Abstract
We previously described a thymus-tropic HIV-1 envelope (R3A Env) from a rapid progressor obtained at the time of transmission. An HIV-1 molecular recombinant with the R3A Env supported extensive replication and pathogenesis in the thymus and did not require Nef. Another Env from the same patient did not display the same thymus-tropic pathogenesis (R3B Env). Here, we show that relative to R3B Env, R3A Env enhances viral entry of T cells, increases fusion-induced cytopathicity, and shows elevated binding efficiency for both CD4 and CXCR4, but not CCR5, in vitro. We created chimeric envelopes to determine the region(s) responsible for each in vitro phenotype and for thymic pathogenesis. Surprisingly, while V1/V2 contributed to enhanced viral entry, CD4 binding efficiency, and cytopathicity in vitro, it made no contribution to thymic pathogenesis. Rather, CXCR4 binding efficiency and V5-gp41-associated activity appear to independently contribute to thymic pathogenesis of the R3A Env. These data highlight the contribution of unique HIV pathogenic factors in the thymic microenvironment and suggest that novel mechanisms may be involved in Env pathogenic activity in vivo.
Keywords: HIV-1, Thymus, Envelope, Pathogenesis, CXCR4, gp41, CD4, Disease progression, Entry
Introduction
Disease progression in HIV-1-infected individuals is influenced by both the virus and the immune system. Progressive disease results in eventual depletion of CD4+ T cells and the onset of AIDS (McCune, 2001). During progression, variants often emerge with Env proteins capable of using CXCR4 as a coreceptor in addition to or instead of CCR5 (Koot et al., 1993; Tersmette et al., 1989). The emergence of CXCR4-tropic Env proteins has been associated with disease progression, loss of T cell homeostasis, and the onset of AIDS (Connor et al., 1993; Koot et al., 1993; Margolick et al., 1995; McCune, 2001; Philpott, 2003; Richman and Bozzette, 1994). In addition to coreceptor usage, affinity for coreceptor (Chakrabarti et al., 2002; Etemad-Moghadam et al., 2000; Gorry et al., 2002; Karlsson et al., 1998, 2004; Si et al., 2004), enhanced fusion in vitro (Chakrabarti et al., 2002; Etemad-Moghadam et al., 2001; Karlsson et al., 1998), enhanced syncytium-induction in vitro (Etemad-Moghadam et al., 2001; Karlsson et al., 1998; Rudensey et al., 1998), and enhanced resistance to neutralizing antibodies (Etemad-Moghadam et al., 1999; Richman et al., 2003; Rudensey et al., 1998; Wei et al., 2003) have all been correlated with viral pathogenesis and disease progression. However, mechanistic details of Env-mediated pathogenesis in vivo and their relationship to disease progression remain to be fully explained.
Many of the insights into mechanisms of viral pathogenesis have been derived from simian studies which allow for experimental manipulation and testing of hypotheses in vivo. In contrast, direct conclusions regarding pathogenesis in humans must largely be drawn by correlation. Model systems to study pathogenesis of HIV-1 include immortalized cell lines, artificially stimulated primary cells, and the more relevant SCID-hu thy/liv and human fetal-thymus organ culture (HF-TOC) models. These thymus models contain primary cells that limit replication of tissue culture-adapted isolates but which support replication and pathogenesis of primary isolates without exogenous stimulation (Meissner et al., 2003; Miller et al., 2001). These models can be used not only to study the effect of HIV-1 on the thymus, but also as general models of lymphoid micro-environments that HIV-1 encounters in vivo.
We previously characterized two env genes (R3A and R3B) isolated at the time of transmission from a patient who progressed rapidly to AIDS (Meissner et al., 2004). While both Env proteins support infection and depletion of stimulated PBMCs, the R3A Env enables elevated replication and pathogenesis in the thymus, even in the absence of Nef. In this report, we demonstrate that the R3A Env displays enhanced virus-cell fusion, fusion-induced cytopathicity, and CXCR4 binding efficiency relative to the R3B Env in a panel of in vitro assays. Furthermore, R3A Env shows enhanced sensitivity to inhibition by soluble CD4 and elevated resistance to Leu3a, a CD4 blocking antibody, suggesting that it has higher affinity for CD4 relative to R3B Env. Using recombinant env genes which allowed for mapping of each phenotype, we dissect the contribution of putative mechanisms to thymic replication and pathogenesis. Surprisingly, elevated CD4 binding efficiency and enhanced viral-cell fusion, both mediated by the V1/V2 region, do not determine thymic replication and pathogenesis. Rather, the data suggest a contribution of CXCR4 binding efficiency and the V5-gp41 region of Env. These data highlight the separation that can exist between in vitro correlates of pathogenesis and factors relevant within a model lymphoid microenvironment.
Results
The R3A Env enables enhanced viral entry of T cells
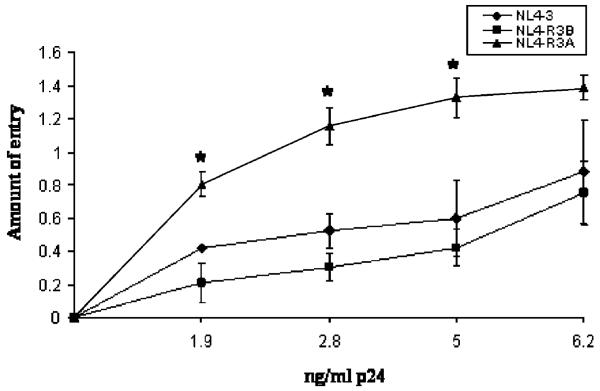
The R3A Env was previously shown to mediate high levels of replication and pathogenesis in the thymus relative to the R3B or NL4-3 envelopes, either in the context of the parental virus or in a recombinant virus lacking Nef (NL4-R3A and NL4-R3B). We previously showed that NL4-luc pseudotyped with R3A Env has increased infectivity for Sup-T1 cells in a single cycle replication assay, which could help explain enhanced thymic replication (Meissner et al., 2004). To extend this finding, we employed the Blam-vpr assay to determine if increased infectivity was correlated with increased viral entry of T cells (Cavrois et al., 2002; Lineberger et al., 2002). When beta-lactamase-expressing virions were used to infect Sup-T1 cells, we found that NL4-R3A was significantly more capable of entering cells than either NL4-R3B or NL4-3 for a given amount of p24 (Fig. 1). Similarly, we found that the R3A Env expressed on A293T cells was more fusogenic towards 1G5 cells in a cell–cell fusion assay (data not shown). Notably, incorporation of R3A and R3B Env into virions and surface expression of each Env on A293T cells were comparable (data not shown).
Fig. 1.
The R3A Env mediates elevated viral fusion with CD4+ T cells. Blam-vpr-containing virions were used to infect Sup-T1 cells by spinoculation. After 2 h of incubation at 37 °C, cells were incubated with flurogenic beta-lactamase substrate for 8 h. The amount of entry was calculated by measuring the ratio of cleaved to uncleaved flurogenic substrate. Shown is a representative of eight independent experiments with error bars derived from triplicate samples and input virus determined by p24 ELISA. (*P <0.05 for R3A vs. either NL4-3 or R3B).
The R3A Env has higher binding efficiency for CD4
Fusion efficiency is determined by interaction of Env with CD4, CCR5 and/or CXCR4, and the nature of the fusion intermediate (Doms, 2000; Eckert and Kim, 2001; Wyatt and Sodroski, 1998). To test which interaction might explain the enhanced entry mediated by the R3A Env, we infected cell lines in the presence of chemical inhibitors to each of these four components. We first tested relative binding efficiency for CD4 by assessing sensitivity of pseudotyped virus to inhibition by soluble CD4 (sCD4) (Beaumont et al., 2004; Kozak et al., 1997; Thali et al., 1991). Because the infectivity of each virus differs (Meissner et al., 2004 and Fig. 1), we normalized infection achieved in the presence of sCD4 to that achieved with no sCD4 for each virus. NL4-3 was found to have the greatest sensitivity to sCD4 (Fig. 2A), consistent with previous findings that tissue culture-adapted viruses typically have increased binding efficiency for CD4 relative to primary envelopes (Kozak et al., 1997; Moore et al., 1992; Platt et al., 2000). Notably, R3A-pseudotyped virus was more sensitive to sCD4 inhibition than R3B-pseudotyped virus, suggesting that R3A has increased binding efficiency for CD4.
Fig. 2.
The R3A Env shows enhanced sensitivity to sCD4 and reduced sensitivity to Leu3a. (A) R3A Env is sensitive to sCD4 relative to R3B Env. TZM-bl cells were infected with virus incubated with sCD4 for 2 h prior to infection. Infection was quantitated by luciferase assay 48 h post-infection and values were normalized to infection in the absence of sCD4. Shown is a representative of 3 experiments with error bars derived from duplicate samples. (B) R3A Env is resistant to Leu-3a relative to R3B Env. Sup-T1 cells were preincubated with the indicated dose of Leu-3a for 1 h prior to infection with pseudotyped NL4-luc. Infection achieved by spinoculation was quantitated by luciferase assay 48 h post-infection and values were normalized to infection in the absence of Leu-3a. Shown is a representative of 3 experiments with error bars derived from quadruplicate samples (*P <0.05 for R3B vs. either NL4-3 or R3A).
To extend these findings, and because other mechanisms may be involved in sCD4 sensitivity, we infected Sup-T1 cells in the presence of Leu3a, an anti-CD4 monoclonal antibody that competitively blocks HIV-1 Env binding to CD4. R3B-pseudotyped virus was found to be significantly more sensitive to inhibition by Leu3a than either NL4-3 or R3A Env (Fig. 2B). Together, these data suggest that R3A Env has higher binding efficiency for CD4 than R3B Env.
The R3A Env has higher binding efficiency for CXCR4 than the R3B Env, but similar CCR5 binding efficiency and sensitivity to T20
When U373-MAGI-CCR5E cells were infected in the presence of the CCR5 antagonist Tak-779 (Baba et al., 1999; Este, 2001), NL4-R3B and NL4-R3A were found to have similar sensitivity to inhibition, suggesting they have similar binding efficiency for CCR5 (Fig. 3A) (Gorry et al., 2002). In contrast, when Sup-T1 cells were infected with pseudotyped virus in the presence of the CXCR4 antagonist AMD-3100, R3A-pseudotyped virions were markedly less inhibited than either R3B- or NL4-3-pseudotyped virions (Fig. 3B). A similar increase in resistance to AMD-3100 was observed for R3A Env when GHOST-CXCR4 cells were used for infection (data not shown). Additionally, infection of CXCR4-shRNA-expressing Sup-T1 cells, which express ~3% of control CXCR4, was much more efficient with R3A Env (Fig. 3C). These data suggest that R3A Env has higher binding efficiency for CXCR4 than R3B Env.
Fig. 3.
The R3A Env shows enhanced binding efficiency for CXCR4, but similar sensitivity to Tak-779 and T20. (A) R3A and R3B Envs show equal sensitivity to a CCR5 antagonist. Infection of U373-MAGI-CCR5E cells was performed in the presence of the CCR5 antagonist TAK-779. Infection was quantitated by counting blue colonies and was normalized to the number of colonies observed with no TAK-779. Shown is a representative of two independent experiments with error bars derived from quadruplicate samples. (B) R3A shows enhanced affinity for CXCR4 relative to R3B. Infection of Sup-T1 cells was performed with NL4-luc-pseudotyped virus in the presence of AMD-3100. Infection was quantitated by luciferase assay after 48 h and was normalized to levels obtained with no AMD-3100. Shown is a representative of seven independent experiments with error bars derived from triplicate samples (*P <0.05 for R3A vs. either NL4-3 or R3B). (C) Sup-T1 cells transduced with vector or X4h shRNA, which reduces CXCR4 surface levels by ~97%, were infected with NL4-luc pseudotyped with the indicated Env. Infection was quantitated by luciferase assay after 48 h and was normalized to levels obtained on vector-transduced cells. Shown is a representative of three independent experiments with error bars derived from triplicate samples (*P <0.05 in comparison to R3A Env or to infection of parental cells). (D) R3A and R3B Envs show equal sensitivity to the fusion inhibitor T20. TZM-bl cells were infected with virus in the presence of the indicated dose of T20. Infection was quantitated and normalized as in A. Shown is a representative of 2 experiments with error bars derived from duplicate samples (*P <0.05 for NL4-R3A vs. either NL4-3 or NL4-R3B).
Because coreceptor affinity has been correlated with resolution of the fusion intermediate and sensitivity to fusion inhibitors (Reeves et al., 2002), we tested the relative sensitivity of each virus to inhibition by the fusion inhibitor T20. As shown in Fig. 3D, NL4-3, NL4-R3B, and NL4-R3A had equal sensitivity to T20. We conclude that R3A Env has enhanced binding efficiency for both CD4 and CXCR4, but not CCR5, relative to R3B Env, and this increase in binding efficiency does not manifest in differential sensitivity to T20. Furthermore, NL4-R3B and NL4-R3A were found to incorporate similar levels of envelope and were equally sensitive to heat denaturation (data not shown), suggesting increased CD4 and CXCR4 affinities are intrinsic properties of the R3A Env.
The R3A Env induces syncytium formation and fusion-induced cytopathicity when expressed in T cell lines
We previously showed that expression of R3A on A293T cells leads to increased depletion of CD4+ thymocytes in a coculture assay (Meissner et al., 2004), which could correlate with thymic replication or pathogenesis. To extend these findings, we assessed whether the R3A Env is cytopathic when expressed in T cells in vitro. Retrovirus vector expressing the Env gene was used to transduce either 1G5 or Sup-T1 cells. Each vector also expresses GFP controlled by the phospho-glucose kinase promoter (Coffield et al., 2003, 2004), allowing for transduction efficiency to be monitored by FACS analysis of GFP expression. At 72 h post-transduction, both 1G5 (Fig. 4A) and Sup-T1 cells (data not shown) showed equivalent transduction efficiencies, with over 90% of cells routinely expressing GFP. Furthermore, the amount of Env expressed in each cell population, as assessed using the anti-gp120 2G12 monoclonal antibody, was comparable across samples in 1G5 cells (Fig. 4A). Similar but lower levels of Env were detected on transduced Sup-T1 cells (data not shown).
Fig. 4.
The R3A Env causes cytopathicity through CXCR4-dependent fusion when expressed in T cells. (A) 1G5 cells were transduced with a retroviral vector expressing HIV Env. Env surface levels were detected 3 days post-transduction using the 2G12 monoclonal antibody or human IgG1 as an isotype control. Shown is a representative stain 3 days post-transduction. The percentage of cells in each quadrant is indicated. (B) R3A Env induces extensive syncytia formation. Syncytia were observed by light microscopy 3 days post-transduction. Shown are representative photos of Sup-T1 cells 3 days post-transduction. (C) The number of live cells surviving 3 days post-transduction was quantitated by trypan blue exclusion. Shown is a representative of 8 independent experiments for Sup-T1 cells. (D and E) R3A-induced cytopathicity is dependent on fusion through CXCR4. Vector or R3A-transduced Sup-T1 cells were incubated with no drug, with T20 (D), or with AMD-3100 (E), and cytopathicity was quantitated as in C. Shown is a representative of 2 independent experiments (*P <0.05 for R3A and R3B vs. vector transduced cells).
Observation of cells 3 days post-transduction showed the R3A Env induced extensive syncytia formation to an extent not observed in vector, NL4-3 Env, or R3B Envexpressing cells (Fig. 4B). Smaller syncytia and clumping were consistently observed in R3B-expressing cells, but never to the extent seen in R3A-expressing cells. To quantitate fusion-induced cytopathicity, the number of living cells surviving 3 days post-transduction was quantitated by trypan blue exclusion. In both 1G5 and Sup-T1 cells, R3A Env was significantly more cytopathic than either NL4-3 or R3B Env (Fig. 4C and data not shown).
R3A-expressing Sup-T1 cells were next cultured in the presence of either T20 or AMD-3100, inhibitors of envelope fusion and CXCR4 interactions. Both drugs significantly ablated R3A Env-induced cytopathicity (Figs. 4D–E), indicating that cytopathicity in this system is dependent on fusion of envelope with target cells and is not simply a byproduct of intracellular envelope toxicity. Collectively, these data indicate that the R3A Env expressed from T cells is more cytopathic per Env molecule than NL4-3 or R3B Env, suggesting that R3A Env may be intrinsically more cytopathic when expressed on infected thymocytes.
Enhanced entry and CD4 binding efficiency of the R3A Env map to the V1/V2 region, while CXCR4 binding efficiency is due to multiple determinants
We previously determined that the R3A and R3B Env proteins show a high degree of sequence homology. The vast majority of sequence variation is in the variable regions, with scattered differences in the gp41 ecto- and intracellular domains (Meissner et al., 2004 and Fig. 5). In an attempt to delineate the regions responsible for the phenotypes observed above, we constructed chimeric envelopes using conserved restriction sites. Eight chimeric envelopes were made by switching the pre-V1, V1/V2, V4, or the V5-gp41 regions between R3A and R3B (Fig. 5). Four of the recombinant env genes were introduced into the NL4-3 backbone (V1/V2 and V5-gp41 recombinants) which, like NL4-R3A and NL4-R3B, resulted in deletion of the nef gene (Meissner et al., 2004). Notably, while the V3 region was switched in tandem with the V1/V2 regions, R3A and R3B have identical V3 loops and thus V3 is not included in the recombinant notation. We tested each of the eight chimeric Env proteins and the four recombinant viruses in the in vitro assays that best distinguish the R3A and R3B Env proteins.
Fig. 5.
Generation of recombinant env genes to map the pathogenic determinants in the R3A Env. Schematic map of the eight recombinant env genes made using conserved restriction sites. Vertical bars indicate specific amino acid differences between R3A and R3B Env proteins. The total number of differences in each region of Env is indicated. The asterisk indicates the additional putative glycosylation site in the R3B Env.
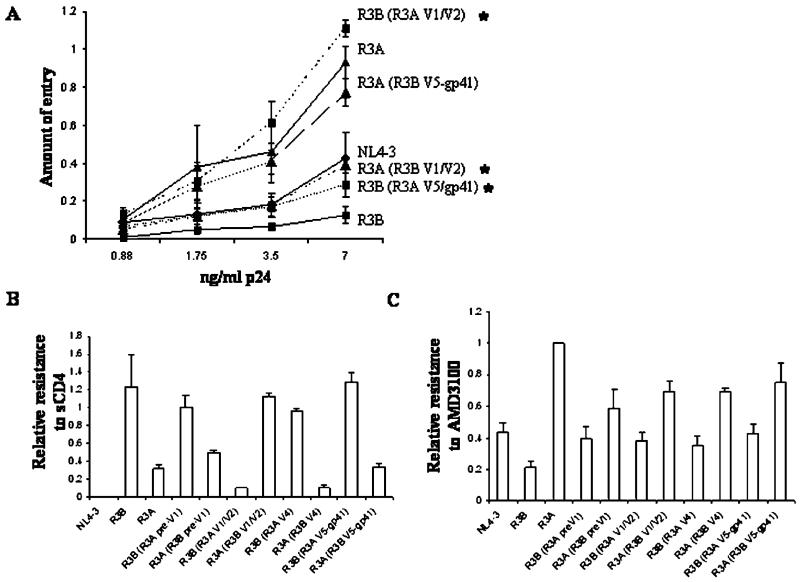
The recombinant viruses were first tested in the Blam-vpr assay to determine the Env region responsible for enhanced viral entry of T cells. As shown in Fig. 6A, switching the V1/V2 region caused an almost complete switch in phenotype, with the R3B (R3A V1/V2) Env showing greater entry than the R3A (R3B V1/V2) Env. In contrast, switching the V5-gp41 region caused little change in entry. Similar results were obtained in an A293T-Env/1G5 cell–cell fusion assay (data not shown). We conclude that the V1/V2 region is the predominant mediator of increased entry for the R3A Env.
Fig. 6.
The V1/V2 region is largely responsible for the enhanced entry and CD4 affinity of R3A Env, while CXCR4 affinity cannot be mapped to one particular region. (A) V1/V2 contributes to enhanced virus fusion of T cells. Blam-vpr-containing virions were used to infect Sup-T1 cells at the dose of p24 indicated. Shown is a representative of 5 independent experiments with error bars derived from quadruplicate samples (*P <0.05 for the indicated recombinant relative to the parental envelope at the highest two doses of p24). (B) Soluble CD4 sensitivity also maps to the V1/V2 region. NL4-luc-pseudotyped virus was pretreated with 4 μg/ml sCD4 prior to infection of GHOST-CXCR4 cells. Infection in the presence of sCD4 was compared to infection without sCD4 treatment. Shown is a representative of 2 experiments with error bars derived from quadruplicate samples. (C) Resistance to AMD-3100 is achieved by multiple determinants. Sup-T1 cells were infected with pseudotyped NL4-luc in the presence of 200 nM AMD-3100. Resistance to inhibition observed with the R3A Env was assigned a value of 1 for each of four experiments, and the relative resistance of each recombinant Env was normalized to this value. Shown are the combined results from four independent experiments with standard error bars indicated.
We next tested the binding efficiency of each chimeric Env for CD4 and CXCR4. As shown in Fig. 6B, the V1/V2 region was the predominant mediator of sensitivity to sCD4, with a complete switch in phenotype between R3B (R3A V1/V2) and R3A (R3B V1/V2). In contrast, the pre-V1, V4, and V5-gp41 regions did not contribute greatly to sCD4 sensitivity. When each of the chimeric Envs was tested for CXCR4 binding efficiency, we were unable to detect the contribution of a distinct region (Fig. 6C). Instead, each R3B recombinant with a single R3A region was still low in resistance and each R3A recombinant with a single R3B region was still high in resistance, suggesting that multiple regions of the R3A Env are responsible for increased CXCR4 binding efficiency. Together, these data suggest that enhanced viral entry mediated by the R3A Env is due to its unique V1/V2 loop which, in turn, determines binding efficiency for the CD4 receptor. Furthermore, based on mapping data, there appears to be no linkage between viral entry and CXCR4 binding efficiency, suggesting that coreceptor engagement is not the limiting step for viral entry.
V1/V2 and V5-gp41 independently contribute to fusion-induced cytopathicity
Each chimeric Env was next tested for fusion-induced cytopathicity in Sup-T1 cells. As above, the R3A Env was more cytopathic to T cells than NL4-3 or R3B Env (Fig. 7). The pre-V1 and V4 regions were not found to contribute to cytopathicity by either syncytia size or cytopathicity quantitation. In contrast, swapping the V1/V2 regions between the Env proteins resulted in the formation of syncytia of intermediate size, while expression of R3B (R3A V5-gp41) caused syncytia that appeared larger than cells expressing R3A (R3B V5-gp41) (data not shown). Quantitation of cytopathicity by live cell counting confirmed an independent contribution of both V1/V2 and V5-gp41 to cytopathicity, with a slightly more impressive contribution of the V5-gp41 region (Fig. 7). These data suggest the contribution of two distinct regions of the R3A Env that additively and independently contribute to T cell cytopathicity. Furthermore, they suggest that CD4 binding efficiency or CXCR4 binding efficiency alone is not solely responsible for cytopathicity, as mapping data for each of these sensitivities do not correlate directly with cytopathicity.
Fig. 7.
V1/V2 and V5-gp41 independently contribute to enhanced fusion-induced cytopathicity of R3A Env. Sup-T1 cells were transduced with Env expression vectors. Cytopathicity was quantitated as in Fig. 2. *Significantly different from the parental envelope, **significantly different from both R3A and R3B Env (P <0.05). Shown are the combined results of 3–6 independent experiments with normalization to vector-transduced cells within each experiment.
The V5-gp41 region, not the V1/V2 region, contributes to enhanced replication and pathogenesis of the R3A Env in the thymus
Based on the in vitro studies, we suspected that either V1/V2 or V5-gp41 would contribute to replication and pathogenesis of the R3A Env in the thymus organ. In contrast, the pre-V1 region, including Vpr, Vpu, Tat, Rev, and part of Env, and the V4 regions did not show a contribution in any in vitro assay and were therefore less likely to play a role. Accordingly, we tested the V1/V2 and V5-gp41 chimeric Env proteins in the context of recombinant NL4-3 viruses for replication and pathogenesis in HF-TOC.
Replication of each recombinant virus was assessed by p24 ELISA of HF-TOC supernatant. Consistent with previous findings, NL4-R3A replicated to higher levels than either NL4-3 or NL4-R3B, a difference most notable at the earliest time point (Fig. 8A). Surprisingly, in spite of the clear contribution of the V1/V2 region to viral entry, CD4 binding efficiency, and cytopathicity in vitro, recombinants with a switch in the V1/V2 region showed an identical replication phenotype as their parental envelope. In contrast, swapping the V5-gp41 region resulted in an intermediate phenotype, with greater replication than NL4-R3B and less replication than NL4-R3A for both NL4-R3A (R3B V5-gp41) and NL4-R3B (R3A V5-gp41).
Fig. 8.
V5-gp41, but not V1/V2, of the R3A Env is uniquely involved in HIV replication and pathogenesis in the thymus model. (A) NL4-3 recombinant viruses expressing the indicated Env were used to infect thymus fragments in the HF-TOC system. Replication was monitored by detection of p24 in the TOC supernatant at the indicated times post-infection. Shown are the combined data from five independent infections. (B) Pathogenesis was assessed by depletion of total CD4+ thymocytes using FACS. Shown are the combined results from five independent experiments. *Indicates significant differences in depletion relative to both NL4-R3B and NL4-R3A at days 11–12 ( P < 0.05).
Depletion of CD4+ thymocytes directly correlated with viral replication. While NL4-R3B was able to cause some depletion of CD4+ thymocytes, NL4-R3A caused almost complete depletion by 11–12 dpi (Fig. 8B). V1/V2 swapping between the Env proteins caused no change in this phenotype, consistent with the lack of change in viral replication. In contrast, switching V5-gp41 resulted in an intermediate phenotype, with NL4-R3B (R3AV5-gp41) and NL4-R3A (R3B V5-gp41) both causing significantly less depletion than NL4-R3A and significantly more depletion than NL4-R3B. We conclude that the V5-gp41 region of R3A is partially, but not completely, responsible for the increased replication and cytopathicity of the R3A and the NL4-R3A viruses, whereas the V1/V2 region makes no contribution. Notably, the Env recombinants with high pathogenic activity in the thymus possessed either the R3A V5-gp41 region or increased CXCR4 binding efficiency, and full pathogenic activity of the R3A Env appears to require both determinants.
Discussion
The ways in which HIV-1 Env contributes to the pathogenesis of AIDS remain to be fully elucidated. In this study, we utilize the human fetal-thymus organ culture to assess the mechanisms of pathogenesis of a primary, transmitted Env associated with rapid disease progression. The R3A and R3B Env proteins, derived from the same patient, both use CXCR4 and CCR5 for infection and can replicate similarly in stimulated PBMCs, yet behave very differently in the context of the thymic microenvironment (Meissner et al., 2004). We show that the R3A Env excels in a number of contexts in vitro, including viral entry, binding efficiency for CD4 and CXCR4, and fusion-induced cytopathicity. Coreceptor affinity (Etemad-Moghadam et al., 2000; Karlsson et al., 1998; Si et al., 2004), syncytium-inducing ability (Etemad-Moghadam et al., 2000; Karlsson et al., 1998), entry (Chakrabarti et al., 2002; Si et al., 2004), and fusogenicity (Chakrabarti et al., 2002; Etemad-Moghadam et al., 2000, 2001; Karlsson et al., 1998) have all been previously correlated with envelope-mediated pathogenesis in vivo and thus were not wholly unexpected. Most significantly, we find that enhanced CD4 binding efficiency, associated with enhanced viral entry and cytopathicity in vitro, makes no contribution to replication and pathogenesis in the thymus organ.
It has been widely observed that tissue-culture-adapted isolates evolve enhanced affinity for CD4 upon passage in vitro (Daar and Ho, 1991; Hoxie et al., 1991; Kozak et al., 1997; Platt et al., 1997), likely because association of virus with CD4 is the rate limiting step for viral infectivity in in vitro systems (Kabat et al., 1994). Consistent with these findings, soluble CD4 sensitivity of each chimeric envelope (Fig. 6B) was directly proportional to viral entry of T cells in vitro (Fig. 6A). Analysis of chimeric envelopes in these assays revealed the clear contribution of the V1/V2 region to both CD4 binding efficiency and viral entry. Furthermore, the V1/V2 region was partially responsible for fusion-induced cytopathicity in the Sup-T1 cell line (Fig. 7). These findings are consistent with the observation that loss of a glycosylation site in V1/V2 of a primary isolate passaged in vivo results in enhanced affinity for CD4 (Pugach et al., 2004). In spite of these V1/V2 contributions in vitro, R3B (R3A V1/V2) and R3A (R3B V1/V2) are indistinguishable from their parental envelope in the thymus (Fig. 8). Thus, the V1/V2 region has no effect on replication and pathogenesis in a representative lymphoid microenvironment. These data raise the intriguing possibility that viral pathogenesis in vivo is not dependent on the efficiency of interaction of envelope with the CD4 receptor, even though CD4 binding is a prerequisite for viral infection. The lack of studies reporting a difference in CD4 binding efficiency of isolates during disease progression or in simian studies of pathogenesis is consistent with this notion (Cayabyab et al., 1999; Chakrabarti et al., 2002; Ivey-Hoyle et al., 1991; Karlsson et al., 1998; Si et al., 2004). Notably, simian studies conclude CD4 binding efficiency is unimportant because pathogenic SHIV Env proteins have the same or lower CD4 affinity than non-pathogenic SHIV Env proteins (Cayabyab et al., 1999; Chakrabarti et al., 2002; Karlsson et al., 1998; Si et al., 2004). Our study is the first to show directly that enhanced CD4 binding efficiency does not contribute to replication and pathogenesis in a human lymphoid organ. That enhanced viral entry in vitro is not relevant for pathogenesis in vivo has support from at least one simian model (Hsu et al., 2003). Rather, it is likely that modulation of CD4 binding efficiency is associated with avoidance of a neutralizing antibody response and immune recognition (Pugach et al., 2004). Minor differences in CD4 binding efficiency among isolates would then merely be a byproduct of this variation (Ivey-Hoyle et al., 1991).
Instead, post-CD4 binding events are likely to modulate viral replication and pathogenesis in vivo. While CCR5 binding efficiency and T20 sensitivity were not found to differ between R3A and R3B in vitro, the R3A Env did show enhanced CXCR4 binding efficiency and fusion-induced cytopathicity in T cells (Fig. 3). While we were unable to map CXCR4 binding efficiency to one particular region (Fig. 6C), the V5-gp41 region clearly contributed to syncytium-induced cytopathicity in vitro (Fig. 7). Our genetic data suggest that increased binding efficiency for CXCR4 and the V5-gp41 region may be independently and additively responsible for R3A Env pathogenesis in the thymus. Thus, while the R3A (R3B V5-gp41) Env maintains high binding efficiency for CXCR4, it loses the R3A V5-gp41 region and yields a partially pathogenic envelope in the thymus (Fig. 8). Similarly, the R3B (R3A V5-gp41) Env maintains low CXCR4 binding efficiency, but gains the R3A V5-gp41 determinant, also yielding a partially pathogenic envelope in the thymus. Only when both CXCR4 binding efficiency and the R3A V5-gp41 region are maintained on one Env (NL4-R3A and NL4-R3A (R3B V1/V2)) is full thymic pathogenesis observed. These results are consistent with previous monkey studies implicating coreceptor binding efficiency and the gp41 region, through as yet unclear mechanisms, to viral pathogenesis (Etemad-Moghadam et al., 2000, 2001; Karlsson et al., 1998). To our knowledge, this is the first support of these conclusions determined experimentally in a human model for HIV-1. Further study will be required to elucidate the potential mechanisms by which coreceptor binding efficiency and the V5-gp41 region modulate viral pathogenesis and to more definitively identify the responsible regions.
Our finding that CXCR4 binding efficiency is correlated with thymic pathogenesis is representative of a number of recent studies that have correlated coreceptor binding efficiency with viral pathogenesis (de Parseval et al., 2004; Etemad-Moghadam et al., 2000; Gorry et al., 2002; Karlsson et al., 1998, 2004) and disease progression (Stalmeijer et al., 2004). Enhanced interaction with CXCR4 could presumably overcome SDF1α-mediated inhibition (Scarlatti et al., 1997) or low CXCR4 expression on certain target cells (Berkowitz et al., 1998; Tokunaga et al., 2001; Zaitseva et al., 1998). Alternatively, this interaction may result in stronger signal transduction which could alter the activation status and/or susceptibility of thymocytes to viral infection or depletion (Arthos et al., 2002; Hernandez-Lopez et al., 2002; Holm et al., 2004; Pedroza-Martins et al., 1998). Additionally, because both R3B and R3A are capable of using CXCR4, the nature of interaction with coreceptor could impact infection, particularly because inclusion of AMD-3100 was able to almost completely block replication and pathogenesis of both NL4-R3A and NL4-R3B in the thymus (data not shown). In support of this hypothesis, while HXB2 and the in vivo-evolved LW isolate both only use CXCR4 as a coreceptor, only the LW Env is permissive for infection in the thymus (Miller et al., 2001; Su et al., 1997), demonstrating that the nature of the Env–CXCR4 interaction may affect viral replication and pathogenesis in vivo.
It is unclear how R3A Env is able to achieve enhanced binding efficiency for CXCR4. No one region is either necessary or sufficient to increase or abrogate CXCR4 binding efficiency (Fig. 6C). Because this is commonly observed in studies of chimeric Env proteins with CXCR4 (Cho et al., 1998; de Vreese et al., 1996; Singh and Collman, 2000; Smyth et al., 1998), coreceptor binding efficiency is likely to be a complex phenotype mediated by specific evolution within diverse regions of each individual Env. Interestingly, while V3 has typically been associated with coreceptor binding efficiency and usage (Etemad-Moghadam et al., 2000; Foda et al., 2001; Hu et al., 2000; Ogert et al., 2001; Si et al., 2004; Su et al., 1997), R3B and R3A have identical V3 loops making a direct contribution of this region to the observed differences improbable. It is possible that the nature of the R3A Env–CXCR4 interaction is conformationally distinct, as has previously been described for other Env proteins (Brelot et al., 1999; Karlsson et al., 2004; Labrosse et al., 2001; Picard et al., 1997), such that Env binding is less sensitive to AMD-3100 inhibition.
The conclusions from our study are limited by the broad regions included in each chimeric Env, particularly the lack of separation between the V5 and gp41 regions. Further separation of pathogenic motifs will help elucidate the exact mechanisms of R3A pathogenesis in the human thymus. In this study, which relies on genetic correlation, we conclude that enhanced CXCR4 binding efficiency and the V5-gp41 region of Env appear to independently contribute to pathogenesis of the R3A Env in the thymus. In contrast, elevated V1/V2-mediated fusion, CD4 binding efficiency, and cytopathicity of R3A do not contribute to thymic pathogenesis. These data suggest that in vitro assessment of CXCR4 binding efficiency and cytopathic potential, as well as replication and pathogenesis in the human thymus, may approximate the pathogenic potential of HIV-1 in vivo. The R3A and R3B Env proteins and their derivatives will be valuable in further understanding the pathogenic activity of HIV-1 Env in the thymus and in disease progression.
Materials and methods
Viral isolates and cell lines
pNL4-3 was used to construct the recombinant viruses NL4-R3A, NL4-R3B, and each chimeric NL4-R3A/R3B construct as previously described using EcoR1 and Xho1 restriction sites (Meissner et al., 2004). A293T and TZM-bl cells (NIAID, NIH) were cultured in DMEM with 10% FBS. GHOST-CXCR4 cells (NIAID, NIH) were cultured in DMEM supplemented with 10% FBS, 500 μg/ml G418, 100 μg/ml hygromycin, and 1 μg/ml puromycin. U373-MAGI-CXCR4CEM and U373-MAGI-CCR5E cells (NIAID, NIH) were cultured in DMEM supplemented with 10% FBS, 0.2 mg/ml G418, 0.1 mg/ml hygromycin B, and 1.0 μg/ml puromycin. Sup-T1 (NIAID, NIH) and 1G5 cells (NIAID, NIH) were cultured in RPMI 1640 with 10% FBS.
Chimeric envelope construction
Chimeric R3A/R3B Envs were created using conserved restriction sites. EcoR1 and DraIII sites were used to create the pre-V1 chimeras which include a switch of the vpr, vpu, rev, tat, and the env gene from amino acids 1–123 including the C1 region and are designated as pre-V1. DraIII and Acc1 were used to create chimeras between amino acids 124 and 319, including regions V1, V2, C2, and part of V3, and are designated as the V1/V2 region. Acc1 and BsaB1 were used to create chimeras from amino acids 320–440 including part of V3, C3, V4, and part of C4, and are designated as the V4 region. BsaB1 and Xho1 were used to create chimeras from amino acids 441–853 including part of C4, V5, C5, and gp41, and are designated as the V5-gp41 region.
Monoclonal antibodies and drugs
The 2G12 monoclonal antibody (NIAID, NIH) was used for cell surface envelope staining. AMD-3100 (NIAID, NIH) was reconstituted in water at a stock concentration of 802 μM. TAK-779 (NIAID, NIH) was reconstituted in DMSO per protocol at a stock concentration of 2 mM. T20 was obtained from Dr. S. Jiang (New York Blood Center, NY) and was reconstituted at a stock concentration of 0.5 mg/ml in 50% ethanol. Recombinant soluble human CD4 was obtained from Progenics Pharmaceuticals. The Leu-3a antibody was obtained from Becton Dickinson. Antiserum to HIV-1SF2 gp120 (NIAID, NIH) and the p24 hybridoma (183-H12-5C) (NIAID, NIH) were used to assess envelope incorporation into virions.
Viral production
VSVg-pseudotyped retrovirus was produced by calcium-phosphate cotransfection of A293T cells with VSVg, Gag/Pol, and MSCV retroviral DNA as previously described (Coffield et al., 2003, 2004; Pear et al., 1993). The MSCV retroviral construct contains GFP under control of the phospho-glucose kinase promoter and envelope under control of the CMV promoter (after transfection) or the MSCV LTR (after transduction). Virus in DMEM (2% FBS) was harvested 2 and 3 days post-transfection, clarified by low speed centrifugation, aliquoted, and stored at −80 °C. NL4-luciferase pseudotyped with HIV-1 Env was produced by calcium-phosphate cotransfection of A293T cells with NL4-luciferase and an MSCV retroviral construct encoding env, and was harvested and stored as above. To produce infectious NL4-3, NL4-R3A, and the other chimeric viruses, proviral DNA was transfected into A293T cells using Effectene (Qiagen) and the supernatant was used to infect PHA-stimulated PBMCs as previously described (Meissner et al., 2004). Virus was harvested from 4 to 9 days post-infection and tittered using U373-MAGI-CXCR4CEM or TZM-bl cells.
shRNA construction and X4 KD cells
We designed our hairpin oligo based on the Shagging protocol developed by Dr. Hannon’s group at Cold Spring Harbor Laboratories (Hannon, 2004): X4h oligo: 5′-AAAAAAGTTATCCGAAGTATACATACTAATCCCCTCAAGCTTCAGGGGATCAGTATATACACTTCAGATAACGGTGTTTCGTCCTTTCCACAA-3′. The oligo was used for PCR to splice the shRNA targeting hCXCR4 onto the hU6 promoter. The PCR product was gel purified and ligated into the PCR2.1 cloning vector (Invitrogen). The cloned product was digested from PCR2.1 using SpeI/XbaI and subsequently cloned into the XbaI site located within the 3′LTR of our MSCV retroviral vector, thus creating a SIN vector. The correct clone was confirmed using the HindIII site incorporated into the loop region of the hairpin oligo. This construct was used to transduce Sup-T1 cells, resulting in ~97% reduction of surface CXCR4 levels without affecting CD4 expression.
Entry assays
For the virus-cell fusion assay, A293T cells were co-transfected using Effectene (Qiagen) with proviral DNA and a Blam-vpr expression construct (pMM310) as previously described (Cavrois et al., 2002; Lineberger et al., 2002). Virus was harvested in phenol red-free DMEM (10% FBS, 10 mM HEPES), clarified by centrifugation, aliquoted, and stored at −80 °C. The amount of virus in each stock was determined by p24 ELISA. 1 × 105 Sup-T1 cells were infected in 96-well plates in a total volume of 150 Al phenolfree DMEM (10% FBS, 10 mM HEPES). Infection was achieved by spinoculation at 2000 × g for 2 h at 22 °C followed by a 2-h incubation at 37 °C (O’Doherty et al., 2000). After centrifugation and supernatant removal, cells were resuspended in 100 μl HBSS and 20 μl 6 × loading dye (GeneBlazer In vivo Detection Kit, 1 μM CCF2-AM, Invitrogen), covered from light, and incubated at 22 °C. After 1 h, the dye solution was aspirated after centrifugation and cells were washed 1 × in PBS. Cells were resuspended in 100 μl PBS and transferred to a clear-bottomed 96-well plate (Costar 3603). After 8–16 h, fluorescence was measured on a fluorometer and relative fusion was determined by taking the ratio of the blue (cleaved) to green (uncleaved) signal after background correction as previously described (Cavrois et al., 2002).
T cell cytopathicity assays
5 × 105 Sup-T1 or 1G5 cells were infected by spinoculation with 500 μl RPMI 1640 (10% FBS), 500 μl of VSVg-pseudotyped retrovirus, and 8 μg/ml polybrene. Cells were incubated at 22 °C for 30 min and then centrifuged at 2000 × g for 2–3 h (22 °C). Transduced cells were cultured in 0.5 ml RPMI 1640 (10% FBS) in a 12-well plate. 2–3 days post-transduction, cells were counted by trypan blue exclusion with blinding of sample identity. Transduction efficiency was monitored by FACS analysis of GFP expression. To detect surface levels of envelope, 5 × 104 cells were stained with 10 μg/ml 2G12 anti-gp120 monoclonal antibody or an isotype control followed by secondary staining with goat F(ab’)2 antihuman IgG (Fc Sp.)-PE (Caltag).
Inhibition of infection
Cell lines and dosage of drug for each particular experiment are indicated in Results. For Tak-779, AMD-3100, and Leu-3a studies, target cells were preincubated with drug for 1 h at 37 °C prior to infection. For sCD4 inhibition studies, virus was preincubated with sCD4 for 2 h at 37 °C prior to addition to target cells. For all studies, relative inhibition was determined by comparing the amount of infection in the presence of each concentration of drug to the amount of infection without drug for each virus.
Human fetal-thymus organ culture
The procedure for HF-TOC has been previously described (Bonyhadi et al., 1995; Meissner et al., 2004; Miller et al., 2001). Briefly, human fetal thymuses (19–24 gestational weeks) were dissected into ~2-mm3 fragments using a dissecting microscope. 5–6 fragments were placed on organotypic culture membranes (Millipore) underlaid by media (RPMI with 10% fetal bovine serum, 50 μg of streptomycin/ml, 50 U of penicillin G/ml, 1 × MEM vitamin solution (Gibco/BRL), 1 × insulin-transferrin-sodium selenite medium supplement (Sigma), and beta-mercaptoethanol) in 6-well tissue culture plates. An equal amount of virus (100–800 IU) in 15 Al of supernatant from infected PHA-stimulated PBMCs or mock supernatant was applied to each fragment. Viral and mock supernatants produced from the same PBMC donor were used within each experiment. Fragments were cultured at 37 °C in 5% CO2 for 10–12 days with daily changes of culture media. Thymocytes were teased out of the fragments using pestles (Bellco Co.) and were stained with CD4-PE and CD8-TC (Caltag) in PBS–2% fetal bovine serum, washed, and resuspended in PBS–1% formaldehyde for FACS analysis. Cytopathicity was quantitated by calculating the total percentage of cells that stained positive for CD4, including CD4+CD8+ and CD4+CD8− thymocytes. To measure viral replication, HF-TOC supernatant prepared in PBS–1% Triton X-100 was measured using an Alliance p24 ELISA kit (Perkin-Elmer).
Statistics
Student’s t test was used for statistical analysis in each experiment. P values of less than 0.05 were considered to be statistically significant. In cases where data from multiple experiments were combined, P values were determined using the averages from each independent experiment.
Acknowledgments
We thank Dr. Ron Swanstrom and Dr. Liguo Zhang for helpful discussions. We thank Dr. Xiao-Fang Yu (Johns Hopkins University) for providing the R3A and R3B isolates. We thank Dr. Michael D. Miller (Merck Laboratories) for providing the Blam-vpr construct (pMM310). We thank Dr. John P. Moore (Cornell University) for providing the bicyclam JM-2987 (hydrobromide salt of AMD-3100). The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: pNL4-3 from Dr. Malcolm Martin, Sup-T1 cells from Dr. James Hoxie, 1G5 cells from Dr. Estuardo Aguilar-Cordova and Dr. John Belmont, TZM-bl from Dr. John C. Kappes, Dr. Xiaoyun Wu, and Tranzyme Inc., GHOST-CXCR4 cells from Dr. Vineet N. KewalRamani and Dr. Dan R. Littman, U373-MAGI-CXCR4CEM and U373-MAGI-CCR5E cells from Dr. Michael Emerman, and HIV-1 gp120 Monoclonal Antibody (2G12) from Dr. Hermann Katinger. We thank the UNC Center for AIDS Research, NIAID, DHHS: P30-A150410 for institutional support. This work was supported by NIH grants AI041356 and AI53804. EM was supported by the NIH training grant T32-AI07419.
References
- Arthos J, Cicala C, Selig SM, White AA, Ravindranath HM, Van Ryk D, Steenbeke TD, Machado E, Khazanie P, Hanback MS, Hanback DB, Rabin RL, Fauci AS. The role of the CD4 receptor versus HIV coreceptors in envelope-mediated apoptosis in peripheral blood mononuclear cells. Virology. 2002;292(1):98–106. doi: 10.1006/viro.2001.1266. [DOI] [PubMed] [Google Scholar]
- Baba M, Nishimura O, Kanzaki N, Okamoto M, Sawada H, Iizawa Y, Shiraishi M, Aramaki Y, Okonogi K, Ogawa Y, Meguro K, Fujino M. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc. Natl. Acad. Sci. U.S.A. 1999;96(10):5698–5703. doi: 10.1073/pnas.96.10.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont T, Quakkelaar E, van Nuenen A, Pantophlet R, Schuitemaker H. Increased sensitivity to CD4 binding site-directed neutralization following in vitro propagation on primary lymphocytes of a neutralization-resistant human immunodeficiency virus IIIB strain isolated from an accidentally infected laboratory worker. J. Virol. 2004;78(11):5651–5657. doi: 10.1128/JVI.78.11.5651-5657.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkowitz RD, Beckerman KP, Schall TJ, McCune JM. CXCR4 and CCR5 expression delineates targets for HIV-1 disruption of T cell differentiation. J. Immunol. 1998;161(7):3702–3710. [PubMed] [Google Scholar]
- Bonyhadi ML, Su L, Auten J, McCune JM, Kaneshima H. Development of a human thymic organ culture model for the study of HIV pathogenesis. AIDS Res. Hum. Retroviruses. 1995;11(9):1073–1080. doi: 10.1089/aid.1995.11.1073. [DOI] [PubMed] [Google Scholar]
- Brelot A, Heveker N, Adema K, Hosie MJ, Willett B, Alizon M. Effect of mutations in the second extracellular loop of CXCR4 on its utilization by human and feline immunodeficiency viruses. J. Virol. 1999;73(4):2576–2586. doi: 10.1128/jvi.73.4.2576-2586.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavrois M, De Noronha C, Greene WC. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat. Biotechnol. 2002;20(11):1151–1154. doi: 10.1038/nbt745. [DOI] [PubMed] [Google Scholar]
- Cayabyab M, Karlsson GB, Etemad-Moghadam BA, Hofmann W, Steenbeke T, Halloran M, Fanton JW, Axthelm MK, Letvin NL, Sodroski JG. Changes in human immunodeficiency virus type 1 envelope glycoproteins responsible for the pathogenicity of a multiply passaged simian-human immunodeficiency virus (SHIV-HXBc2) J. Virol. 1999;73(2):976–984. doi: 10.1128/jvi.73.2.976-984.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti LA, Ivanovic T, Cheng-Mayer C. Properties of the surface envelope glycoprotein associated with virulence of simian-human immunodeficiency virus SHIV (SF33A) molecular clones. J. Virol. 2002;76(4):1588–1599. doi: 10.1128/JVI.76.4.1588-1599.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho MW, Lee MK, Carney MC, Berson JF, Doms RW, Martin MA. Identification of determinants on a dualtropic human immunodeficiency virus type 1 envelope glycoprotein that confer usage of CXCR4. J. Virol. 1998;72(3):2509–2515. doi: 10.1128/jvi.72.3.2509-2515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffield VM, Jiang Q, Su L. A genetic approach to inactivating chemokine receptors using a modified viral protein. Nat. Biotechnol. 2003;21(11):1321–1327. doi: 10.1038/nbt889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffield VM, Helms WS, Jiang Q, Su L. G{alpha}13 mediates a signal that is essential for proliferation and survival of thymocyte progenitors. J. Exp. Med. 2004;200(10):1315–1324. doi: 10.1084/jem.20040944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor RI, Mohri H, Cao Y, Ho DD. Increased viral burden and cytopathicity correlate temporally with CD4+ T-lymphocyte decline and clinical progression in human immunodeficiency virus type 1-infected individuals. J. Virol. 1993;67(4):1772–1777. doi: 10.1128/jvi.67.4.1772-1777.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daar ES, Ho DD. Relative resistance of primary HIV-1 isolates to neutralization by soluble CD4. Am. J. Med. 1991;90(4A):22S–26S. doi: 10.1016/0002-9343(91)90407-o. [DOI] [PubMed] [Google Scholar]
- de Parseval A, Ngo S, Sun P, Elder JH. Factors that increase the effective concentration of CXCR4 dictate feline immunodeficiency virus tropism and kinetics of replication. J. Virol. 2004;78(17):9132–9143. doi: 10.1128/JVI.78.17.9132-9143.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vreese K, Kofler-Mongold V, Leutgeb C, Weber V, Vermeire K, Schacht S, Anne J, de Clercq E, Datema R, Werner G. The molecular target of bicyclams, potent inhibitors of human immunodeficiency virus replication. J. Virol. 1996;70(2):689–696. doi: 10.1128/jvi.70.2.689-696.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doms RW. Beyond receptor expression: the influence of receptor conformation, density, and affinity in HIV-1 infection. Virology. 2000;276(2):229–237. doi: 10.1006/viro.2000.0612. [DOI] [PubMed] [Google Scholar]
- Eckert DM, Kim PS. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- Este JA. TAK-779 (Takeda) Curr. Opin. Investig Drugs. 2001;2(3):354–356. [PubMed] [Google Scholar]
- Etemad-Moghadam B, Sun Y, Nicholson EK, Karlsson GB, Schenten D, Sodroski J. Determinants of neutralization resistance in the envelope glycoproteins of a simian-human immuno-deficiency virus passaged in vivo. J. Virol. 1999;73(10):8873–8879. doi: 10.1128/jvi.73.10.8873-8879.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemad-Moghadam B, Sun Y, Nicholson EK, Fernandes M, Liou K, Gomila R, Lee J, Sodroski J. Envelope glycoprotein determinants of increased fusogenicity in a pathogenic simian-human immunodeficiency virus (SHIV-KB9) passaged in vivo. J. Virol. 2000;74(9):4433–4440. doi: 10.1128/jvi.74.9.4433-4440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemad-Moghadam B, Rhone D, Steenbeke T, Sun Y, Manola J, Gelman R, Fanton JW, Racz P, Tenner-Racz K, Axthelm MK, Letvin NL, Sodroski J. Membrane-fusing capacity of the human immunodeficiency virus envelope proteins determines the efficiency of CD+ T-cell depletion in macaques infected by a simian-human immunodeficiency virus. J. Virol. 2001;75(12):5646–5655. doi: 10.1128/JVI.75.12.5646-5655.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foda M, Harada S, Maeda Y. Role of V3 independent domains on a dualtropic human immunodeficiency virus type 1 (HIV-1) envelope gp120 in CCR5 coreceptor utilization and viral infectivity. Microbiol. Immunol. 2001;45(7):521–530. doi: 10.1111/j.1348-0421.2001.tb02653.x. [DOI] [PubMed] [Google Scholar]
- Gorry PR, Taylor J, Holm GH, Mehle A, Morgan T, Cayabyab M, Farzan M, Wang H, Bell JE, Kunstman K, Moore JP, Wolinsky SM, Gabuzda D. Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J. Virol. 2002;76(12):6277–6292. doi: 10.1128/JVI.76.12.6277-6292.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon G. 2004 http://katahdin.cshl.org:9331/portal/scripts/main2.pl?link=protocolsandcontent=protocols.html.
- Hernandez-Lopez C, Varas A, Sacedon R, Jimenez E, Munoz JJ, Zapata AG, Vicente A. Stromal cell-derived factor 1/CXCR4 signaling is critical for early human T-cell development. Blood. 2002;99(2):546–554. doi: 10.1182/blood.v99.2.546. [DOI] [PubMed] [Google Scholar]
- Holm GH, Zhang C, Gorry PR, Peden K, Schols D, De Clercq E, Gabuzda D. Apoptosis of bystander T cells induced by human immunodeficiency virus type 1 with increased envelope/receptor affinity and coreceptor binding site exposure. J. Virol. 2004;78(9):4541–4551. doi: 10.1128/JVI.78.9.4541-4551.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoxie JA, Brass LF, Pletcher CH, Haggarty BS, Hahn BH. Cytopathic variants of an attenuated isolate of human immunodeficiency virus type 2 exhibit increased affinity for CD4. J. Virol. 1991;65(9):5096–5101. doi: 10.1128/jvi.65.9.5096-5101.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu M, Harouse JM, Gettie A, Buckner C, Blanchard J, Cheng-Mayer C. Increased mucosal transmission but not enhanced pathogenicity of the CCR5-tropic, simian AIDS-inducing simian/human immunodeficiency virus SHIV (SF162P3) maps to envelope gp120. J. Virol. 2003;77(2):989–998. doi: 10.1128/JVI.77.2.989-998.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu QX, Barry AP, Wang ZX, Connolly SM, Peiper SC, Greenberg ML. Evolution of the human immunodeficiency virus type 1 envelope during infection reveals molecular corollaries of specificity for coreceptor utilization and AIDS pathogenesis. J. Virol. 2000;74(24):11858–11872. doi: 10.1128/jvi.74.24.11858-11872.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey-Hoyle M, Culp JS, Chaikin MA, Hellmig BD, Matthews TJ, Sweet RW, Rosenberg M. Envelope glycoproteins from biologically diverse isolates of immunodeficiency viruses have widely different affinities for CD4. Proc. Natl. Acad. Sci. U. S. A. 1991;88(2):512–516. doi: 10.1073/pnas.88.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabat D, Kozak SL, Wehrly K, Chesebro B. Differences in CD4 dependence for infectivity of laboratory-adapted and primary patient isolates of human immunodeficiency virus type 1. J. Virol. 1994;68(4):2570–2577. doi: 10.1128/jvi.68.4.2570-2577.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson GB, Halloran M, Schenten D, Lee J, Racz P, Tenner-Racz K, Manola J, Gelman R, Etemad-Moghadam B, Desjardins E, Wyatt R, Gerard NP, Marcon L, Margolin D, Fanton J, Axthelm MK, Letvin NL, Sodroski J. The envelope glycoprotein ectodomains determine the efficiency of CD4+ T lymphocyte depletion in simian-human immunodeficiency virus-infected macaques. J. Exp. Med. 1998;188(6):1159–1171. doi: 10.1084/jem.188.6.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson I, Antonsson L, Shi Y, Oberg M, Karlsson A, Albert J, Olde B, Owman C, Jansson M, Fenyo EM. Coevolution of RANTES sensitivity and mode of CCR5 receptor use by human immunodeficiency virus type 1 of the R5 phenotype. J. Virol. 2004;78(21):11807–11815. doi: 10.1128/JVI.78.21.11807-11815.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koot M, Keet IP, Vos AH, de Goede RE, Roos MT, Coutinho RA, Miedema F, Schellekens PT, Tersmette M. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann. Intern. Med. 1993;118(9):681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- Kozak SL, Platt EJ, Madani N, Ferro FE, Jr., Peden K, Kabat D. CD4, CXCR-4, and CCR-5 dependencies for infections by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. J. Virol. 1997;71(2):873–882. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrosse B, Treboute C, Brelot A, Alizon M. Cooperation of the V1/V2 and V3 domains of human immunodeficiency virus type 1 gp120 for interaction with the CXCR4 receptor. J. Virol. 2001;75(12):5457–5464. doi: 10.1128/JVI.75.12.5457-5464.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lineberger JE, Danzeisen R, Hazuda DJ, Simon AJ, Miller MD. Altering expression levels of human immunodeficiency virus type 1 gp120μgp41 affects efficiency but not kinetics of cellμcell fusion. J. Virol. 2002;76(7):3522–3533. doi: 10.1128/JVI.76.7.3522-3533.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolick JB, Munoz A, Donnenberg AD, Park LP, Galai N, Giorgi JV, O’Gorman MR, Ferbas J. Failure of T-cell homeostasis preceding AIDS in HIV-1 infection. The Multicenter AIDS Cohort Study. Nat. Med. 1995;1(7):674–680. doi: 10.1038/nm0795-674. [DOI] [PubMed] [Google Scholar]
- McCune JM. The dynamics of CD4+ T-cell depletion in HIV disease. Nature. 2001;410(6831):974–979. doi: 10.1038/35073648. [DOI] [PubMed] [Google Scholar]
- Meissner EG, Duus KM, Loomis R, D’Agostin R, Su L. HIV-1 replication and pathogenesis in the human thymus. Curr. HIV Res. 2003;1(3):275–285. doi: 10.2174/1570162033485258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner EG, Duus KM, Gao F, Yu XF, Su L. Characterization of a thymus-tropic HIV-1 isolate from a rapid progressor: role of the envelope. Virology. 2004;328(1):74–88. doi: 10.1016/j.virol.2004.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller ED, Duus KM, Townsend M, Yi Y, Collman R, Reitz M, Su L. Human immunodeficiency virus type 1 IIIB selected for replication in vivo exhibits increased envelope glycoproteins in virions without alteration in coreceptor usage: separation of in vivo replication from macrophage tropism. J. Virol. 2001;75(18):8498–8506. doi: 10.1128/JVI.75.18.8498-8506.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JP, McKeating JA, Huang YX, Ashkenazi A, Ho DD. Virions of primary human immunodeficiency virus type 1 isolates resistant to soluble CD4 (sCD4) neutralization differ in sCD4 binding and glycoprotein gp120 retention from sCD4-sensitive isolates. J. Virol. 1992;66(1):235–243. doi: 10.1128/jvi.66.1.235-243.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Doherty U, Swiggard WJ, Malim MH. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 2000;74(21):10074–10080. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogert RA, Lee MK, Ross W, Buckler-White A, Martin MA, Cho MW. N-linked glycosylation sites adjacent to and within the V1/V2 and the V3 loops of dualtropic human immunodeficiency virus type 1 isolate DH12 gp120 affect coreceptor usage and cellular tropism. J. Virol. 2001;75(13):5998–6006. doi: 10.1128/JVI.75.13.5998-6006.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. U.S.A. 1993;90(18):8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedroza-Martins L, Gurney KB, Torbett BE, Uittenbogaart CH. Differential tropism and replication kinetics of human immunodeficiency virus type 1 isolates in thymocytes: coreceptor expression allows viral entry, but productive infection of distinct subsets is determined at the postentry level. J. Virol. 1998;72(12):9441–9452. doi: 10.1128/jvi.72.12.9441-9452.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott SM. HIV-1 coreceptor usage, transmission, and disease progression. Curr. HIV Res. 2003;1(2):217–227. doi: 10.2174/1570162033485357. [DOI] [PubMed] [Google Scholar]
- Picard L, Wilkinson DA, McKnight A, Gray PW, Hoxie JA, Clapham PR, Weiss RA. Role of the amino-terminal extracellular domain of CXCR-4 in human immunodeficiency virus type 1 entry. Virology. 1997;231(1):105–111. doi: 10.1006/viro.1997.8506. [DOI] [PubMed] [Google Scholar]
- Platt EJ, Madani N, Kozak SL, Kabat D. Infectious properties of human immunodeficiency virus type 1 mutants with distinct affinities for the CD4 receptor. J. Virol. 1997;71(2):883–890. doi: 10.1128/jvi.71.2.883-890.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt EJ, Kozak SL, Kabat D. Critical role of enhanced CD4 affinity in laboratory adaptation of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses. 2000;16(9):871–882. doi: 10.1089/08892220050042819. [DOI] [PubMed] [Google Scholar]
- Pugach P, Kuhmann SE, Taylor J, Marozsan AJ, Snyder A, Ketas T, Wolinsky SM, Korber BT, Moore JP. The prolonged culture of human immunodeficiency virus type 1 in primary lymphocytes increases its sensitivity to neutralization by soluble CD4. Virology. 2004;321(1):8–22. doi: 10.1016/j.virol.2003.12.012. [DOI] [PubMed] [Google Scholar]
- Reeves JD, Gallo SA, Ahmad N, Miamidian JL, Harvey PE, Sharron M, Pohlmann S, Sfakianos JN, Derdeyn CA, Blumenthal R, Hunter E, Doms RW. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. U.S.A. 2002;99(25):16249–16254. doi: 10.1073/pnas.252469399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman DD, Bozzette SA. The impact of the syncytium-inducing phenotype of human immunodeficiency virus on disease progression. J. Infect. Dis. 1994;169(5):968–974. doi: 10.1093/infdis/169.5.968. [DOI] [PubMed] [Google Scholar]
- Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. U.S.A. 2003;100(7):4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudensey LM, Kimata JT, Long EM, Chackerian B, Overbaugh J. Changes in the extracellular envelope glycoprotein of variants that evolve during the course of simian immunodeficiency virus SIVMne infection affect neutralizing antibody recognition, syncytium formation, and macrophage tropism but not replication, cytopathicity, or CCR-5 coreceptor recognition. J. Virol. 1998;72(1):209–217. doi: 10.1128/jvi.72.1.209-217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarlatti G, Tresoldi E, Bjorndal A, Fredriksson R, Colognesi C, Deng HK, Malnati MS, Plebani A, Siccardi AG, Littman DR, Fenyo EM, Lusso P. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat. Med. 1997;3(11):1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- Si Z, Gorry P, Babcock G, Owens CM, Cayabyab M, Phan N, Sodroski J. Envelope glycoprotein determinants of increased entry in a pathogenic simian-human immunodeficiency virus (SHIV-HXBc2P 3.2) passaged in monkeys. AIDS Res. Hum. Retroviruses. 2004;20(2):163–173. doi: 10.1089/088922204773004888. [DOI] [PubMed] [Google Scholar]
- Singh A, Collman RG. Heterogeneous spectrum of coreceptor usage among variants within a dualtropic human immunodeficiency virus type 1 primary-isolate quasispecies. J. Virol. 2000;74(21):10229–10235. doi: 10.1128/jvi.74.21.10229-10235.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth RJ, Yi Y, Singh A, Collman RG. Determinants of entry cofactor utilization and tropism in a dualtropic human immunodeficiency virus type 1 primary isolate. J. Virol. 1998;72(5):4478–4484. doi: 10.1128/jvi.72.5.4478-4484.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalmeijer EH, Van Rij RP, Boeser-Nunnink B, Visser JA, Naarding MA, Schols D, Schuitemaker H. In vivo evolution of X4 human immunodeficiency virus type 1 variants in the natural course of infection coincides with decreasing sensitivity to CXCR4 antagonists. J. Virol. 2004;78(6):2722–2728. doi: 10.1128/JVI.78.6.2722-2728.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L, Kaneshima H, Bonyhadi ML, Lee R, Auten J, Wolf A, Du B, Rabin L, Hahn BH, Terwilliger E, McCune JM. Identification of HIV-1 determinants for replication in vivo. Virology. 1997;227(1):45–52. doi: 10.1006/viro.1996.8338. [DOI] [PubMed] [Google Scholar]
- Tersmette M, Gruters RA, de Wolf F, de Goede RE, Lange JM, Schellekens PT, Goudsmit J, Huisman HG, Miedema F. Evidence for a role of virulent human immunodeficiency virus (HIV) variants in the pathogenesis of acquired immunodeficiency syndrome: studies on sequential HIV isolates. J. Virol. 1989;63(5):2118–2125. doi: 10.1128/jvi.63.5.2118-2125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thali M, Olshevsky U, Furman C, Gabuzda D, Li J, Sodroski J. Effects of changes in gp120-CD4 binding affinity on human immunodeficiency virus type 1 envelope glycoprotein function and soluble CD4 sensitivity. J. Virol. 1991;65(9):5007–5012. doi: 10.1128/jvi.65.9.5007-5012.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga K, Greenberg ML, Morse MA, Cumming RI, Lyerly HK, Cullen BR. Molecular basis for cell tropism of CXCR4-dependent human immunodeficiency virus type 1 isolates. J. Virol. 2001;75(15):6776–6785. doi: 10.1128/JVI.75.15.6776-6785.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. Antibody neutralization and escape by HIV-1. Nature. 2003;422(6929):307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280(5371):1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- Zaitseva MB, Lee S, Rabin RL, Tiffany HL, Farber JM, Peden KW, Murphy PM, Golding H. CXCR4 and CCR5 on human thymocytes: biological function and role in HIV-1 infection. J. Immunol. 1998;161(6):3103–3113. [PubMed] [Google Scholar]