Abstract
Small disulfide-bonded proteins (SDPs) are rich sources for therapeutic drugs. Designing drugs from these proteins requires three-dimensional structural information, which is only available for a subset of these proteins. SDPMOD addresses this deficit in structural information by providing a freely available automated comparative modeling service to the research community. For expert users, SDPMOD offers a manual mode that permits the selection of a desired template as well as a semi-automated mode that allows users to select the template from a suggested list. Besides the selection of templates, expert users can edit the target–template alignment, thus allowing further customization of the modeling process. Furthermore, the web service provides model stereochemical quality evaluation using PROCHECK. SDPMOD is freely accessible to academic users via the web interface at http://proline.bic.nus.edu.sg/sdpmod.
INTRODUCTION
Small disulfide-bonded proteins (SDPs) are a special class of proteins that are relatively small in size (length ≤100 residues) and have disulfide bonds within their three-dimensional (3D) structures (1). SDPs include many secretory proteins which serve predatory, defensive or regulatory roles (such as toxins, inhibitors and hormones), and they are rich source for therapeutic drugs (2) and pesticides (3). The 3D structures of SDPs are essential for understanding the functions of SDPs and for drug design. However, 3D structure determination through experimental methods such as X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy are still both time-consuming and expensive. This results in a gap between the number of known 3D structures and the number of primary sequences that could be narrowed using large-scale automated protein structure prediction.
Among current structure prediction methods, comparative modeling is the most reliable method for generating 3D models. Comparative modeling of protein structures often requires expert knowledge and proficiency in specialized methods. In the mid-1990s, Peitsch and coworkers developed the first automated modeling server SWISS-MODEL (4), which is currently the most widely used server of this genre. Recently, several other automated comparative modeling servers have also been developed, such as CPHmodels (5), 3D-JIGSAW (6), ModWeb (7) and ESyPred3D (8).
Although so many automated comparative modeling servers are available, most of them do not work well on small SDPs for two reasons. Most of the automated servers are primarily designed for globular protein domains, making it difficult to discriminate small-sized SDPs from background noise. Taking as an example the sequence of α-conotoxin PnIA (9) (PDB id: 1PEN; 16 residues; 2 disulfide bridges in its structure), we note that both SWISS-MODEL and ModWeb report that they do not cover the modeling of sequences <25 or ≤30 amino acid residues in length, respectively, while the other three servers state that no suitable templates can be identified for this sequence.
The second reason is that SDPs have distinct characteristics from medium-sized and large globular proteins. They usually do not have a compact hydrophobic core, which is a major factor in stabilizing protein structure. Their side chains are more likely to be exposed to solvent and their conformations are more flexible. The 3D structures of small proteins are usually dominated by disulfide bridges, metal or ligand (according to SCOP classification) (10) and tend to bind or interact with large molecules. In small disulfide-rich proteins, the effects of disulfide bridges and constrained residues such as prolines are more significant than sequence similarity. As such, the comparative modeling rules for such proteins are highly specific and different from those adopted for large globular proteins. These distinct features require specific methods and datasets to be developed for the comparative modeling of SDPs.
To address these problems, we have first developed special strategies and rules for large-scale automated comparative modeling of the entire family of conotoxins (L. Kong and S. Ranganathan, unpublished data). Subsequently these rules were extended to other SDPs. Here, we present SDPMOD, a comprehensive comparative modeling server that is designed specifically for SDPs with specialized rules and datasets.
MATERIALS AND METHODS
Non-redundant SDP structure dataset
Before the modeling can proceed, a non-redundant dataset for SDPs needs to be created to serve as the template repository. Structures containing protein chains of length <100 amino acids with at least two cysteines were retrieved from the Protein Data Bank (PDB) (11) and loaded into MySQL, a relational database management system for flexible query and manipulation. The redundancy in SDP structures was removed at two levels. First, for NMR structures which have multiple monomer models, the representative monomers were selected using NMRCLUST (12). Second, when multiple structures exist for the same sequence, the representative structure was chosen according to its structural qualities. The structural qualities are ranked by the following criteria (adopted from PDB): (i) X-ray structures over NMR structures, (ii) higher-quality factor (1/resolution − R-value) for X-ray structures and higher restraint per residue for NMR, (iii) better geometry, (iv) fewer missing atoms and non-standard residues and (v) later deposition date. Based on the above strategy, a non-redundant structure database for SDPs was generated. Currently it contains >1300 non-redundant protein chains and their coordinates. The database will be automatically updated once a month.
Modeling procedure
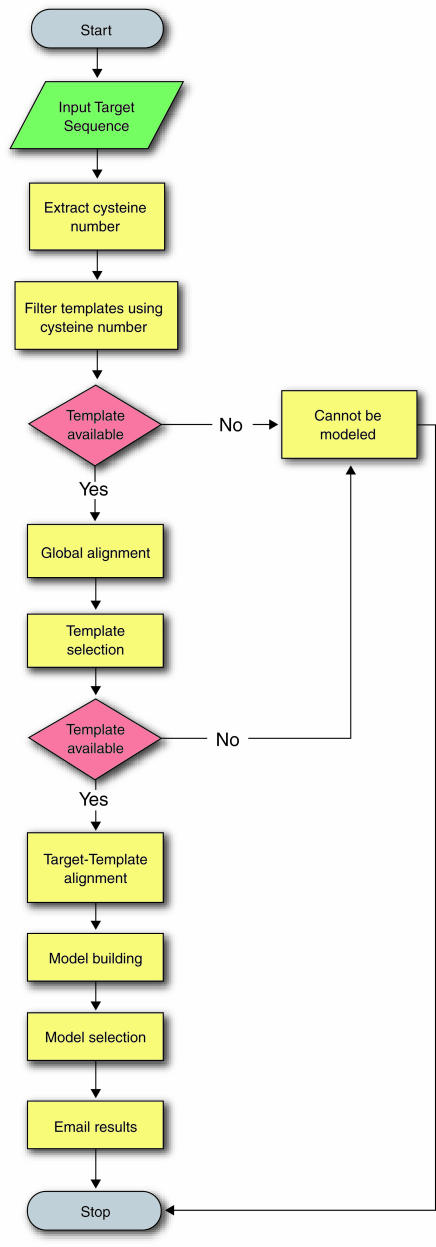
The SDPMOD server performs comparative modeling in four steps: (i) template selection, (ii) target–template alignment, (iii) model building and (iv) model evaluation (13). Figure 1 shows the detailed modeling procedure for automated modeling. The non-redundant dataset is first filtered using the number of cysteine residues, and the resulting template sequences are globally aligned to the target sequence using a modified scoring matrix derived from the non-redundant SDP dataset. The best templates are then selected based on the alignment scores. Target–template alignment and model building are achieved by MODELLER (14) (http://salilab.org/modeller/modeller.html), using a customized matrix to ensure that all the cysteine residues are well aligned. The final models are chosen according to the MODELLER objective function score, which reflects low energy and least stereochemical violations. Finally, the overall structural quality of the generated models is evaluated against stereochemical parameters derived from high-quality experimental structures by PROCHECK (15) (http://www.biochem.ucl.ac.uk/~roman/procheck/procheck.html).
Figure 1.
The SDPMOD methodology for automatic comparative modeling of small disulfide-bonded proteins.
Benchmarking
A large-scale benchmarking excercise was completed using the fully automated mode of the SDPMOD server. A control set of 664 sequences (a subset of our non-redundant SDP dataset) with known structures was used to evaluate the reliability of the server. The Cα root mean square deviation (RMSD) values between models and their actual experimental structures were calculated. The benchmarking results show SDPMOD can predict 3D models with a reasonable accuracy. For example, in the 40–70% sequence identity range, 64% of models have Cα RMSD values <1.5 Å. The detailed analysis of the accuracy of our modeling protocol is available from http://proline.bic.nus.edu.sg/sdpmod/accuracy.html.
WEB SERVICE
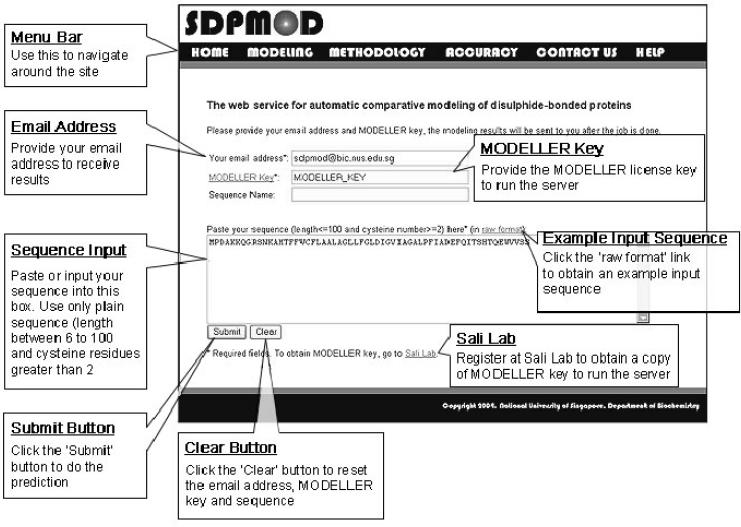
SDPMOD is freely accessible to academic or non-profit users via a web interface (shown in Figure 2) at http://proline.bic.nus.edu.sg/sdpmod. SDPMOD is primarily designed as a fully automated procedure for ease of use. However, due to the complexity of comparative modeling, human intervention and expert knowledge may be required for optimal modeling of some proteins at two critical stages, namely template selection and target–template alignment (6). To allow for human intervention, the current version of the SDPMOD server provides three modes of modeling (fully automated, semi-automated and manual) to meet the different needs of the expert users.
Figure 2.
Example of the SDPMOD input page.
The ‘fully automated’ mode presents an easy-to-use interface. Users can simply submit a target sequence with their email address and their MODELLER license key, obtained from the MODELLER registration page http://salilab.org/modeller/registration.shtml, and the modeling will be carried out automatically according to the procedure described in Figure 1. In the ‘semi-automated’ mode, a ranked list of potential templates will be returned after the target sequence is submitted. Users can then choose the best template and adjust the target–template alignment using expert knowledge. In the ‘manual’ mode, users are allowed to propose a template from our non-redundant SDP structure dataset and modify the target–template alignment where necessary.
After the modeling process is completed, a link with the prediction results will be returned via email. Users can refer to the link to view the prediction result and download the models. The prediction results consist of (i) a summary of the selected template(s), (ii) the predicted model based on each template in PDB format and (iii) a brief report for each modeling attempt that includes the target–template alignment used in model building, a comparison of the model against the template by means of RMSD and a PROCHECK report on the stereochemical quality of the models.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank our colleagues at the Department of Biochemistry, National University of Singapore for their helpful comments and discussions. We are especially grateful to Professor Andrej Sali for permitting us to use MODELLER as a part of the server and Dr Ben Webb for useful suggestions. L.K. and B.L. would also like to thank the National University of Singapore for the award of Agency for Science, Technology and Research, Singapore (ASTAR) scholarships that made this work possible.
REFERENCES
- 1.Harrison P.M. and Sternberg,M.J. (1996) The disulphide beta-cross: from cystine geometry and clustering to classification of small disulphide-rich protein folds. J. Mol. Biol., 264, 603–623. [DOI] [PubMed] [Google Scholar]
- 2.Shen G.S., Layer,R.T. and McCabe,R.T. (2000) Conopeptides: from deadly venoms to novel therapeutics. Drug Discov. Today, 5, 98–106. [DOI] [PubMed] [Google Scholar]
- 3.Richardson M. (1977) The proteinase inhibitors of plants and micro-organisms. Phytochemistry, 16, 159–169. [Google Scholar]
- 4.Peitsch M.C. (1996) ProMod and Swiss-Model: internet-based tools for automated comparative protein modelling. Biochem. Soc. Trans., 24, 274–279. [DOI] [PubMed] [Google Scholar]
- 5.Lund O., Frimand,K., Gorodkin,J., Bohr,H., Bohr,J., Hansen,J. and Brunak,S. (1997) Protein distance constraints predicted by neural networks and probability density functions. Protein Eng., 10, 1241–1248. [DOI] [PubMed] [Google Scholar]
- 6.Bates P.A., Kelley,L.A., MacCallum,R.M. and Sternberg,M.J. (2001) Enhancement of protein modeling by human intervention in applying the automatic programs 3D-JIGSAW and 3D-PSSM. Proteins, Suppl 5, 39–46. [DOI] [PubMed] [Google Scholar]
- 7.Pieper U., Eswar,N., Stuart,A.C., Ilyin,V.A. and Sali,A. (2002) MODBASE, a database of annotated comparative protein structure models. Nucleic Acids Res., 30, 255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lambert C., Leonard,N., De Bolle,X. and Depiereux,E. (2002) ESyPred3D: prediction of proteins 3D structures. Bioinformatics, 18, 1250–1256. [DOI] [PubMed] [Google Scholar]
- 9.Hu S.H., Gehrmann,J., Guddat,L.W., Alewood,P.F., Craik,D.J. and Martin,J.L. (1996) The 1.1 Å crystal structure of the neuronal acetylcholine receptor antagonist, alpha-conotoxin PnIA from Conus pennaceus. Structure, 4, 417–423. [DOI] [PubMed] [Google Scholar]
- 10.Murzin A.G., Brenner,S.E., Hubbard,T. and Chothia,C. (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol., 247, 536–540. [DOI] [PubMed] [Google Scholar]
- 11.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelley L.A., Gardner,S.P. and Sutcliffe,M.J. (1996) An automated approach for clustering an ensemble of NMR-derived protein structures into conformationally related subfamilies. Protein Eng., 9, 1063–1065. [DOI] [PubMed] [Google Scholar]
- 13.Marti-Renom M.A., Stuart,A.C., Fiser,A., Sanchez,R., Melo,F. and Sali,A. (2000) Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct., 29, 291–325. [DOI] [PubMed] [Google Scholar]
- 14.Sali A. and Blundell,T.L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol., 234, 779–815. [DOI] [PubMed] [Google Scholar]
- 15.Laskowski R.A., Moss,D.S. and Thornton,J.M. (1993) Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol., 231, 1049–1067. [DOI] [PubMed] [Google Scholar]