by the structural genomics project (9) was selected as the query surface because it contains potential catalytic residues based on inspection using the CASTp web server (Figure 1A). A significant surface hit was found from a member protein of the peroxidase family (PDB id = 1a8s, CASTp pocket id = 33) (10) (Figure 1B). A closer examination of the matched surface patterns reveals a Ser–His–Asp catalytic triad on 1a8s that is similar to the one in BioH.
Figure 1.
Interactive environment for the visualization of pvSOAR search results. This example depicts a pocket surface pattern of (A) protein BioH from E.coli (CASTp pocket id = 35, PDB id = 1m33) and (D) haloperoxidase (CASTp pocket id = 33, PDB id = 1a8s) from Pseudomonas fluorescens. The superposition of conserved pocket residues and the superposition after projection on to a unit sphere are shown in (B) and (C), respectively. The alignment of sequence patterns of the pocket residues is shown in (E). These two surface patterns share strong similarity (cRMSD P-value = 1.073 × 10−3, oRMSD P-value = 3.836 × 10−6), indicating that there may be functional similarity between BioH protein and haloperoxidase.
Querying a whole structure against a database
Comparing the surface patterns on an entire structure with a database can be a very effective strategy for locating the functional site of a protein structure and for postulating a hypothesis of its biochemical role for proteins with unknown biological role, such as those generated in the structural genomics project. pvSOAR accommodates this type of search with an option for an interactive whole structure search. A screenshot is shown in Figure 2.
Figure 2.
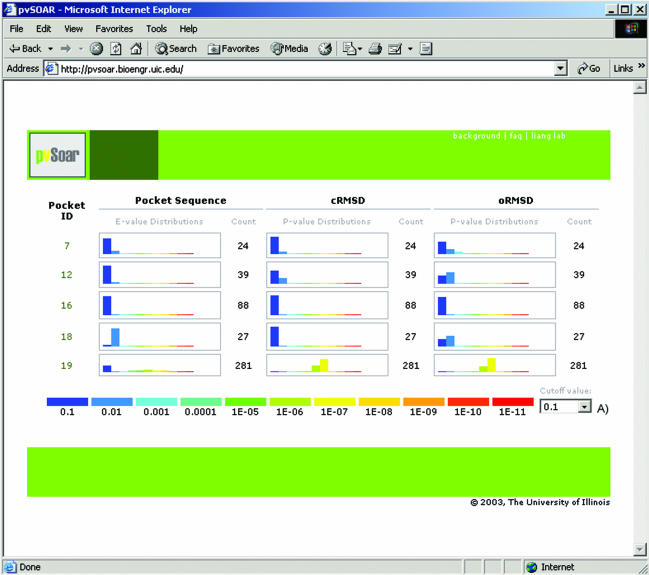
Screenshot of statistically significant search results between all surface patterns on myoglobin from Physeter catodon (PDB id = 109 m) and the CASTp database. Each surface has three histograms, reporting the distribution of significance score for E-values, P-value for cRMSD, and P-value for oRMSD. The drop-down box (A) allows for interactively setting up a threshold value to filter hits presented.
After selecting a query structure, all surface patterns derived from pockets and voids on that structure are searched against a chosen database. Significant surface matches are then returned and organized in ascending order by pocket identification number, which is roughly the same as sorting by volume of a pocket or void. A histogram displays the number of hits for each surface pattern at various significance levels by E-value, cRMSD P-value and oRMSD P-value. An interactive drop-down tab allows the user to filter the results based on a user-specified cutoff value. Clicking on a pocket id will then take the user to a search result table as described above. Only results below the current cutoff threshold value are presented.
Pairwise comparison of surface patterns
This option allows a convenient comparison of a surface pattern against another surface pattern. It bypasses the database search step and provides a quick visualization of surface comparison between two known protein structures (Figure 1).
EFFICIENT SEARCH STRATEGIES
Because of the rapidly increasing size of the Protein Data Bank, efficient search strategies are helpful in extracting the most relevant information rapidly from the pvSOAR server. Careful decision should be made in selecting the query pattern/structure and the database for identifying similar surfaces between proteins. In general, it is much faster to check a surface pattern rather than a whole structure against a database. It is also much faster to search PDBSELECT90 than the full pvSOAR database, and searching the PDBSelect databases often produces a more manageable list of results for manual inspection. In most cases, using the full CASTp database of surfaces is undesirable because of the overwhelming number of homologous structures for many proteins in the PDB, unless this is the interest of user's investigation.
The search strategy for the example of BioH presented earlier is as follows: (i) identifying key residues from the literature, (ii) identifying which pocket contains these residues by visualization using CASTp, (iii) using pvSOAR to check the surface pattern containing these residues against a database. More detailed examples of how pvSOAR can help to discover relationships between protein surfaces can be found in reference (1).
AVAILABILITY
pvSOAR can be freely accessed on the World Wide Web at http://pvsoar.bioengr.uic.edu.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Andrzej Joachimiak for suggesting the example of BioH. This work is supported by grants from the National Science Foundation (CAREER DBI0133856 and DBI0078270) and National Institute of Health (GM68958).
REFERENCES
- 1.Binkowski T.A., Adamian,L. and Liang,J. (2003) Inferring functional relationships of proteins from local sequence and spatial surface patterns. J. Mol. Biol., 332, 505–526. [DOI] [PubMed] [Google Scholar]
- 2.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binkowski T.A., Naghibzadeh,S. and Liang,J. (2003) Castp: computed atlas of surface topography of proteins. Nucleic Acids Res., 31(13), 3352–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Umeyama S. (1991) Least-squares estimation of transformation parameters between two point patterns. IEEE Trans. Pattern Anal. Mach. Intell., 13, 376–380. [Google Scholar]
- 5.Murzin A.G., Brenner,S.E., Hubbard,T. and Chothia,C. (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol., 247, 536–540. [DOI] [PubMed] [Google Scholar]
- 6.Orengo C.A., Michie,A.D., Jones,S., Jones,D.T., Swindells,M.B. and Thornton,J.M. (1997) CATH—a hierarchic classification of protein domain structures. Structure, 5, 1093–1108. [DOI] [PubMed] [Google Scholar]
- 7.Webb E. and NC-IUBMB (1992) Enzyme Nomenclature. Academic Press. [Google Scholar]
- 8.Hobohm U. and Sander,C. (1992) Selection of a representative set of structures from the Brookhaven protein data bank. Protein Sci., 1, 409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanishvili R., Yahunin,A.F., Laskowski,R.A., Skarina,T., Evdokimova,E., Doherty-Kirby,A., Lajoie,G.A., Thornton,J.M., Arrowsmith,C.H., Savchenko,A. et al. (2003) Integrating structure, bioinformatics, and enzymology to discover function. J. Biol. Comp., 278(28), 26039–26045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hofmann B., Tolzer,S., Pelletier,I., Altenbuchner,J., van Pee,K.H. and Hecht,H.J. (1998) Structural investigation of the cofactor-free chloroperoxidases. J. Mol. Biol., 279, 889–900. [DOI] [PubMed] [Google Scholar]