Abstract
A typical gene contains two levels of information: a sequence that encodes a particular protein and a host of other signals that are necessary for the correct expression of the transcript. While much attention has been focused on the effects of sequence variation on the amino acid sequence, variations that disrupt gene processing signals can dramatically impact gene function. A variation that disrupts an exonic splicing enhancer (ESE), for example, could cause exon skipping which would result in the exclusion of an entire exon from the mRNA transcript. RESCUE-ESE, a computational approach used in conjunction with experimental validation, previously identified 238 candidate ESE hexamers in human genes. The RESCUE-ESE method has recently been implemented in three additional species: mouse, zebrafish and pufferfish. Here we describe an online ESE analysis tool (http://genes.mit.edu/burgelab/rescue-ese/) that annotates RESCUE-ESE hexamers in vertebrate exons and can be used to predict splicing phenotypes by identifying sequence changes that disrupt or alter predicted ESEs.
INTRODUCTION
The splicing machinery requires the presence of several cis-acting sequence elements to accurately recognize and remove introns from pre-mRNA. While the splice sites themselves are located in the introns, additional sequences that enhance splicing from an exonic location are also known. These exonic splicing enhancers (ESEs) are short oligonucleotide sequences that occur frequently in both constitutively and alternatively spliced exons (1–5). ESEs are often recognized by proteins of the SR family, which function by recruiting components of the core splicing machinery to nearby splice sites (6) or by counteracting the effects of nearby silencing elements (7).
A variety of selection schemes have been used to determine which sequences are capable of functioning as ESEs (2–5). These SELEX methods start with a complex pool of random sequence and invoke an iterative selection–amplification protocol which progressively enriches the fraction of molecules in the total pool that possess ESE activity. Functional SELEX experiments, performed in vitro, have described the binding specificity of four SR proteins, and these motifs have been incorporated into an online ESE annotation tool called ESEFinder (8).
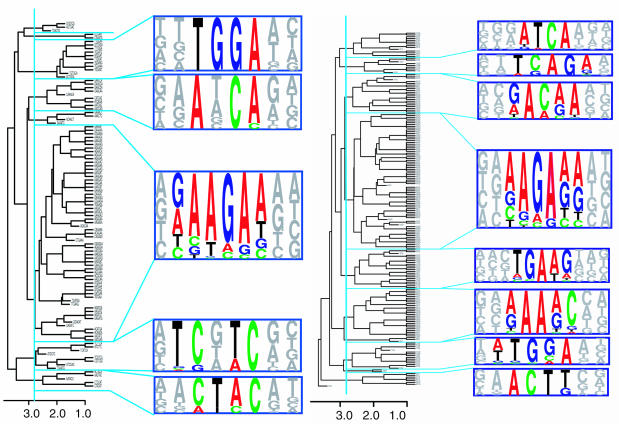
Previously, we reported a computational method, RESCUE-ESE, which identifies ESEs in human genomic sequences by searching for hexanucleotides that satisfy the following two criteria: (i) they are significantly enriched in human exons relative to introns, and (ii) they are significantly more frequent in exons with weak (non-consensus) splice sites than in exons with strong (consensus) splice sites (1). This method identified a set of 238 hexamers (of the 4096 possible hexamers) that were clustered into 10 groups: two groups of 5′ss ESEs, five groups of 3′ss ESEs and three additional groups that appear to be common to both the 3′ss and 5′ss (Figure 1A, B). In vivo tests of splicing enhancer activity, performed on 10 test sequences that were chosen to represent each of the 10 non-redundant ESE motifs (Figure 1), confirmed activity in all 10 cases, and point mutations that were chosen to disrupt the predicted ESE hexamer within the test sequence reduced exon inclusion by more than 2-fold in 9 of the 10 cases (1).
Figure 1.
ESE hexamers for the 3′ss and the 5′ss have been clustered into 10 distinct motifs. The RESCUE-ESE protocol identified (A) 103 hexamers from the 5′ss and (B) 198 hexamers from the 3′ss which, when clustered on the basis of sequence similarity (penalizing 1 point for a mismatch or shift), could be aligned to form five ESE motifs for the 5′ss and eight motifs for the 3′ss. The dissimilarity between hexamers (or average dissimilarity between clusters of hexamers) is shown on the bottom scale, where the blue line represents the threshold dissimilarity of 2.7 that was used to define the clusters. The weight matrices that define the ESE motifs are represented as pictograms where the size of each letter is proportional to the nucleotide frequency for each of the 7–10 positions in the ESE motif.
The importance of considering the possibility of RNA-processing phenotypes in the analysis of mutations has been emphasized by the growing list of exonic variations that cause or modify disease without altering the amino acid sequence of the protein [see the list of 23 disease genes reviewed in (9)]. While these particular variations were implicated in exon skipping, pre-mRNA processing phenotypes are assayed infrequently, and many other synonymous mutations detected in genetic screens and presumed to be neutral may also turn out to have splicing phenotypes.
In order to facilitate the analysis of point mutations we have created an online ESE annotation tool (http://genes.mit.edu/burgelab/rescue-ese/). The predictive power of the set of RESCUE-ESE hexamers used on this server was previously demonstrated on a set of published mutations that cause exon skipping in the human HPRT gene. About one-half of the base changes found in the set of splicing mutants completely eliminated RESCUE-predicted ESEs from the wild-type HPRT sequence (1). It has also been inferred, from a large-scale analysis of the single nucleotide polymorphism database, that natural selection has eliminated approximately one-fifth of all mutations that disrupt RESCUE-predicted ESEs, further supporting the idea that predicted ESEs are frequently functional in human genes (W. G. Fairbrother, D. Holste, C. B. Burge and P. A. Sharp, submitted for publication).
Repeating the RESCUE-ESE protocol on different species revealed substantial overlap between hexamers predicted to be ESEs in vertebrates (G. W. Yeo, S. Hoon, B. Venkatesh and C. B. Burge, submitted for publication). Approximately three-quarters of the human RESCUE-ESE set (172 hexamers) were also predicted as ESEs in mouse. About one-third of human ESEs (73 hexamers) are also predicted to have ESE activity in human, mouse and fish. This smaller core set of hexamers tended to be purine rich and to resemble the classical GAR motif. Additional analysis revealed that most of the hexamers that were predicted to be ESEs in humans had similar, non-uniform, distribution across exons in datasets derived from three additional vertebrate species (mouse, zebrafish and pufferfish). The density of ESEs along vertebrate exons increases near the 3′ss and 5′ss (W. G. Fairbrother, D. Holste, C. B. Burge and P. A. Sharp, submitted for publication; G. W. Yeo, S. Hoon, B. Venkatesh and C. B. Burge, submitted for publication). Taken together, this work argues that the predicted ESEs are functional sequences that have been conserved throughout the vertebrate lineage and will be useful in the analysis of mutations in several species.
DESCRIPTION
The entry page to the RESCUE-ESE server contains a brief description of the RESCUE-ESE method and the details of how particular ESEs were experimentally validated (Figure 2). The web server allows a sequence to be checked for presence of candidate ESE hexamers. A text file containing the non-redundant list of 238 human ESEs can be downloaded from the entry page. An input sequence can be pasted directly into the window or uploaded in multi-FASTA format and annotated with the ESEs from the selected species (human, mouse, pufferfish and zebrafish are the currently available options).
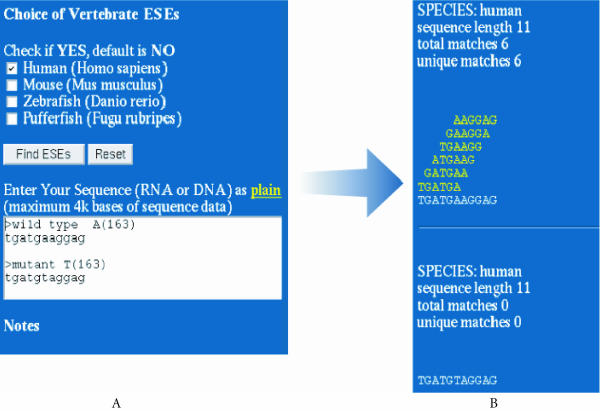
Figure 2.
The RESCUE-ESE web server. An exon skipping mutation from the human HPRT gene was analyzed by RESCUE-ESE. (A) Multi FASTA format files containing the wild type and mutant sequences of an A to T mutation at HPRT position 163 were pasted into the input window on the entry page of http://genes.mit.edu/burgelab/rescue-ese. (B) Species buttons allow the user to select from multiple sets of RESCUE-ESE hexamers. The output displays the input sequence with the ESEs drawn above in yellow. Additional links include ESE references, the original RESCUE-ESE paper and download options for ESE hexamers.
The server is case insensitive and accepts either DNA (T) or RNA (U) sequences as input (up to 4 kb in length). Although ambiguous nucleotides (e.g. R, Y or N) are accepted, the program will not predict ESE hexamers that overlap these positions. The effect of sequence variation on ESEs can be analyzed by entering both the wild type and variant sequence into the input window as two sequences in FASTA format. This application is illustrated (in Figure 2A) with an A to T mutation in the HPRT gene that disrupts six RESCUE-ESE hexamers (Figure 2B) and is known to cause exon skipping (Fairbrother et al., submitted for publication). ESEs are annotated on an output page as yellow hexamers drawn above the white input sequence against a dark-blue background (Figure 2B).
DISCUSSION
Since ESE disruption or alteration carries with it the potential to cause disease, effective ESE annotation will be useful in analyzing sequence variation. Mutations that are analyzed in this manner can fall into one of four categories: (i) ESE disruption, where one or more predicted ESE hexamers present in the wild-type sequence are disrupted by the mutation; (ii) ESE alteration, where ESEs are present in both the wild-type and mutant version of the sequence; (iii) ESE neutral, where ESEs are present in neither the wild-type nor mutant sequence and (iv) ESE creation, where predicted ESE hexamers are present in the mutant but not wild-type sequence. While the relatively small number of mutations that have been discovered within an exon (at least 5 nt from the splice site) and shown to be associated with exon skipping limits our ability to measure the sensitivity of ESE annotation, about one-half of the known exon skipping mutants in the human HPRT gene were classified as ESE disruption mutations by our method. This value is approximately three times higher than would be expected by chance [P < 0.01 (1)]. Conversely, ESE disruption events were found to be under-represented within the public database of human single nucleotide polymorphisms (dbSNP), a set of variations that is presumed to be predominantly selectively neutral (W. G. Fairbrother, D. Holste, C. B. Burge and P. A. Sharp, submitted for publication). The annotation of SNPs revealed that both ESE disruption events and, to a lesser degree, ESE alteration events were selected against in the human population. This result suggests that both ESE disruption and ESE alteration mutations could alter splicing phenotypes. By this criterion, RESCUE-ESE hexamers would correctly predict a splicing defect in 53% of the exonic mutations in HPRT that are associated with exon skipping.
While the analysis of known splicing mutants suggests a sensitivity of at least 50%, a quantitative measure of specificity is more elusive. Without studying the splicing phenotypes of a large number of exonic mutations, it is difficult to reliably determine the true incidence of ESE disruption within the set of mutations predicted to disrupt ESEs. Site-directed mutagenesis studies designed to disrupt predicted ESEs in wild-type exonic sequences reduced splicing in vivo for 9 of the 10 cases tested in a splicing reporter construct (1). While 90% of these ESE disruption mutations reduced splicing in a reporter system, our recent analysis of natural variations (SNPs) in human exons suggested that only ∼20% are eliminated by natural selection. This selection was much stronger (∼50%) for mutations located within 25 nt of splice sites (Fairbrother et al., submitted for publication). These differences suggest, (i) a small amount of exon skipping may be tolerated in certain genes/exons and (ii) that the specificity of splicing phenotype predictions based on RESCUE-ESE annotations would be higher for mutations that fall near splice sites. In addition to the location of an ESE within an exon, there are other external variables that could modulate the function of an ESE. Context effects, such as secondary structure or adjacent negative elements, could limit the accessibility and, hence, activity of a predicted ESE in a particular location.
An additional benefit of a computational approach such as RESCUE-ESE is that it can be readily implemented in other genomes as they become available. We present a tool capable of annotating ESEs from several species. This allows the user to identify elements that can function as ESEs across several species. This feature may facilitate ESE analysis in other organisms, ensure the correct processing of fish or human genes in transgenic mice or serve as an indicator of predicted ESE quality. Although representative hexamers were found to have ESE activity in an in vivo splicing assay, some fraction of the candidate ESE hexamers are likely to be false positives of the RESCUE method. While it is possible that certain recognition sequences could be restricted to particular lineages, hexamers that are predicted to be ESEs in several species are very likely to have a lower false positive rate than species-specific ESEs. In summary, we expect that this cross-species approach will provide useful insight into the functional relevance of gene processing signals. We plan to add other species options (such as Drosophila melanogaster and Carnorhabditis elegans) to the server as we complete our ESE analysis of additional species.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank the researchers who have used the website and reviewers of this manuscript for providing the feedback that has greatly improved the service. This work was supported by a Pharmaceutical Research and Manufacturers of America postdoctoral fellowship (W.G.F.) and by a grant from the NIH (C.B.B.), NIH grant R37-GM34277 (P.A.S.), NSF grant 0218506 (P.A.S.), NCI grant P30-CA14051 to the Cancer Center Support (P.A.S.) and a Functional Genomics Innovation award from the Burrough Wellcome fund (C.B.B. and P.A.S.).
REFERENCES
- 1.Fairbrother W.G., Yeh,R.F., Sharp,P.A. and Burge,C.B. (2002) Predictive identification of exonic splicing enhancers in human genes. Science, 297, 1007–1013. [DOI] [PubMed] [Google Scholar]
- 2.Liu H.X., Zhang,M. and Krainer,A.R. (1998) Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev., 12, 1998–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tian H. and Kole,R. (1995) Selection of novel exon recognition elements from a pool of random sequences. Mol. Cell Biol., 15, 6291–6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaal T.D. and Maniatis,T. (1999) Selection and characterization of pre-mRNA splicing enhancers: identification of novel SR protein-specific enhancer sequences. Mol. Cell Biol., 19, 1705–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coulter L.R., Landree,M.A. and Cooper,T.A. (1997) Identification of a new class of exonic splicing enhancers by in vivo selection. Mol. Cell Biol., 17, 2143–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graveley B.R. (2000) Sorting out the complexity of SR protein functions. RNA, 6, 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kan J.L. and Green,M.R. (1999) Pre-mRNA splicing of IgM exons M1 and M2 is directed by a juxtaposed splicing enhancer and inhibitor. Genes Dev., 13, 462–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cartegni L., Wang,J., Zhu,Z., Zhang,M.Q. and Krainer,A.R. (2003) ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res., 31, 3568–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cartegni L., Chew,S.L. and Krainer,A.R. (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nature Rev. Genet., 3, 285–298. [DOI] [PubMed] [Google Scholar]
- 10.Valentine C.R. (1998) The association of nonsense codons with exon skipping. Mutat. Res., 411, 87–117. [DOI] [PubMed] [Google Scholar]