Abstract
Study objectives
Sleep disturbances are common in military personnel and are associated with increased risk for psychiatric morbidity, including posttraumatic stress disorder (PTSD) and depression, as well as inflammation. Improved sleep quality is linked to reductions in inflammatory bio-markers; however, the underlying mechanisms remain elusive.
Methods
In this study, we examine whole genome expression changes related to improved sleep in 68 military personnel diagnosed with insomnia. Subjects were classified into the following groups and then compared: improved sleep (n = 46), or non-improved sleep (n = 22) following three months of standard of care treatment for insomnia. Within subject differential expression was determined from microarray data using the Partek Genomics Suite analysis program and the ingenuity pathway analysis (IPA) was used to determine key regulators of observed expression changes. Changes in symptoms of depression and PTSD were also compared.
Results
At baseline, both groups were similar in demographics, clinical characteristics, and gene-expression profiles. The microarray data revealed that 217 coding genes were differentially expressed at the follow-up-period compared to baseline in the participants with improved sleep. Expression of inflammatory cytokines were reduced including IL-1β, IL-6, IL-8, and IL-13, with fold changes ranging from −3.19 to −2.1, and there were increases in the expression of inflammatory regulatory genes including toll-like receptors 1, 4, 7, and 8 in the improved sleep group. IPA revealed six gene networks, including ubiquitin, which was a major regulator in these gene-expression changes. The improved sleep group also had a significant reduction in the severity of depressive symptoms.
Conclusion
Interventions that restore sleep likely reduce the expression of inflammatory genes, which relate to ubiquitin genes and relate to reductions in depressive symptoms.
Keywords: gene expression, insomnia, inflammation, depression, military personnel
Introduction
Military personnel are often exposed to stressful conditions and erratic sleep schedules during deployment, thereby increasing their risk of developing a sleep disorder, most notably insomnia (1, 2). Deployment also increases the risk for psychiatric morbidity, including symptoms of posttraumatic stress disorder (PTSD) and depression, which are often comorbid with insomnia (1, 3–5). Insomnia is the most common symptom for referral in military personnel returning from deployment in Operation Enduring Freedom and Operation Iraqi Freedom (3). Not only is insomnia common but it also results in substantial costs to the individual and society. In the United States, total costs in productivity loss for chronic insomnia (e.g., employment disruption, insomnia-related accidents) are estimated to be between $77 and $92 million each year (6) and total annual costs are estimated to be beyond $100 billion (7). In addition, more than half of military personnel with sleep disturbances present with at least one other medical comorbidity, and those with insomnia are two times more likely to have PTSD (8). Therefore, there is a complex interplay between sleep quality and maintenance of physical and psychiatric health in military personnel, which likely has underlying biological foundations.
Recent studies have begun to determine the relationship of sleep to psychiatric morbidity in those who are under extreme stress (9). For example, pre-deployment sleep disturbance is linked to the onset of depression and PTSD following deployment (10). PTSD includes the symptom of nightmares, which is highly related to insomnia onset and non-remittance (11). Depression also includes symptoms of insomnia including difficulty falling or staying asleep, and these sleep disturbances often do not remit with standard of care treatment for depression (12). Therefore, current studies suggest that there is a shared vulnerability among PTSD, depression, and insomnia, and that these symptoms are often highly related in military personnel who deploy. Seeking care for sleep is often less stigmatizing than mental health care, resulting in military personnel seeking care for insomnia at almost five times the rate as mental health care (13, 14). Therefore, a better understanding of the underlying shared vulnerability of PTSD, depression and insomnia symptoms following deployment may inform interventions to address these comorbidities.
Sleep disturbance, PTSD, and depression are all associated with higher concentrations of inflammatory bio-markers, as well as a greater risk for inflammatory-related morbidities including: metabolic syndrome, cardiovascular disease, obesity, and type II diabetes (15–17). Even short-term reductions in sleep duration in healthy participants increases inflammatory cytokine production, including interleukin-6 (IL-6) (18–20). Chronic insomnia (21), as well as PTSD and depression are associated with increased concentrations of inflammatory cytokines (22). We previously reported that PTSD, depression, and insomnia in military personnel are associated with higher concentrations of inflammatory proteins including C-reactive protein (CRP) (23), and that improved sleep resulted in reductions in inflammation (24). Although these findings support the relationship between sleep and inflammation, the underlying molecular mechanisms of changes in inflammation following sleep improvement remain elusive (2). Gene-expression changes lead to differential protein activity in general; however, there are many other mechanisms that can contribute to the regulation of protein production, and availability, including epigenetic modifications. Therefore, gene-expression changes are one of the first investigations that can explain molecular mechanisms related to proteomic activity changes, and provide novel insights into complex inter-relationships among gene-regulatory mechanisms.
Examination of gene-expression profiles provides insight into how altered gene-activity relates to stress exposure and symptoms. Expression of a gene can increase or decrease and is the initial step to producing proteins that communicate cell function changes, including those active in the immune response system. Combat-related stressors, such as irregular work schedules, traumatic brain injuries, and other life threatening events, are linked to greater concentrations of inflammatory proteins, which are likely a result of gene-expression changes (25). Peripheral blood gene expression is comparable to post-mortem expression in more than 60% of genes, providing a minimally invasive method to investigate gene expression changes in living individuals (26, 27). Peripheral blood has been used to determine that reductions in the expression of circadian rhythm genes, including Clock, brain and muscle Arnt-like protein 1, and Period1, are implicated in symptoms of depression (28). PTSD is also linked to altered gene expression, including reductions in the expression of immune-regulating genes (29); however, the role of sleep and depression were not determined. Identification of inflammatory gene-expression changes related to sleep disturbance might allow for an understanding of the impact of insomnia on other systems and may inform development of targeted interventions to help prevent the development of comorbid disorders and inflammation.
To investigate this critical issue, we used an observational study of military personnel following deployment to determine global gene-expression changes related to improved sleep following 3 months of standard of care treatment. These findings may provide the insight needed to understand relationships between sleep, gene regulation of inflammation, and mental health.
Materials and Methods
Study design
This investigation is part of a large ongoing study of U.S. military personnel attending for an initial evaluation of sleep disturbance at the Madigan Army Medical Center Sleep Medicine Clinic in Tacoma, WA, USA. The institutional review board at Madigan Army Medical Center approved the study, and informed consent was obtained from each participant (5). Sixty-eight participants met the study’s inclusion criteria for a sleep diagnosis and completed both pre- and post-treatment assessments; they are included in this analysis. Study participants who met the inclusion criteria were active-duty military personnel who deployed within the previous 18 months with a current diagnosis of insomnia or comorbid insomnia with obstructive sleep apnea (OSA), as previously described (5). Participants were excluded from the study if they had a recent history of drug or alcohol abuse or current diagnosis of bipolar disorder, schizophrenia, or other psychotic disorder. Symptoms related to sleep quality, depression, and PTSD were evaluated through validated instruments at pre-treatment and post-treatment. Participants were classified into one of two groups (improved sleep or non-improved sleep) based on changes in sleep quality from pre- to post-treatment. Both groups were compared on gene-expression changes in inflammatory genes, as well as changes in symptoms of PTSD and depression.
Assessment and treatment of insomnia and OSA
All participants underwent a clinical evaluation and overnight polysomnography as part of a sleep medicine evaluation using standardized techniques previously reported (5). Depending on their sleep disorder diagnosis, patients were then assigned to a standard of care treatment group. Those with an insomnia diagnosis received 3 months of sleep-focused cognitive behavioral treatment for insomnia (CBT-i), which included aspects of cognitive therapy, stimulus control, sleep restriction, sleep practice (hygiene) education, and time monitoring behavior. Participants diagnosed with comorbid insomnia and OSA received either sleep education or 3 months of CBT-i for their insomnia, and autotitrating positive airway pressure therapy (APAP) for their OSA. A psychologist certified in behavioral sleep medicine or a sleep medicine physician with expertise in CBT-I administered and oversaw all treatments.
Defining improved sleep
The Pittsburgh sleep quality index (PSQI) was administered to assess participants’ sleep quality and sleep dysfunction over the previous month. This self-report questionnaire has strong diagnostic sensitivity in differentiating good and poor sleepers. Based on a scale of 0 (healthy sleep quality) to 21 (unhealthy sleep quality), a cut-off score of more than 5 is consistent with an insomnia diagnosis (30) and is both reliable (Cronbach’s α = 0.83) and valid (κ = 0.75, p < 0.001). For analysis purposes, participants with a reduction in their PSQI score from pre- to post-treatment were assigned to the improved sleep group (n = 46). Meanwhile, participants with an increase or no change in PSQI score from pre- to post-treatment were assigned to the non-improved sleep group (n = 22).
Assessment of PTSD and depression
Symptoms of depression were measured using the quick inventory of depressive symptomatology (QIDS). This 16-item inventory has a total possible score range from 0 (lowest severity) to 27 (highest severity). A score of 11, which indicates a moderate severity of depression, was used as the diagnosis cut-off (31). The PTSD checklist-military version (PCL-M) was used to determine symptoms of PTSD (32). A total possible score ranges from 17 (lowest severity) to 85 (highest severity). A total score of 50 and above resulted in a positive PTSD diagnostic screen. The PCL is both reliable and valid compared to the gold standard of the Structured Clinical Interview for DSM-III-R (33).
mRNA acquisition, quantitation, and hybridization
Blood samples were collected in PAXgene blood RNA tubes and processed with PAXgene™ Blood RNA Kits (PreAnalytiX, Qiagen) for RNA extraction. Quality and quantity of extracted RNA were evaluated with the NanoDrop DN-1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA) and the Agilent Bioanalyzer 2100 eukaryotic total RNA Nano assay (Agilent Technologies, Inc., Santa Clara, CA, USA). The 260/280 ratio ranged from 2.03 to 2.34 and the RNA integrity numbers (RIN) were >7.0 in all samples. According to standards (34) a RIN of 6.0 or greater is indicative of admissible quality, and no samples were excluded based on this criterion. Using the GeneChip (GC) 3′ IVT Plus Expression kit, each RNA (100 ng) sample was reverse transcribed, converted to biotinylated cRNA, and hybridized to Affymetrix HG-U133 Plus 2.0 microarrays (Affymetrix, Santa Clara, CA, USA), which contain 54,675 probe-sets representing more than 38,500 specific genes. After staining with streptavidin-phycoerythrin and thorough washing, the raw data were obtained by laser scanning imaging. All assays were undertaken based on standard Affymetrix protocols.
Statistical analysis
Partek Genomics Suite software, version 6.6 (Partek Inc., St. Louis, MO, USA), was used for all analytic procedures performed on microarray data. Interrogating probes were imported, and corrections for background signal were applied using the robust multi-array average (RMA) method, with additional corrections applied for the GC-content of probes. The probe-sets were standardized using quantile normalization, and expression levels of each probe underwent log-2 transformation to yield distributions of data that more closely approximated normality.
Parameters for identifying differentially expressed genes over time were then identified using analysis of variance (ANOVA) of each probe set’s expression level as a function of time: pre-treatment compared to post-treatment while adjusting for batch effect. A repeated measures comparison was made in each group. Restricted maximum likelihood (REML) method was employed to fit the fixed and random effects of the design separately. Significant differentially expressed gene lists were generated based on criteria of 1.5-fold (up or down) change magnitude and a p-value corrected for multiple comparisons using a false discovery rate (FDR) <0.05, for each group.
Two-tailed chi-square tests and independent t-tests were used to investigate any pre-treatment group differences that might affect the main analysis. Fisher’s exact tests were used when expected cell counts were <5. The main analysis investigated the relationship between sleep quality improvements or declines and symptoms of PTSD and depression, and gene expression changes following 3 months of sleep treatment. Separate paired t-tests were used to examine changes in outcome variables from pre- to post-treatment for both groups. Independent t-tests were used to examine between-group differences on outcome variables. The confidence level was set at p = 0.05 for all analyses.
QIAGEN’s Ingenuity Pathway Analysis (IPA, QIAGEN Redwood City, www.qiagen.com/ingenuity) was used to determine which diseases and disorders related to observed gene-expression changes, as well as top-gene networks implicated in these gene-expression changes.
Results
Demographics and clinical characteristics
The demographic, military, and clinical characteristics of the 68 participants are described in Table 1. As a whole, the sample was primarily male (97%) and Caucasian (62%) with a mean age of 33.8 years (7.8 SD) and 13.8 years (1.8 SD) of education. At pre-treatment, 47% of participants scored positive for depression and 28% scored positive for PTSD. Current use of relevant medications was reported by 40% of the participants. They include antidepressants (28%), pain medications (18%), benzodiazepines (6%), non-benzodiazepines (6%), and the alpha-blocker, prazosin (4%). At pre-treatment, all participants reported a score of at least six on the PSQI, which is a valid indication of poor sleep. There were no between-group differences in demographic variables [i.e., age, gender, race, education, body mass index (BMI), medication use, and military characteristics] with the exception of military rank, for which higher level ranks were more prevalent in the improved sleep group (p = 0.015). The prevalence of insomnia, OSA, traumatic brain injury (TBI), depression, and PTSD diagnoses were comparable between groups at baseline and both groups were characterized with high rates of these comorbid symptoms. There was no difference in treatment assignment, either CBT or APAP, between the improved sleep and non-improved sleep groups. The mean score in the PSQI was significantly different, with the improved sleep group having a mean score change of −4.8, whereas the non-improved group had an increase of 2.8 on the PSQI, p < 0.01 (see Table 2).
Table 1.
Demographic, military, and clinical characteristics.
| Improved sleep (n = 46) | Non-improved sleep (n = 22) | x2/t | p | |
|---|---|---|---|---|
| Age: mean (SD) | 33.5 (7.5) | 34.3 (8.7) | 0.358 | 0.722 |
| Males: n (%) | 45 (97.8) | 21 (95.5) | 0.293 | 0.546 |
| Race: n (%) | 3.663 | 0.056 | ||
| Caucasian | 32 (69.6) | 10 (45.5) | ||
| All others | 14 (30.4) | 12 (54.5) | ||
| Education in years: mean (SD) | 14.0 (2.1) | 13.4 (1.3) | −1.202 | 0.234 |
| Rank: n (%) | 5.921 | 0.015 | ||
| Lower | 26 (56.5) | 19 (86.4) | ||
| Senior or Officer | 20 (43.5) | 3 (13.6) | ||
| Front line military exposure: n (%) | 17 (37.0) | 9 (40.9) | 0.098 | 0.754 |
| Number of deployments: n (%) | 0.718 | 0.397 | ||
| 1 or 2 | 30 (65.2) | 12 (54.5) | ||
| 3 or more | 16 (34.8) | 10 (45.5) | ||
| Time since deployment: n (%) | 0.729 | 0.393 | ||
| <6 months | 14 (30.4) | 9 (40.9) | ||
| >6 months | 32 (69.6) | 13 (59.1) | ||
| BMI: mean (SD) | 30.2 (4.3) | 30.5 (3.4) | 0.330 | 0.743 |
| Medication use: n (%) | 16 (34.8) | 11 (50.0) | 1.440 | 0.230 |
| OSA diagnosis: n (%) | 12 (26.1) | 5 (22.7) | 0.090 | 0.765 |
| TBI diagnosis: n (%) | 18 (39.1) | 9 (40.9) | 0.020 | 0.888 |
| Probable depression diagnosis: n (%) | 21 (45.7) | 11 (50.0) | 0.113 | 0.737 |
| Probable PTSD diagnosis: n (%) | 11 (23.9) | 8 (36.4) | 1.146 | 0.284 |
BMI, body mass index; OSA, obstructive sleep apnea; PTSD, posttraumatic stress disorder; TBI, traumatic brain injury.
Fisher’s exact test was used when expected cell counts were <5.
Table 2.
Pre- and post-treatment symptom severity mean (SD).
| Improved sleep (n = 46) |
Non-improved sleep (n = 22) |
Between-group |
||||||
|---|---|---|---|---|---|---|---|---|
| Pre-treatment | Post-treatment | p | Pre-treatment | Post-treatment | p | Baseline | Changea | |
| p | p | |||||||
| Sleep quality | 12.8 (4.1) | 8.0 (3.9) | <0.001 | 12.1 (3.6) | 14.9 (3.1) | <0.001 | 0.484 | <0.001 |
| Depression severity | 10.5 (5.3) | 8.1 (5.0) | 0.001 | 11.00 (4.7) | 12.2 (5.0) | 0.134 | 0.732 | 0.002 |
| PTSD severity | 38.7 (15.3) | 35.6 (16.5) | 0.067 | 43.5 (16.2) | 46.5 (17.6) | 0.223 | 0.237 | 0.040 |
aPre- to post-treatment change-score. Bold font indicates findings with significant change.
Psychological symptom change
Sleep quality, depression, and PTSD outcomes were assessed before and after 3 months of sleep treatment. Table 2 details the pre- and post-treatment symptom severity of both groups. Briefly, from pre- to post-treatment, the improved sleep group reported significant reductions in depression [t(45) = 3.463, p = 0.001] and a trend for decreases in posttraumatic symptoms [t(45) = 1.875, p = 0.067]. These improvements were significant between groups (all p-values <0.05). Meanwhile, the non-improved sleep group reported no significant change in depression (p = 0.134) or posttraumatic symptoms (p = 0.223).
Gene-expression profiles
The microarray data revealed that 217 coding genes (Table 3) were differentially expressed at the follow-up-period compared to baseline in the participants who improved in sleep. Of these 217 genes, 113 genes were down-regulated (Table 4) and 104 were up-regulated (Table 5). None of these genes changed significantly in the non-improved sleep group.
Table 3.
All differentially expressed genes.
| Gene symbol | Gene title | Fold-change | p-Value |
|---|---|---|---|
| CCL4 | Chemokine (C—C motif) ligand 4 | −3.38991 | 0.000400502 |
| EGR3 | Early growth response 3 | −3.33259 | 4.37E-05 |
| IL1B | Interleukin 1, beta | −3.19348 | 0.00054254 |
| TIGIT | T cell immunoreceptor with Ig and ITIM domains | −2.999421 | 0.000116146 |
| CD96 | CD96 molecule | −2.9992104 | 0.000593266 |
| CCL3///CCL3L1///CCL3L3 | Chemokine (C—C motif) ligand 3///chemokine (C—C motif) ligand 3-like 1///chemokine | −2.99786 | 1.32E-05 |
| CCL5 | Chemokine (C—C motif) ligand 5 | −2.96621 | 0.000882838 |
| TNF | Tumor necrosis factor | −2.90578 | 0.000536776 |
| DUSP1 | Dual specificity phosphatase 1 | −2.89657 | 1.73E-05 |
| GATA3 | GATA binding protein 3 | −2.889336 | 0.000852941 |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | −2.86159 | 0.000116328 |
| BZRAP1 | Benzodiazepine receptor (peripheral) associated protein 1 | −2.84583 | 0.000429033 |
| KIR3DL1///KIR3DL2///LOC727787 | Killer cell immunoglobulin-like receptor, three domains, long cytoplasmic tail, 1///k | −2.79571 | 0.00111701 |
| IL18R1 | Interleukin 18 receptor 1 | −2.79239 | 0.00131446 |
| NFATC2 | Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 | −2.77617 | 0.000468791 |
| IFITM1 | Interferon Interferon-induced transmembrane protein 1 (9—27) | −2.67642 | 0.000796847 |
| IL6 | Interleukin 6 (interferon, beta 2) | −2.66587 | 0.000169597 |
| IFITM1 | Interferon Interferon-induced transmembrane protein 1 (9—27) | −2.59115 | 0.00120357 |
| GLCCI1 | Glucocorticoid Glucocorticoid-induced transcript 1 | −2.57928 | 0.000863656 |
| PER1 | Period homolog 1 (Drosophila) | −2.34547 | 0.000172781 |
| TARP///TRGC2 | TCR gamma alternate reading frame protein///T cell receptor gamma constant 2 | −2.31319 | 0.000858636 |
| TARP | TCR gamma alternate reading frame protein | −2.2425 | 0.00123683 |
| TP53INP2 | Tumor protein p53 inducible nuclear protein 2 | −2.23152 | 2.18E-04 |
| CDC14A | CDC14 cell division cycle 14 homolog A (S. cerevisiae) | −2.22332 | 0.00055414 |
| ZNF831 | Zinc finger protein 831 | −2.21191 | 0.000940436 |
| BCL10 | B-cell CLL/lymphoma 10 | −2.19636 | 1.92E-04 |
| TARP///TRGC2 | TCR gamma alternate reading frame protein///T cell receptor gamma constant 2 | −2.18635 | 0.00138229 |
| TARP///TRGC2 | TCR gamma alternate reading frame protein///T cell receptor gamma constant 2 | −2.17218 | 0.00136634 |
| STAT3 | Signal transducer and activator of transcription 3 (acute-phase response factor) | −2.17143 | 2.79E-05 |
| IL13 | Interleukin 13 | −2.0998 | 0.000453301 |
| ZNF831 | Zinc finger protein 831 | −2.07393 | 0.000998177 |
| GZMB | Granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) | −2.07291 | 0.000106599 |
| KLF12 | Kruppel-like factor 12 | −2.06742 | 0.000474026 |
| SPON2 | Spondin 2, extracellular matrix protein | −2.06256 | 0.000562071 |
| MBNL2 | Muscleblind-like 2 (Drosophila) | −2.06215 | 0.000146908 |
| KRAS | v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog | −2.04976 | 0.000826814 |
| CRA | Complement factor C4 | −2.04778987 | 7.09143E-05 |
| FCAR | Fc fragment of IgA, receptor for | −2.03608 | 1.89E-04 |
| NCL | Nucleolin | −2.0312 | 4.39101E-07 |
| IFRD1 | Interferon-related developmental regulator 1 | −2.02534 | 7.16E-06 |
| S1PR5 | Sphingosine-1-phosphate receptor 5 | −1.99987 | 0.000877998 |
| NAP1L5 | Nucleosome assembly protein 1-like 5 | −1.9825 | 0.00103436 |
| FGFBP2 | Fibroblast growth factor binding protein 2 | −1.95856 | 0.00097833 |
| TRD@ | T cell receptor delta locus | −1.95283 | 0.00111344 |
| PYHIN1 | Pyrin and HIN domain family, member 1 | −1.93462 | 0.000986752 |
| GFI1 | Growth factor independent 1 transcription repressor | −1.93194 | 0.00076425 |
| GZMM | Granzyme M (lymphocyte met-ase 1) | −1.93125 | 0.00085383 |
| TRD@ | T cell receptor delta locus | −1.9207 | 0.00111727 |
| TSC22D3 | TSC22 domain family, member 3 | −1.91096 | 2.28E-06 |
| KLRB1 | Killer cell lectin-like receptor subfamily B, member 1 | −1.90964 | 0.00136588 |
| IL18RAP | Interleukin 18 receptor accessory protein | −1.90962 | 0.000221487 |
| ICOS | Inducible co-stimulator | −1.903002 | 0.000880293 |
| MYLIP | Myosin regulatory light chain interacting protein | −1.89332 | 8.32E-06 |
| TGFBR3 | Transforming growth factor, beta receptor III | −1.8931 | 0.00121075 |
| ZAP70 | Zeta-chain (TCR) associated protein kinase 70 kDa | −1.89061 | 0.000662326 |
| CTSW | Cathepsin W | −1.88397 | 0.00129928 |
| CEP78 | Centrosomal protein 78 kDa | −1.88364 | 0.000127057 |
| ARL4C | ADP-ribosylation factor-like 4C | −1.8832 | 0.000160201 |
| ARL4C | ADP-ribosylation factor-like 4C | −1.87226 | 0.000419378 |
| KLRD1 | Killer cell lectin-like receptor subfamily D, member 1 | −1.86593 | 0.00124982 |
| PRKCQ | Protein kinase C, theta | −1.86552 | 0.00113571 |
| NR4A1 | Nuclear receptor subfamily 4, group A, member 1 | −1.86138 | 8.90E-05 |
| SYTL2 | Synaptotagmin-like 2 | −1.85889 | 0.00135142 |
| ARL4C | ADP-ribosylation factor-like 4C | −1.85424 | 0.000286602 |
| AGAP1 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 | −1.80837 | 0.000287414 |
| SH2D2A | SH2 domain containing 2A | −1.80828 | 0.000300214 |
| KLRK1 | Killer cell lectin-like receptor subfamily K, member 1 | −1.78448 | 0.00102739 |
| RUNX3 | Runt-related transcription factor 3 | −1.77892 | 0.00013984 |
| KIR3DL3 | Killer cell immunoglobulin-like receptor, three domains, long cytoplasmic tail, 3 | −1.70915 | 0.000885828 |
| PER1 | Period homolog 1 (Drosophila) | −1.70146 | 0.000444082 |
| CHD2 | Chromodomain helicase DNA DNA-binding protein 2 | −1.70091 | 0.000257214 |
| DTX3 | Deltex homolog 3 (Drosophila) | −1.70068 | 0.000829046 |
| PRKCH | Protein kinase C, eta | −1.68436 | 0.00106358 |
| FAM179A | Family with sequence similarity 179, member A | −1.68265 | 0.000135121 |
| MGAT4A | Mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A | −1.67906 | 0.000326335 |
| GBP5 | Guanylate binding protein 5 | −1.67871 | 0.000452814 |
| MLC1 | Megalencephalic leukoencephalopathy with subcortical cysts 1 | −1.66993 | 0.000433367 |
| ANKRD36 | Ankyrin repeat domain 36 | −1.63932 | 0.000866547 |
| NFATC2 | Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 | −1.63898 | 0.000941657 |
| SYNE2 | Spectrin repeat containing, nuclear envelope 2 | −1.63663 | 1.19E-05 |
| ID2 | Inhibitor of DNA DNA-binding 2, dominant negative helix—loop—helix protein | −1.60803 | 1.67E-06 |
| KIF21A | Kinesin family member 21A | −1.60799 | 0.00136965 |
| CTNNB1 | Catenin (cadherin-associated protein), beta 1, 88 kDa | −1.6076 | 4.09E-05 |
| TSEN54 | tRNA splicing endonuclease 54 homolog (S. cerevisiae) | −1.60547 | 0.000604728 |
| ZNF451 | Zinc finger protein 451 | −1.60463 | 6.28E-06 |
| TSPAN5 | Tetraspanin 5 | −1.60208 | 0.00114212 |
| RASGEF1A | RasGEF domain family, member 1A | −1.60149 | 0.000447691 |
| TNIK | TRAF2 and NCK interacting kinase | −1.59894 | 2.50E-05 |
| CBLB | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | −1.5979 | 0.000320734 |
| MLLT11 | Myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocate | −1.59317 | 0.000831119 |
| TAF9B | TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31 kDa | −1.5885 | 0.000321819 |
| HNRNPL | Heterogeneous nuclear ribonucleoprotein L | −1.5871 | 3.77E-06 |
| SESN2 | Sestrin 2 | −1.57849 | 6.97E-05 |
| LOC100130872 | Hypothetical LOC100130872 | −1.56568 | 1.48E-05 |
| SESN2 | Sestrin 2 | −1.56417 | 0.00050745 |
| CCL5 | Chemokine (C—C motif) ligand 5 | −1.56409 | 0.000430274 |
| PIK3R1 | Phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | −1.56311 | 0.00143535 |
| TFDP2 | Transcription factor Dp-2 (E2F dimerization partner 2) | −1.55293 | 0.000804861 |
| PDE4B | Phosphodiesterase 4B, cAMP-specific | −1.55035 | 0.00085501 |
| RBM14 | RNA binding motif protein 14 | −1.54638 | 2.35E-05 |
| FAM100B | Family with sequence similarity 100, member B | −1.53662 | 7.63E-05 |
| PDE4D | Phosphodiesterase 4D, cAMP-specific | −1.53463 | 0.000469774 |
| GRPEL1 | GrpE-like 1, mitochondrial (E. coli) | −1.53133 | 1.23E-05 |
| ZNF507 | Zinc finger protein 507 | −1.52977 | 0.000202111 |
| NUP153 | Nucleoporin 153 kDa | −1.52758 | 0.000367042 |
| FUS | Fused in sarcoma | −1.52544 | 0.0010451 |
| PTTG1 | Pituitary tumor-transforming 1 | −1.52511 | 0.000818797 |
| WAC | WW domain containing adaptor with coiled-coil | −1.52328 | 0.000534293 |
| EIF1 | Eukaryotic translation initiation factor 1 | −1.52229 | 6.21E-06 |
| TSEN54 | tRNA splicing endonuclease 54 homolog (S. cerevisiae) | −1.50904 | 0.000620767 |
| MYLIP | Myosin regulatory light chain interacting protein | −1.50894 | 3.24E-05 |
| CALU | Calumenin | −1.50768 | 0.000736822 |
| EHD4 | EH-domain containing 4 | −1.50216 | 0.000209267 |
| ATP11B | ATPase, class VI, type 11B | 1.50127 | 0.000202173 |
| FKBP15 | FK506 binding protein 15, 133 kDa | 1.50442 | 0.000270622 |
| MOCS3 | Molybdenum cofactor synthesis 3 | 1.5075 | 4.38E-06 |
| FAM13A | Family with sequence similarity 13, member A | 1.50801 | 0.00135619 |
| CDC42EP3 | CDC42 effector protein (Rho GTPase binding) 3 | 1.50849 | 2.59E-05 |
| DACH1 | Dachshund homolog 1 (Drosophila) | 1.51638 | 0.000969487 |
| HHEX | Hematopoietically expressed homeobox | 1.51675 | 6.21E-05 |
| PDE7B | Phosphodiesterase 7B | 1.51869 | 2.37E-05 |
| OBFC2A | Oligonucleotide/oligosaccharide-binding fold containing 2A | 1.52002 | 0.000343088 |
| CR1 | Complement component (3b/4b) receptor 1 (Knops blood group) | 1.52311 | 9.05E-05 |
| PHF23 | PHD finger protein 23 | 1.53301 | 3.41E-06 |
| ZNF780A | Zinc finger protein 780A | 1.53325 | 0.000505801 |
| GPR27 | G protein-coupled receptor 27 | 1.53487 | 0.0011538 |
| CDC42EP3 | CDC42 effector protein (Rho GTPase binding) 3 | 1.53614 | 1.72E-05 |
| ADPRH | ADP-ribosylarginine hydrolase | 1.53673 | 0.000301124 |
| TLR4 | Toll-like receptor 4 | 1.54384 | 0.000155809 |
| TLR10 | Toll-like receptor 10 | 1.54391 | 0.000150659 |
| SIRPB2 | Signal-regulatory protein beta 2 | 1.55351 | 9.39E-05 |
| FUBP1 | Far upstream element (FUSE) binding protein 1 | 1.55437 | 3.43E-05 |
| NLRC4 | NLR family, CARD domain containing 4 | 1.5579 | 0.0010619 |
| OBFC2A | Oligonucleotide/oligosaccharide-binding fold containing 2A | 1.55822 | 6.88E-05 |
| TRPS1 | Trichorhinophalangeal syndrome I | 1.56073 | 0.00117806 |
| CLIC4 | Chloride intracellular channel 4 | 1.56137 | 0.00142631 |
| ANKRD57 | Ankyrin repeat domain 57 | 1.56891 | 0.000496592 |
| FOSL2 | FOS-like antigen 2 | 1.57091 | 0.00103799 |
| FBXO30 | F-box protein 30 | 1.57898 | 3.37E-05 |
| S1PR3 | Sphingosine-1-phosphate receptor 3 | 1.57953 | 0.000766403 |
| MFAP3 | Microfibrillar-associated protein 3 | 1.58049 | 8.30E-05 |
| CPD | Carboxypeptidase D | 1.58561 | 0.000270503 |
| RNF24 | Ring finger protein 24 | 1.5887 | 0.000779404 |
| PIGM | Phosphatidylinositol glycan anchor biosynthesis, class M | 1.58922 | 3.93E-06 |
| SAP30L | SAP30-like | 1.599 | 3.48E-05 |
| SERPINI2 | Serpin peptidase inhibitor, clade I (pancpin), member 2 | 1.60236 | 0.000725023 |
| BACH1 | BTB and CNC homology 1, basic leucine zipper transcription factor 1 | 1.60519 | 5.89E-06 |
| ZNF322B | Zinc finger protein 322B | 1.60589 | 0.000435523 |
| ZEB2 | Zinc finger E-box binding homeobox 2 | 1.61326 | 0.0010863 |
| GPATCH2 | G patch domain containing 2 | 1.61607 | 0.000829918 |
| KCNE3 | Potassium voltage-gated channel, Isk-related family, member 3 | 1.62682 | 0.000871206 |
| ENTPD1 | Ectonucleoside triphosphate diphosphohydrolase 1 | 1.62899 | 0.000688504 |
| FAM114A1 | Family with sequence similarity 114, member A1 | 1.63805 | 0.000957221 |
| ZNF697 | Zinc finger protein 697 | 1.63815 | 0.000362559 |
| SIPA1L1 | Signal-induced proliferation-associated 1 1-like 1 | 1.63837 | 0.000200843 |
| MGST1 | Microsomal glutathione S-transferase 1 | 1.63932 | 0.000155808 |
| CENPBD1 | CENPB DNA-binding domains containing 1 | 1.63975 | 0.000598446 |
| LOC100505956 | Hypothetical LOC100505956 | 1.6413 | 0.00103297 |
| CTTNBP2NL | CTTNBP2 N-terminal like | 1.6428 | 0.00126079 |
| CLLU1 | Chronic lymphocytic leukemia up-regulated 1 | 1.65206 | 0.000787124 |
| ST8SIA4 | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 | 1.66224 | 0.000130372 |
| FAR2 | Fatty acyl CoA reductase 2 | 1.68735 | 5.72E-05 |
| DAPP1 | Dual adaptor of phosphotyrosine and 3-phosphoinositides | 1.68966 | 0.000609128 |
| PLD1 | Phospholipase D1, phosphatidylcholine-specific | 1.69193 | 0.000517971 |
| MGST1 | Microsomal glutathione S-transferase 1 | 1.69246 | 0.00020858 |
| MGST1 | Microsomal glutathione S-transferase 1 | 1.69458 | 7.19E-06 |
| DSC2 | Desmocollin 2 | 1.69681 | 0.000254904 |
| GPATCH2 | G patch domain containing 2 | 1.70199 | 0.000112569 |
| ST6GALNAC3 | ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2 | 1.70328 | 0.000428429 |
| FBN2 | Fibrillin 2 | 1.721 | 0.000252441 |
| FCGR1B | Fc fragment of IgG, high affinity Ib, receptor (CD64) | 1.72442 | 0.000706171 |
| HP | Haptoglobin | 1.72582 | 0.000718758 |
| FRAT2 | Frequently rearranged in advanced T-cell lymphomas 2 | 1.72608 | 0.000190515 |
| HHEX | Hematopoietically expressed homeobox | 1.7329 | 4.10E-06 |
| P2RY13 | Purinergic receptor P2Y, G-protein coupled, 13 | 1.73405 | 0.000133887 |
| MGST1 | Microsomal glutathione S-transferase 1 | 1.74784 | 7.56E-05 |
| LOC100506828 | Hypothetical LOC100506828 | 1.75228 | 7.44E-05 |
| TFEC | Transcription factor EC | 1.75494 | 0.000135064 |
| WLS | Wntless homolog (Drosophila) | 1.76165 | 0.000227731 |
| FCGR1A///FCGR1C | Fc fragment of IgG, high affinity Ia, receptor (CD64)///Fc fragment of IgG, high affinity | 1.76282 | 0.00095328 |
| SGMS2 | Sphingomyelin synthase 2 | 1.76728 | 0.000618004 |
| ATP6V1A | ATPase, H + transporting, lysosomal 70 kDa, V1 subunit A | 1.77021 | 3.21E-05 |
| FPR2 | Formyl peptide receptor 2 | 1.77041 | 8.98E-05 |
| ADAM9 | ADAM metallopeptidase domain 9 | 1.82176 | 2.72E-05 |
| TMEM49 | Transmembrane protein 49 | 1.8221 | 0.000516735 |
| WSB1 | WD repeat and SOCS box-containing 1 | 1.82444 | 0.000178707 |
| NR3C1 | Promoter 1B of the GR | 1.83029221 | 0.00019027 |
| ADAMTS5 | ADAM metallopeptidase with thrombospondin type 1 motif, 5 | 1.83321 | 0.000299013 |
| FPR2 | Formyl peptide receptor 2 | 1.85476 | 0.000499535 |
| EMP1 | Epithelial membrane protein 1 | 1.8738 | 0.000128519 |
| SPATA5L1 | Spermatogenesis associated 5-like 1 | 1.87522 | 0.00102786 |
| PLA2G4A | Phospholipase A2, group IVA (cytosolic, calcium-dependent) | 1.87895 | 0.000196578 |
| TLR1 | Toll-like receptor 1 | 1.93858 | 2.14E-07 |
| LOC203274 | Hypothetical protein LOC203274 | 1.94801 | 5.09E-05 |
| NFATC2 | Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 | 1.9776 | 5.93E-05 |
| FBXO30 | F-box protein 30 | 1.98992 | 0.000899931 |
| ATP6V1A | ATPase, H + transporting, lysosomal 70 kDa, V1 subunit A | 1.99 | 0.000810192 |
| CYP1B1 | Cytochrome P450, family 1, subfamily B, polypeptide 1 | 1.99311 | 9.37E-06 |
| STEAP4 | STEAP family member 4 | 2.01298 | 0.000150426 |
| IL8R1 | Interleukin 8 receptor 1 | 2.03E + 00 | 3.92893E-05 |
| IL2 | interleukin 2 | 2.03419987 | 3.10905E-06 |
| FOLR3 | Folate receptor 3 (gamma) | 2.05781 | 0.00017189 |
| FKBP15 | FK506 binding protein 15, 133 kDa | 2.094500914 | 4.30009E-05 |
| SLITRK4 | SLIT and NTRK-like family, member 4 | 2.16832 | 6.47E-05 |
| ADAMTS5 | ADAM metallopeptidase with thrombospondin type 1 motif, 5 | 2.18224 | 0.00016302 |
| PTPRO | Protein tyrosine phosphatase, receptor type, O | 2.21369 | 0.000260765 |
| FAM198B | Family with sequence similarity 198, member B | 2.22299 | 0.000661349 |
| FKBP506 | FKBP506 binding protein | 2.223221099 | 1.90294E-06 |
| IFNGR1 | Interferon gamma receptor 1 | 2.51823 | 0.000195788 |
| TLR7 | Toll-like receptor 7 | 2.59205 | 0.00115974 |
| RIN2 | Ras and Rab interactor 2 | 2.62017 | 0.000313438 |
| TLR8 | Toll-like receptor 8 | 2.62471 | 0.00112645 |
| TLR4 | Toll-like receptor 4 | 2.66149 | 0.000182505 |
| MIR21 | MicroRNA 21 | 2.68409 | 0.000265287 |
| CYP1B1 | Cytochrome P450, family 1, subfamily B, polypeptide 1 | 2.83233 | 0.000146054 |
| PDK4 | Pyruvate dehydrogenase kinase, isozyme 4 | 2.99062 | 7.43E-07 |
| PDK4 | Pyruvate dehydrogenase kinase, isozyme 4 | 3.1238 | 0.000118651 |
Table 4.
Down-regulated gene expression (top 50).
| Gene symbol | Gene title | Fold-change | p-Value |
|---|---|---|---|
| CCL4 | Chemokine (C–C motif) ligand 4 | −3.39 | 0.0004 |
| EGR3 | Early growth response 3 | −3.333 | 4.37E-05 |
| IL1B | Interleukin 1, beta | −3.193 | 0.00054 |
| TIGIT | T cell immunoreceptor with Ig and ITIM domains | −2.999 | 0.00012 |
| CD96 | CD96 molecule | −2.999 | 0.00059 |
| CCL3///CCL3L1///CCL3L3 | Chemokine (C–C motif) ligand 3///chemokine (C–C motif) ligand 3-like 1///chemokine | −2.998 | 1.32E-05 |
| CCL5 | Chemokine (C–C motif) ligand 5 | −2.966 | 0.00088 |
| TNF | Tumor necrosis factor | −2.906 | 0.00054 |
| DUSP1 | Dual specificity phosphatase 1 | −2.897 | 1.73E-05 |
| GATA3 | GATA binding protein 3 | −2.889 | 0.00085 |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | −2.862 | 0.00012 |
| BZRAP1 | Benzodiazepine receptor (peripheral) associated protein 1 | −2.846 | 0.00043 |
| KIR3DL1///KIR3DL2///LOC727787 | Killer cell immunoglobulin-like receptor, three domains, long cytoplasmic tail, 1///k | −2.796 | 0.00112 |
| IL18R1 | Interleukin 18 receptor 1 | −2.792 | 0.00131 |
| NFATC2 | Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 | −2.776 | 0.00047 |
| IFITM1 | Interferon-induced transmembrane protein 1 (9–27) | −2.676 | 0.0008 |
| IL6 | Interleukin 6 (interferon, beta 2) | −2.666 | 0.00017 |
| IFITM1 | Interferon-induced transmembrane protein 1 (9–27) | −2.591 | 0.0012 |
| GLCCI1 | Glucocorticoid induced transcript 1 | −2.579 | 0.00086 |
| PER1 | Period homolog 1 (Drosophila) | −2.345 | 0.00017 |
| TARP///TRGC2 | TCR gamma alternate reading frame protein///T cell receptor gamma constant 2 | −2.313 | 0.00086 |
| TARP | TCR gamma alternate reading frame protein | −2.243 | 0.00123683 |
| TP53INP2 | Tumor protein p53 inducible nuclear protein 2 | −2.232 | 2.18E-04 |
| CDC14A | CDC14 cell division cycle 14 homolog A (S. cerevisiae) | −2.223 | 0.00055 |
| ZNF831 | Zinc finger protein 831 | −2.212 | 0.00094 |
| BCL10 | B-cell CLL/lymphoma 10 | −2.196 | 1.92E-04 |
| TARP///TRGC2 | TCR gamma alternate reading frame protein///T cell receptor gamma constant 2 | −2.186 | 0.00138 |
| TARP///TRGC2 | TCR gamma alternate reading frame protein///T cell receptor gamma constant 2 | −2.172 | 0.00137 |
| STAT3 | Signal transducer and activator of transcription 3 (acute-phase response factor) | −2.171 | 2.79E-05 |
| IL13 | Interleukin 13 | −2.1 | 0.00045 |
| ZNF831 | Zinc finger protein 831 | −2.074 | 0.001 |
| GZMB | Granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) | −2.073 | 0.00011 |
| KLF12 | Kruppel-like factor 12 | −2.067 | 0.00047 |
| SPON2 | Spondin 2, extracellular matrix protein | −2.063 | 0.00056 |
| MBNL2 | Muscleblind-like 2 (Drosophila) | −2.062 | 0.00015 |
| KRAS | v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog | −2.05 | 0.00083 |
| CRA | Complement factor C4 | −2.048 | 7.09E-05 |
| FCAR | Fc fragment of IgA, receptor for | −2.036 | 1.89E-04 |
| NCL | Nucleolin | −2.031 | 4.39E-07 |
| IFRD1 | Interferon-related developmental regulator 1 | −2.025 | 7.16E-06 |
| S1PR5 | Sphingosine-1-phosphate receptor 5 | −2.00 | 0.00088 |
| NAP1L5 | Nucleosome assembly protein 1-like 5 | −1.983 | 0.00103 |
| FGFBP2 | Fibroblast growth factor binding protein 2 | −1.959 | 0.00098 |
| TRD | T cell receptor delta locus | −1.953 | 0.00111 |
| PYHIN1 | Pyrin and HIN domain family, member 1 | −1.935 | 0.00099 |
| GFI1 | Growth factor independent 1 transcription repressor | −1.932 | 0.00076 |
| GZMM | Granzyme M (lymphocyte met-ase 1) | −1.931 | 0.00085 |
| TRD | T cell receptor delta locus | −1.921 | 0.00112 |
| TSC22D3 | TSC22 domain family, member 3 | −1.911 | 2.28E-06 |
| KLRB1 | Killer cell lectin-like receptor subfamily B, member 1 | −1.91 | 0.00137 |
Table 5.
Up-regulated gene expression (top 50).
| Gene symbol | Gene title | Fold-change | p-Value |
|---|---|---|---|
| PDK4 | Pyruvate dehydrogenase kinase, isozyme 4 | 3.1238 | 0.00012 |
| CYP1B1 | Cytochrome P450, family 1, subfamily B, polypeptide 1 | 2.83233 | 0.00015 |
| MIR21 | MicroRNA 21 | 2.68409 | 0.00027 |
| TLR4 | Toll-like receptor 4 | 2.66149 | 0.00018 |
| TLR8 | Toll-like receptor 8 | 2.62471 | 0.00113 |
| RIN2 | Ras and Rab interactor 2 | 2.62017 | 0.00031 |
| TLR7 | Toll-like receptor 7 | 2.59205 | 0.00116 |
| IFNGR1 | Interferon gamma receptor 1 | 2.51823 | 0.0002 |
| FKBP506 | FKBP506 binding protein | 2.22322 | 1.90E-06 |
| FAM198B | Family with sequence similarity 198, member B | 2.22299 | 0.00066 |
| PTPRO | Protein tyrosine phosphatase, receptor type, O | 2.21369 | 0.00026 |
| ADAMTS5 | ADAM metallopeptidase with thrombospondin type 1 motif, 5 | 2.18224 | 0.00016 |
| SLITRK4 | SLIT and NTRK-like family, member 4 | 2.16832 | 6.47E-05 |
| FKBP15 | FK506 binding protein 15, 133 kDa | 2.0945 | 4.30E-05 |
| FOLR3 | Folate receptor 3 (gamma) | 2.05781 | 0.00017 |
| IL2 | Interleukin 2 | 2.0342 | 3.10E-06 |
| IL8R1 | Interleukin 8 receptor 1 | 2.03E + 00 | 3.90E-05 |
| STEAP4 | STEAP family member 4 | 2.01298 | 0.00015 |
| ATP6V1A | ATPase, H+ transporting, lysosomal 70 kDa, V1 subunit A | 1.99 | 0.00081 |
| FBXO30 | F-box protein 30 | 1.98992 | 0.0009 |
| NFATC2 | Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 | 1.9776 | 5.90E-05 |
| LOC203274 | Hypothetical protein LOC203274 | 1.94801 | 5.09E-05 |
| TLR1 | Toll-like receptor 1 | 1.93858 | 2.14E-07 |
| PLA2G4A | Phospholipase A2, group IVA (cytosolic, calcium-dependent) | 1.87895 | 0.0002 |
| SPATA5L1 | Spermatogenesis associated 5-like 1 | 1.87522 | 0.00103 |
| EMP1 | Epithelial membrane protein 1 | 1.8738 | 0.00013 |
| FPR2 | Formyl peptide receptor 2 | 1.85476 | 0.0005 |
| ADAMTS5 | ADAM metallopeptidase with thrombospondin type 1 motif, 5 | 1.83321 | 0.0003 |
| NR3C1 | Promoter 1B of the GR | 1.83029 | 0.00019 |
| WSB1 | WD repeat and SOCS box-containing 1 | 1.82444 | 0.00018 |
| TMEM49 | Transmembrane protein 49 | 1.8221 | 0.00052 |
| ADAM9 | ADAM metallopeptidase domain 9 | 1.82176 | 2.72E-05 |
| FPR2 | Formyl peptide receptor 2 | 1.77041 | 8.98E-05 |
| ATP6V1A | ATPase, H+ transporting, lysosomal 70 kDa, V1 subunit A | 1.77021 | 3.21E-05 |
| SGMS2 | Sphingomyelin synthase 2 | 1.76728 | 0.00062 |
| FCGR1A///FCGR1C | Fc fragment of IgG, high affinity Ia, receptor (CD64)///Fc fragment of IgG, high affinity | 1.76282 | 0.00095 |
| WLS | Wntless homolog (Drosophila) | 1.76165 | 0.00023 |
| TFEC | Transcription factor EC | 1.75494 | 0.00014 |
| LOC100506828 | Hypothetical LOC100506828 | 1.75228 | 7.44E-05 |
| MGST1 | Microsomal glutathione S-transferase 1 | 1.74784 | 7.56E-05 |
| P2RY13 | Purinergic receptor P2Y, G-protein coupled, 13 | 1.73405 | 0.00013 |
| HHEX | Hematopoietically expressed homeobox | 1.7329 | 4.10E-06 |
| FRAT2 | Frequently rearranged in advanced T-cell lymphomas 2 | 1.72608 | 0.00019 |
| HP | Haptoglobin | 1.72582 | 0.00072 |
| FCGR1B | Fc fragment of IgG, high affinity Ib, receptor (CD64) | 1.72442 | 0.00071 |
| FBN2 | Fibrillin 2 | 1.721 | 0.00025 |
| ST6GALNAC3 | ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2 | 1.70328 | 0.00043 |
| GPATCH2 | G patch domain containing 2 | 1.70199 | 0.00011 |
| DSC2 | Desmocollin 2 | 1.69681 | 0.00025 |
| MGST1 | Microsomal glutathione S-transferase 1 | 1.69458 | 7.19E-06 |
Expression of inflammatory cytokines were substantially reduced in the improved sleep group, including interleukin 1 beta (IL-1β), interleukin 6 (IL-6), interleukin 8 (IL-8), and interleukin 13 (IL-13), with fold changes ranging from −3.19 to −2.10. We also report significant reductions in chemokine genes including chemokine (C–C motif) ligand 4 (CCL4), which was the most substantially reduced gene, with a fold change of −3.39, as well as chemokine (C–C motif) ligand 3 (CCL3), and chemokine (C–C motif) ligand 5 (CCL5), with fold-changes of −3.00 and −2.97, respectively, in the improved sleep group.
Genes with increases in expression in the improved sleep group were predominately inflammatory regulatory genes including toll-like receptors 4 and 8 (TLR4 and TLR8), with a 2.66- and 2.62-fold change, respectively, as well as toll-like receptors 7 and 1 (TLR7 and TLR1), with fold changes of 2.6 and 1.9, respectively. Stress-related genes of cytochrome P450, family 1, subfamily B, polypeptide 1 (CYP1B1), FKBP506 binding protein (FKBP506), and FK506 binding protein 15, 133 kDa (FKBP15) exhibited fold changes >2.0. The pyruvate dehydrogenase kinase, isozyme 4 genes had the greatest increase in expression, with a fold change of 3.12.
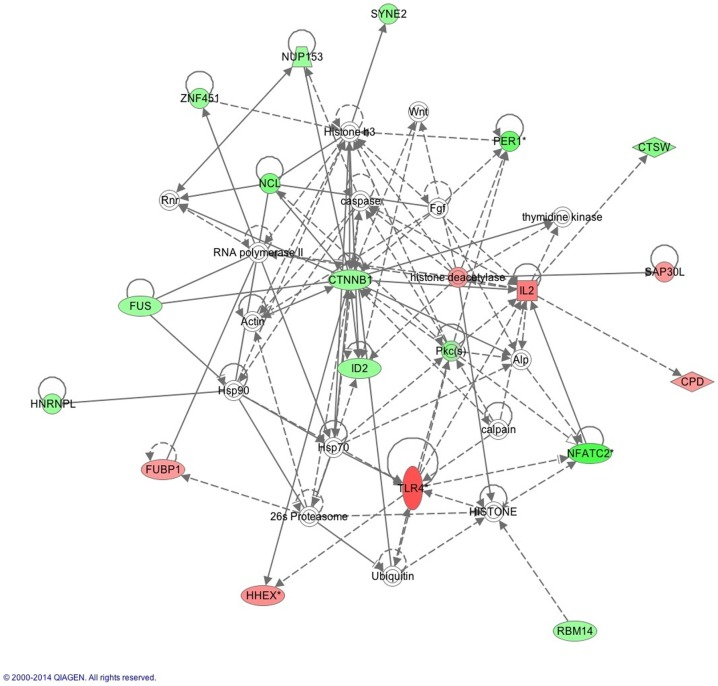
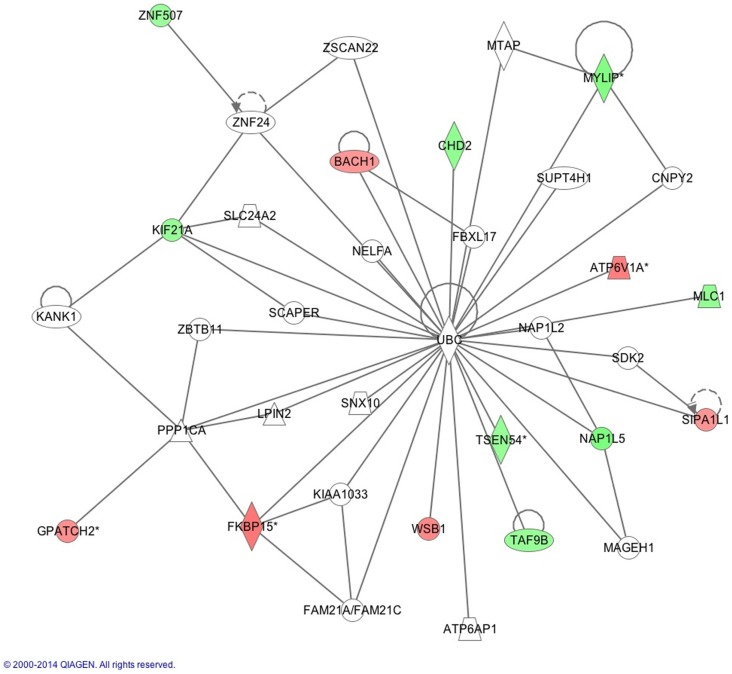
The IPA report shows conical pathways were altered and related to improved sleep. The pathways include the inflammatory response, infectious disease, inflammatory disease, and organismal injury and abnormalities. There were six significant gene-networks identified, the scores of which are reported in Table 6. Networks 1 and 2 are provided in Figures 1 and 2, and the remaining Figures S1–S4 are provided in the Supplementary Material. With the exception of the third IPA network, the other five contained genes above a fold change of 2 in the networks reported. Network 1 includes genes related to cellular development and tissue disorders, and has a score of 29. In this network, there are three main hubs: Catenin (cadherin-associated protein), beta 1, 88 kDa (CTNNβ1), toll-like receptors (TLR), and interleukin 2 (IL-2), as well as minor hubs of ubiquitin (UBC), polyketide synthase (PKS), and nuclear factor of activated T-cells 2 (NFATC2). Network 2 has a score of 23, and includes genes related to cancer, and tissue and developmental disorders. This network has a primary hub of UBC, which is related to an increase in expression of FKBP15 and ATPase, H+ transporting, lysosomal 70 kDa, and V1 subunit A (ATP6V1A), and reductions in the expression of TSEN54 tRNA splicing endonuclease and megalencephalic leukoencephalopathy with subcortical cysts 1.
Table 6.
IPA network scores.
| Networks | Score |
|---|---|
| 1 | 29 |
| 2 | 14 |
| 3 | 14 |
| 4 | 14 |
| 5 | 13 |
| 6 | 14 |
This table lists the scores of the six networks (Figures 1 and 2; Figures S1–S4 in Supplementary Material) as reported by ingenuity pathway analysis (IPA). Following the compilation of networks, based on the interconnectivity of each gene, IPA finalizes the report by ranking each network based on a calculation of p-scores [p-score = −log10(p-value)]. This score accounts for the number of genes versus the size of the network to estimate how valid this network is to the original list of genes. This allows for the networks to be prioritized as seen in Table 6, where a score above 20 is seen as more significant.
Figure 1.
This network shows genes related to cellular development and connective tissue. Generated molecular network based on differential expression in the improved sleep group from baseline to follow-up using ingenuity knowledge database. Coloring is based on the expression values of the genes (fold changes shown): down-regulation in green and up-regulation in red. Genes with no coloring are added from ingenuity knowledge database. Direct and indirect relationships are shown by solid and dashed lines, respectively. The arrow indicates specific directionality of interactions. Genes with an asterisk indicate that multiple identifiers (probesets) map to the gene in the molecular network. Reprinted with permission from QIAGEN’s Ingenuity® Pathway Analysis (IPA®) (http://www.ingenuity.com/).
Figure 2.
This network shows genes related to developmental disorders, as reported by IPA. Reprinted with permission from QIAGEN’s Ingenuity® Pathway Analysis (IPA®) (http://www.ingenuity.com/).
Discussion
The primary aim of the current study is to examine the relationship between changes in self-reported sleep quality and changes in expression of inflammatory genes and symptoms of depression and PTSD following 3 months of sleep treatment in military personnel with insomnia and comorbid psychiatric disorders. At post-treatment, participants with improved sleep have significantly reduced depression symptoms and changes in gene expression, which most notably reflect reductions in inflammatory cytokine genes. These novel findings provide unique insights into our previous findings, which indicated improved sleep quality is associated with reductions in concentrations of CRP and improvements in health-related quality of life in military personnel (23). These results suggest that sleep-focused treatments effectively reduce inflammatory gene activity, and may attenuate the risk for inflammatory-related morbidity in this population.
Specifically, we report that gene activity is reduced in 113 genes following improved sleep, including significant reductions in the expression of the following inflammatory cytokine genes: CCL4, IL-1β, tumor necrosis factor (TNF), IL-6, and IL-13 (Table 4). Elevated concentrations of IL-6 have been associated with symptoms of insomnia, PTSD, and depression (22, 35). The reduction in IL-6 secretion has significant ramifications as IL-6 has a pertinent role in the inflammatory response, including initiation of the acute-phase response and increasing metabolic risks as well as vascular plaque formation (36). Peripherally, IL-6 activates the production of inflammatory cytokines by initiating intracellular signaling with TLR (37). IL-6 is linked to substantial morbidity risks through arterial plaque formation, which can lead to cardiovascular disease and risks, high BMI, chronic pain, and an elevated risk for diabetes (38–42). Therefore, our findings suggest that improved sleep quality plays an important role in decreasing inflammation through gene-expression networks, which may also have a health-promoting role over time.
IL-6 also crosses the blood–brain barrier and contributes to central inflammation, and higher activity can contribute to mood alterations including symptoms of depression and anxiety (36). Here, we report depression symptoms decrease following improved sleep, and there is a trend for decreases in PTSD symptoms. Prior studies link inflammation to the onset of depressive symptoms, suggesting a complex bi-directional relationship that contributes to mental well-being (43, 44). Higher concentrations of IL-6 as well as increased gene-activity are established bio-markers for depression (45), yet, less is known about the inflammatory mechanisms that result from the interaction of insomnia and depression, and the resolution of symptoms. Previously, we reported that depression and insomnia in military personnel are associated with higher concentrations of CRP (23), and that improved sleep reduces inflammation (24). Here, we extend these findings by linking gene expression to sleep improvement and reduction in depression symptoms. In addition, we report a significant reduction in the expression of the CCL4 gene, which was reduced by −3.39 fold in the improved sleep group. CCL4 is an initiator of the inflammatory processes, suggesting inflammatory pathways are reduced in activity following sleep restoration and reduction in depression symptoms. Greater CCL4 activity is linked to major depressive disorder in both adolescents and adults (46, 47); however, it is not clear if the activity of this gene or other inflammatory initiating genes change following sleep improvement and mood change. Therefore, our findings of reductions in CCL4 following sleep improvement are novel and provide some initial insights into complex relationships among genes that regulate inflammation, sleep, and mood.
The current study also found a significant increase in the expression of 104 genes, with most being related to the regulation of inflammatory activity (Table 5). Of these up-regulated genes, the highest fold change is in pyruvate dehydrogenase kinase, isozyme 4 (PDK4), with a 3.12-fold change in the improved sleep group. This gene is of interest as it is located in the matrix of the mitochondria. PDK4 reduces the conversion of pyruvate, which is produced from the oxidation of glucose and amino acids, to acetyl-CoA. It also contributes to the regulation of glucose metabolism and has ramifications for overall health and morbidity. PDK4 has not been previously linked to sleep disturbance or mood. Here, we report that this gene increases expression in the context of reduced expression of genes that influence inflammation and contribute significantly to metabolic risk. Exercise alters the activity of PDK4, which relates to reductions in IL-6 activity, suggesting that PDK4 is involved in metabolic processes that are important to maintaining health (48). PDK4 is also implicated in the maintenance of body weight during periods of fasting, as it is required to maximize glucose metabolism; however, when animals are exposed to high-caloric diets that are unrestricted, PDK4 increases in expression (49). Therefore, PDK4 has complex relations to comorbid conditions of type 2 diabetes, cardiovascular disease, and other conditions that are common in military personnel and impact their morbidity. For these reasons, additional studies are needed to determine how this gene relates to sleep and mood, as well as morbidity risk in future studies.
In this study, we report two main gene-networks that are implicated in sleep improvement. The first involves the key regulator CTNNβ1, a gene that relates to the activity of both IL-2 and TLR4, as well as ubiquitin genes (Figure 1). The CTNNβ1 is most often implicated in vulnerability to and recovery from cancer (50–52); however, the role of this gene in sleep quality has not yet been determined. This gene encodes for the β-catenin protein, which regulates cell development and adhesion between cells, in turn creating and preserving epithelial cell layers, which is related to inflammatory gene activity. Activation of the Wnt signaling pathway, which is a pertinent system for cell-to-cell communications and which becomes activated during inflammatory responses, is essential in maintaining immune function system balance between inflammatory and anti-inflammatory activities. The result is increased concentrations of the protein that is coded by CTNNβ1 gene, which is linked to both increases in symptoms of depression and anxiety in clinical samples (53). Of importance, we also report an increase in IL-2 gene expression, which is a primary anti-inflammatory cytokine and is important for regulating inflammatory cytokine activity including IL-1β, IL-6, and TNF. Therefore, an increase in the expression of the IL-2 gene in conjunction with other inflammatory regulators suggests that regulatory gene networks are altered following sleep restoration, and likely contribute to our previous finding of reductions in CRP following improved sleep (23). Thus, these gene-expression findings together suggest that there is an alteration in the activity of the acute-phase response, which is responsible for the initiation of CRP production. Although we did not find that CRP significantly reduced in activity, our finding of reductions in the activity of these precursor genes that are more easily reduced in activity suggests that changes in CRP gene-activity may require more time to observe. Although our findings provide some key insights into the role of inflammation following sleep change, much less is known about the long-term impact of sleep disturbance on morbidity risks in military personnel and veterans.
We also report that TLRs, and specifically TLR4 gene expression is increased in the improved sleep group, and is also a key gene-network regulator of inflammatory activities. TLR4 recognizes pathogen-associated molecular patterns that are expressed by infectious agents, and mediate the production of cytokines necessary for the development of effective immunity, thereby regulating inflammatory cytokine activities following a stimulus. TLR4 has been linked to repeated hypoxia events in a pre-clinical model, suggesting that acute sleep deprivation may increase TLR activity (54). Our findings of an increase in TLR4 following improved sleep in conjunction with reduced activities of inflammatory cytokines suggests that the activities of TLR4 are impacted, contributing to a restoration of inflammatory immune cell function. Since UBC is tightly related to TLR4 in our gene-network, it may be that the protein-interaction properties of UBC that results in lessened gene activity may contribute to this immune profile in gene expression. It may also be that different molecular regulation mechanisms, including DNA methylation or other gene-silencing processes, are occurring. Nonetheless, the presence of TLR4 in this sample suggests it aided in the reduction of inflammation, as well as interacting with other pathways within the improved sleep group.
Our findings also implicated the protein UBC pathway in gene expression following improved sleep (Figure 2). Protein UBC is required for the removal of oxidized or misfolded proteins that are a consequence of neuronal activity that occurs during waking hours; it also alters the location of the protein in the cell or prevents protein interactions. In pre-clinical models, substantial sleep deprivation results in an accumulation of these bi-products within neurons, and subsequent neuronal damage (55). Accumulation of over-oxidized proteins is linked to cognitive impairments in insomnia patients (56). Therefore, there is an important role for ubiquitin in sleep regulation, which has been recently described in pre-clinical models including Drosophila, linking a knockdown of the E3 ubiquitin ligase Cul3 to sleep disruption (57). Reductions in the activity of E3 ubiquitin ligase Cul3 in neurons resulted in the insomniac phenotype (55). In mice with a spontaneous deletion in the ubiquitin carboxyl-terminal hydrolase L1 gene, there was a lower number of orexin A-immunoreactive neurons in the lateral hypothalamus and greater disruption in circadian rhythm compared to wild-type mice (58). Thus, our findings, within the context of previous studies, suggest that UBC genes are key mediators of sleep-related gene-activity changes.
Our findings have clinical relevance, as previous studies report that insomnia tends to precede depression and PTSD and that treatment of sleep disturbances decreases disease severity (59). Second, because of the stigma associated with mental illness, military personnel are more likely to seek treatment for sleep disturbances than for PTSD (14). Treatment for depression and PTSD symptoms results in substantial financial cost that may be mitigated by providing sleep interventions. In support of this, a comprehensive study undertaken by the RAND corporation estimates that treatment for either major depression or PTSD costs between $5,000 and $15,000 for each military personnel treated over 1 year (60). Lastly, our finding that even short-term improvement in sleep has a significant impact on inflammatory gene-activity, as well as reductions in depression symptoms, suggests that early assessment and treatment for insomnia following deployment is warranted, especially in military personnel with mood declines.
While our study has strengths, including a validated sleep measure, evaluation of comorbid symptoms, and a longitudinal design, there are some notable limitations. These include a relatively small sample of military personnel recruited from the same treatment facility. In addition, our sample primarily consisted of male participants and therefore cannot be generalized to women. Although we used validated questionnaires to determine PTSD and depression symptoms, these instruments did not provide a definitive clinical diagnosis. Lastly, other variations such as prescribed medications used by patients prior to the study and dose of treatment, including the number of treatment sessions attended and other interventional methods could have further impact on gene expression; however, the aim of this study is to focus on symptom and gene-expression changes, not to validate a standard of care treatment. In the future, a larger sample size and better estimate of treatment adherence and fidelity among participants are needed to confirm these findings.
Our novel findings suggest improved sleep quality has a biological correlate as well as a correlation with psychological symptoms. Less is known regarding the long-term impact of sleep disturbance on morbidity risks in military personnel and veterans; however, our findings suggest that improving sleep quality reduces inflammatory risks and psychiatric symptom severity. Therefore, our findings provide fundamental insights related to the connections between sleep improvement and gene expression, including inflammatory gene-activity, which may be used to reduce morbidity risks in military personnel following deployment. Improving assessment and treatment of sleep disturbance in this at-risk group may provide a novel health promotion opportunity.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/article/10.3389/fpsyt.2015.00059/abstract
This network shows genes related to cell-mediated immune responses, as reported by IPA. Reprinted with permission from QIAGEN’s Ingenuity® Pathway Analysis (IPA®) (http://www.ingenuity.com/).
This network shows genes related to connective tissue disorders, as reported by IPA. Reprinted with permission from QIAGEN’s Ingenuity® Pathway Analysis (IPA®) (http://www.ingenuity.com/).
This network shows genes related to cellular function and maintenance and cellular development, as reported by IPA. Reprinted with permission from QIAGEN’s Ingenuity® Pathway Analysis (IPA®) (http://www.ingenuity.com/).
This network shows genes related to organismal injury and abnormalities, as reported by IPA. Reprinted with permission from QIAGEN’s Ingenuity® Pathway Analysis (IPA®) (http://www.ingenuity.com/).
Abbreviations
BMI, body mass index; CBT, cognitive behavioral therapy; CCL4, chemokine (C-C motif) ligand 4; CRP, C-reactive protein; CTNNβ1, catenin (cadherin-associated protein), beta 1, 88 kDa; FDR, false discovery rate; GC, GeneChip; IL-1β, interleukin 1 beta; IL-6, interleukin 6; IL-8, interleukin 8; IL-13, interleukin 13; IPA, ingenuity pathway analysis; NFATC2, nuclear factor of activated T-cells; OSA, obstructive sleep apnea; PCL-M, posttraumatic stress disorder checklist-military version; PDK4, pyruvate dehydrogenase kinase, isozyme 4; PKS, polyketide synthase; PSQI, the Pittsburg sleep quality index; PTSD, posttraumatic stress disorder; QIDS, quick inventory of depressive symptomatology; REML, restricted maximum likelihood; RIN, RNA integrity numbers; RMA, robust multi-array average; TLR1, toll-like receptor 1; TLR4, toll-like receptor 4; TLR7, toll-like receptor 7; TLR8, toll-like receptor 8; TNF, tumor necrosis factor; UBC, ubiquitin.
References
- 1.Seelig AD, Jacobson IG, Smith B, Hooper TI, Boyko EJ, Gackstetter GD, et al. Sleep patterns before, during, and after deployment to Iraq and Afghanistan. Sleep (2010) 33(12):1615–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McLay RN, Klam WP, Volkert SL. Insomnia is the most commonly reported symptom and predicts other symptoms of post-traumatic stress disorder in U.S. service members returning from military deployments. Mil Med (2010) 175(10):759–62. 10.7205/MILMED-D-10-00193 [DOI] [PubMed] [Google Scholar]
- 3.National Center for Post-Traumatic Stress Disorder (US) aWRAMC. Iraq War Clinician Guide. 2nd ed National Center for PTSD and the Department of Veterans Affairs. White River Junction, VT: National Center for Post-Traumatic Stress Disorder; (2004). [Google Scholar]
- 4.Plumb TR, Peachey JT, Zelman DC. Sleep disturbance is common among service members and veterans of Operations Enduring Freedom and Iraqi Freedom. Psychol Serv (2014) 11(2):209–19. 10.1037/a0034958 [DOI] [PubMed] [Google Scholar]
- 5.Mysliwiec V, Gill J, Lee H, Baxter T, Pierce R, Barr TL, et al. Sleep disorders in US military personnel: a high rate of comorbid insomnia and obstructive sleep apnea. Chest (2013) 144(2):549–57. 10.1378/chest.13-0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin SA, Aikens JE, Chervin RD. Toward cost-effectiveness analysis in the diagnosis and treatment of insomnia. Sleep Med Rev (2004) 8(1):63–72. 10.1016/j.smrv.2003.08.001 [DOI] [PubMed] [Google Scholar]
- 7.Stoller MK. Economic effects of insomnia. Clin Ther (1994) 16(5):873–97. [PubMed] [Google Scholar]
- 8.Mysliwiec V, McGraw L, Pierce R, Smith P, Trapp B, Roth BJ. Sleep disorders and associated medical comorbidities in active duty military personnel. Sleep (2013) 36(2):167–74. 10.5665/sleep.2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gehrman P, Seelig AD, Jacobson IG, Boyko EJ, Hooper TI, Gackstetter GD, et al. Predeployment sleep duration and insomnia symptoms as risk factors for new-onset mental health disorders following military deployment. Sleep (2013) 36(7):1009–18. 10.5665/sleep.2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koffel E, Polusny MA, Arbisi PA, Erbes CR. Pre-deployment daytime and nighttime sleep complaints as predictors of post-deployment PTSD and depression in National Guard troops. J Anxiety Disord (2013) 27(5):512–9. 10.1016/j.janxdis.2013.07.003 [DOI] [PubMed] [Google Scholar]
- 11.Pigeon WR, Campbell CE, Possemato K, Ouimette P. Longitudinal relationships of insomnia, nightmares, and PTSD severity in recent combat veterans. J Psychosom Res (2013) 75(6):546–50. 10.1016/j.jpsychores.2013.09.004 [DOI] [PubMed] [Google Scholar]
- 12.Cho HJ, Lavretsky H, Olmstead R, Levin MJ, Oxman MN, Irwin MR. Sleep disturbance and depression recurrence in community-dwelling older adults: a prospective study. Am J Psychiatry (2008) 165:1543–50. 10.1176/appi.ajp.2008.07121882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoge CW, Castro CA, Messer SC, McGurk D, Cotting DI, Koffman RL. Combat duty in Iraq and Afghanistan, mental health problems and barriers to care. US Army Med Dep J (2008):7–17. [PubMed] [Google Scholar]
- 14.Fear NT, Seddon R, Jones N, Greenberg N, Wessely S. Does anonymity increase the reporting of mental health symptoms? BMC Public Health (2012) 12:797. 10.1186/1471-2458-12-797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luxton DD, Greenburg D, Ryan J, Niven A, Wheeler G, Mysliwiec V. Prevalence and impact of short sleep duration in redeployed OIF soldiers. Sleep (2011) 34(9):1189–95. 10.5665/SLEEP.1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Church D, Hawk C, Brooks AJ, Toukolehto O, Wren M, Dinter I, et al. Psychological trauma symptom improvement in veterans using emotional freedom techniques: a randomized controlled trial. J Nerv Ment Dis (2013) 201(2):153–60. 10.1097/NMD.0b013e31827f6351 [DOI] [PubMed] [Google Scholar]
- 17.Lopresti AL, Drummond PD. Obesity and psychiatric disorders: commonalities in dysregulated biological pathways and their implications for treatment. Prog Neuropsychopharmacol Biol Psychiatry (2013) 45:92–9. 10.1016/j.pnpbp.2013.05.005 [DOI] [PubMed] [Google Scholar]
- 18.Prather AA, Marsland AL, Hall M, Neumann SA, Muldoon MF, Manuck SB. Normative variation in self-reported sleep quality and sleep debt is associated with stimulated pro-inflammatory cytokine production. Biol Psychol (2009) 82(1):12–7. 10.1016/j.biopsycho.2009.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chennaoui M, Sauvet F, Drogou C, Van Beers P, Langrume C, Guillard M, et al. Effect of one night of sleep loss on changes in tumor necrosis factor alpha (TNF-alpha) levels in healthy men. Cytokine (2011) 56(2):318–24. 10.1016/j.cyto.2011.06.002 [DOI] [PubMed] [Google Scholar]
- 20.Axelsson J, Rehman JU, Akerstedt T, Ekman R, Miller GE, Hoglund CO, et al. Effects of sustained sleep restriction on mitogen-stimulated cytokines, chemokines and T helper 1/T helper 2 balance in humans. PLoS One (2013) 8(12):e82291. 10.1371/journal.pone.0082291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vgontzas AN, Zoumakis M, Papanicolaou DA, Bixler EO, Prolo P, Lin HM, et al. Chronic insomnia is associated with a shift of interleukin-6 and tumor necrosis factor secretion from nighttime to daytime. Metabolism (2002) 51(7):887–92. 10.1053/meta.2002.33357 [DOI] [PubMed] [Google Scholar]
- 22.Gill JM, Saligan L, Woods S, Page G. PTSD is associated with an excess of inflammatory immune activities. Perspect Psychiatr Care (2009) 45(4):262–77. 10.1111/j.1744-6163.2009.00229.x [DOI] [PubMed] [Google Scholar]
- 23.Heinzelmann M, Lee H, Rak H, Livingston W, Barr T, Baxter T, et al. Sleep restoration is associated with reduced plasma C-reactive protein and depression symptoms in military personnel with sleep disturbance after deployment. Sleep Med (2014) 15(12):1565–70. 10.1016/j.sleep.2014.08.004 [DOI] [PubMed] [Google Scholar]
- 24.Gill J, Lee H, Barr T, Baxter T, Heinzelmann M, Rak H, et al. Lower health related quality of life in U.S. military personnel is associated with service-related disorders and inflammation. Psychiatry Res (2014) 216(1):116–22. 10.1016/j.psychres.2014.01.046 [DOI] [PubMed] [Google Scholar]
- 25.Heinzelmann M, Gill J. Epigenetic mechanisms shape the biological response to trauma and risk for PTSD: a critical review. Nurs Res Pract (2013) 2013:10. 10.1155/2013/417010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rollins B, Martin MV, Morgan L, Vawter MP. Analysis of whole genome biomarker expression in blood and brain. Am J Med Genet B Neuropsychiatr Genet (2010) 153B(4):919–36. 10.1002/ajmg.b.31062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohane IS, Valtchinov VI. Quantifying the white blood cell transcriptome as an accessible window to the multiorgan transcriptome. Bioinformatics (2012) 28(4):538–45. 10.1093/bioinformatics/btr713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gouin JP, Connors J, Kiecolt-Glaser JK, Glaser R, Malarkey WB, Atkinson C, et al. Altered expression of circadian rhythm genes among individuals with a history of depression. J Affect Disord (2010) 126(1–2):161–6. 10.1016/j.jad.2010.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uddin M, Galea S, Chang SC, Aiello AE, Wildman DE, de los Santos R, et al. Gene expression and methylation signatures of MAN2C1 are associated with PTSD. Dis Markers (2011) 30(2–3):111–21. 10.3233/DMA-2011-0750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buysse DJ, Reynolds CF, III, Monk TH, Berman SR, Kupfer DJ. The Pittsburgh sleep quality index: a new instrument for psychiatric practice and research. Psychiatry Res (1989) 28(2):193–213. 10.1016/0165-1781(89)90047-4 [DOI] [PubMed] [Google Scholar]
- 31.Trivedi MH, Rush AJ, Ibrahim HM, Carmody TJ, Biggs MM, Suppes T, et al. The Inventory of Depressive Symptomatology, Clinician Rating (IDS-C) and Self-Report (IDS-SR), and the Quick Inventory of Depressive Symptomatology, Clinician Rating (QIDS-C) and Self-Report (QIDS-SR) in public sector patients with mood disorders: a psychometric evaluation. Psychol Med (2004) 34(1):73–82. 10.1017/S0033291703001107 [DOI] [PubMed] [Google Scholar]
- 32.Weathers FW, Keane TM, Davidson JR. Clinician-administered PTSD scale: a review of the first ten years of research. Depress Anxiety (2001) 13(3):132–56. 10.1002/da.1029 [DOI] [PubMed] [Google Scholar]
- 33.Wilkins KC, Lang AJ, Norman SB. Synthesis of the psychometric properties of the PTSD checklist (PCL) military, civilian, and specific versions. Depress Anxiety (2011) 28(7):596–606. 10.1002/da.20837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol (2006) 7:3. 10.1186/1471-2199-7-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song Y, Zhou D, Guan Z, Wang X. Disturbance of serum interleukin-2 and interleukin-8 levels in posttraumatic and non-posttraumatic stress disorder earthquake survivors in Northern China. Neuroimmunomodulation (2007) 14(5):248–54. 10.1159/000112050 [DOI] [PubMed] [Google Scholar]
- 36.Öz-Arslan D, Rüscher W, Myrtek D, Ziemer M, Jin Y, Damaj BB, et al. IL-6 and IL-8 release is mediated via multiple signaling pathways after stimulating dendritic cells with lysophospholipids. J Leukoc Biol (2006) 80(2):287–97. 10.1189/jlb.1205751 [DOI] [PubMed] [Google Scholar]
- 37.Gu Q, Shi Q, Yang H. The role of TLR and chemokine in wear particle-induced aseptic loosening. J Biomed Biotechnol (2012) 2012:596870. 10.1155/2012/596870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kablak-Ziembicka A, Przewlocki T, Stepien E, Pieniazek P, Rzeznik D, Sliwiak D, et al. Relationship between carotid intima-media thickness, cytokines, atherosclerosis extent and a two-year cardiovascular risk in patients with arteriosclerosis. Kardiol Pol (2011) 69(10):1024–31. [PubMed] [Google Scholar]
- 39.Sahin-Efe A, Katsikeris F, Mantzoros CS. Advances in adipokines. Metabolism (2012) 61(12):1659–65 10.1016/j.metabol.2012.09.001 [DOI] [PubMed] [Google Scholar]
- 40.Boscarino JA, Forsberg CW, Goldberg J. A twin study of the association between PTSD symptoms and rheumatoid arthritis. Psychosom Med (2010) 72(5):481–6. 10.1097/PSY.0b013e3181d9a80c [DOI] [PubMed] [Google Scholar]
- 41.Xue Y, Taub PR, Iqbal N, Fard A, Wentworth B, Redwine L, et al. Cardiac biomarkers, mortality, and post-traumatic stress disorder in military veterans. Am J Cardiol (2012) 109(8):1215–8. 10.1016/j.amjcard.2011.11.063 [DOI] [PubMed] [Google Scholar]
- 42.Boyko EJ, Jacobson IG, Smith B, Ryan MA, Hooper TI, Amoroso PJ, et al. Risk of diabetes in U.S. military service members in relation to combat deployment and mental health. Diabetes Care (2010) 33(8):1771–7. 10.2337/dc10-0296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Capuron L, Ravaud A, Gualde N, Bosmans E, Dantzer R, Maes M, et al. Association between immune activation and early depressive symptoms in cancer patients treated with interleukin-2-based therapy. Psychoneuroendocrinology (2001) 26(8):797–808. 10.1016/S0306-4530(01)00030-0 [DOI] [PubMed] [Google Scholar]
- 44.Wium-Andersen MK, Orsted DD, Nielsen SF, Nordestgaard BG. Elevated C-reactive protein levels, psychological distress, and depression in 73, 131 individuals. JAMA Psychiatry (2013) 70(2):176–84. 10.1001/2013.jamapsychiatry.102 [DOI] [PubMed] [Google Scholar]
- 45.Anderson G, Kubera M, Duda W, Lason W, Berk M, Maes M. Increased IL-6 trans-signaling in depression: focus on the tryptophan catabolite pathway, melatonin and neuroprogression. Pharmacol Rep (2013) 65(6):1647–54. 10.1016/S1734-1140(13)71526-3 [DOI] [PubMed] [Google Scholar]
- 46.Lehto SM, Niskanen L, Herzig K-H, Tolmunen T, Huotari A, Viinamäki H, et al. Serum chemokine levels in major depressive disorder. Psychoneuroendocrinology (2010) 35(2):226–32 10.1016/j.psyneuen.2009.06.007 [DOI] [PubMed] [Google Scholar]
- 47.Lu S, Peng H, Wang L, Vasish S, Zhang Y, Gao W, et al. Elevated specific peripheral cytokines found in major depressive disorder patients with childhood trauma exposure: a cytokine antibody array analysis. Compr Psychiatry (2013) 54(7):953–61. 10.1016/j.comppsych.2013.03.026 [DOI] [PubMed] [Google Scholar]
- 48.Wan Z, Ritchie I, Beaudoin MS, Castellani L, Chan CB, Wright DC. IL-6 indirectly modulates the induction of glyceroneogenic enzymes in adipose tissue during exercise. PLoS One (2012) 7(7):e41719. 10.1371/journal.pone.0041719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frier BC, Jacobs RL, Wright DC. Interactions between the consumption of a high-fat diet and fasting in the regulation of fatty acid oxidation enzyme gene expression: an evaluation of potential mechanisms. Am J Physiol Regul Integr Comp Physiol (2011) 300(2):R212–21. 10.1152/ajpregu.00367.2010 [DOI] [PubMed] [Google Scholar]
- 50.Gilles C, Polette M, Mestdagt M, Nawrocki-Raby B, Ruggeri P, Birembaut P, et al. Transactivation of vimentin by beta-catenin in human breast cancer cells. Cancer Res (2003) 63(10):2658–64. 10.1371/journal.pone.0081584 [DOI] [PubMed] [Google Scholar]
- 51.Chen ZS, Ling DJ, Zhang YD, Feng JX, Zhang XY, Shi TS. Octamer-binding protein 4 affects the cell biology and phenotypic transition of lung cancer cells involving beta-catenin/E-cadherin complex degradation. Mol Med Rep (2015) 11(3):1851–8. 10.3892/mmr.2014.2992 [DOI] [PubMed] [Google Scholar]
- 52.Li K, Hu C, Mei C, Ren Z, Vera JC, Zhuang Z, et al. Sequential combination of decitabine and idarubicin synergistically enhances anti-leukemia effect followed by demethylating Wnt pathway inhibitor promoters and downregulating Wnt pathway nuclear target. J Transl Med (2014) 12:167. 10.1186/1479-5876-12-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gould TD, Dow ER, O’Donnell KC, Chen G, Manji HK. Targeting signal transduction pathways in the treatment of mood disorders: recent insights into the relevance of the Wnt pathway. CNS Neurol Disord Drug Targets (2007) 6(3):193–204. 10.2174/187152707780619308 [DOI] [PubMed] [Google Scholar]
- 54.Smith SM, Friedle SA, Watters JJ. Chronic intermittent hypoxia exerts CNS region-specific effects on rat microglial inflammatory and TLR4 gene expression. PLoS One (2013) 8(12):e81584. 10.1371/journal.pone.0081584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stavropoulos N, Young MW. Insomniac and Cullin-3 regulate sleep and wakefulness in Drosophila. Neuron (2011) 72(6):964–76. 10.1016/j.neuron.2011.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stenfors CU, Magnusson Hanson L, Oxenstierna G, Theorell T, Nilsson LG. Psychosocial working conditions and cognitive complaints among Swedish employees. PLoS One (2013) 8(4):e60637. 10.1371/journal.pone.0060637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pfeiffenberger C, Allada R. Cul3 and the BTB adaptor insomniac are key regulators of sleep homeostasis and a dopamine arousal pathway in Drosophila. PLoS Genet (2012) 8(10):e1003003. 10.1371/journal.pgen.1003003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pfeffer M, Plenzig S, Gispert S, Wada K, Korf HW, Von Gall C. Disturbed sleep/wake rhythms and neuronal cell loss in lateral hypothalamus and retina of mice with a spontaneous deletion in the ubiquitin carboxyl-terminal hydrolase L1 gene. Neurobiol Aging (2012) 33(2):393–403. 10.1016/j.neurobiolaging.2010.02.019 [DOI] [PubMed] [Google Scholar]
- 59.Wright KM, Britt TW, Bliese PD, Adler AB, Picchioni D, Moore D. Insomnia as predictor versus outcome of PTSD and depression among Iraq combat veterans. J Clin Psychol (2011) 67(12):1240–58. 10.1002/jclp.20845 [DOI] [PubMed] [Google Scholar]
- 60.Eibner C, et al. The cost of post-deployment mental health and cognitive conditions. In: Tanielian T, Jaycox LH, editors. Invisible Wounds of War: Psychological and Cognitive Injuries, their Consequences and Services to Assist Recovery. Vol. 1–2 (2008). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This network shows genes related to cell-mediated immune responses, as reported by IPA. Reprinted with permission from QIAGEN’s Ingenuity® Pathway Analysis (IPA®) (http://www.ingenuity.com/).
This network shows genes related to connective tissue disorders, as reported by IPA. Reprinted with permission from QIAGEN’s Ingenuity® Pathway Analysis (IPA®) (http://www.ingenuity.com/).
This network shows genes related to cellular function and maintenance and cellular development, as reported by IPA. Reprinted with permission from QIAGEN’s Ingenuity® Pathway Analysis (IPA®) (http://www.ingenuity.com/).
This network shows genes related to organismal injury and abnormalities, as reported by IPA. Reprinted with permission from QIAGEN’s Ingenuity® Pathway Analysis (IPA®) (http://www.ingenuity.com/).