Abstract
Because molecular mechanisms of development are extraordinarily complex, the understanding of these processes requires the integration of pertinent research data. Using the Gene Expression Database for Mouse Development (GXD) as an example, we illustrate the progress made towards this goal, and discuss relevant issues that apply to developmental databases and developmental research in general. Since its first release in 1998, GXD has served the scientific community by integrating multiple types of expression data from publications and electronic submissions and by making these data freely and widely available. Focusing on endogenous gene expression in wild-type and mutant mice and covering data from RNA in situ hybridization, in situ reporter (knock-in), immunohistochemistry, RT-PCR, northern blot and western blot experiments, the database has grown tremendously over the years in terms of data content and search utilities. Currently, GXD includes over 1.4 million annotated expression results and over 260,000 images. All these data and images are readily accessible to many types of database searches. Here we describe the data and search tools of GXD; explain how to use the database most effectively; discuss how we acquire, curate, and integrate developmental expression information; and describe how the research community can help in this process.
Keywords: literature curation, data integration, online resource, in situ hybridization, immunohistochemistry, anatomy ontology
INTRODUCTION
Gene expression data provide crucial insights into the molecular mechanisms of development, differentiation, and disease. However, the data are voluminous, complex, and heterogeneous. They are generated by many different laboratories and scattered through thousands of publications. Without the help of centralized databases, it is impossible to keep abreast of all this information, let alone to access and search these data in a cohesive and integrated way.
The Gene Expression Database for Mouse Development (GXD) was one of the first databases to address these critical issues (Ringwald et al., 1994; Ringwald et al., 1999). As a mammalian model system, the mouse is heavily used for developmental research. Tissues from all developmental stages and from different mouse strains and mutants are subject to detailed expression studies. For many years, the GXD project has been curating mouse developmental expression data from the published literature, as well as acquiring data through direct submissions and collaboration with efforts that generate pertinent expression data at a large-scale. For example, GXD has incorporated the in situ hybridization data from the EurExpress (Diez-Roux et al., 2011), GenePaint (Visel et al., 2004), GUDMAP (Genitourinary Molecular Anatomy Project; Harding et al., 2011), and BGEM (Brain Gene Expression Map; Magdaleno et al., 2006) projects. GXD integrates data from all these different sources and, as a major component of the Mouse Genome Informatics (MGI) resource (www.informatics.jax.org), combines the expression information with genetic, functional, and phenotypic data. Therefore, these expression data are readily accessible to many types of database searches (Smith et al., 2014; Finger et al., 2011; Blake et al., 2014).
Here we describe the current status of GXD with a particular emphasis on its search utilities. Further, we illustrate issues of data curation and integration that apply to developmental research in general.
CONCEPTS, SCOPE, AND EXPRESSION DATA CONTENT
GXD covers all developmental stages and all organ systems and comprises expression data from wild-type and mutant mice. The main focus is on endogenous gene expression data during development. As data accumulate, GXD aims to provide increasingly complete information about which RNA and protein products are made from a given gene, where and when these products are expressed, and how their expression varies in different mouse strains and mutants. Because there is no single assay type that can provide answers to all these questions, GXD is designed as a system that can integrate different types of expression data. At this point, GXD captures data from RNA in situ hybridization, in situ reporter (knock-in), immunohistochemistry, RT-PCR, northern blot and western blot experiments.
Expression patterns (i.e. the time and space of gene expression) are described in a standardized way by using an extensive anatomical ontology that has been developed in collaboration with the eMouseAtlas (EMAP) project (Hayamizu et al., 2013; Bard et al., 1998). The ontology is structured hierarchically, allowing the integrated description of expression patterns from experiments with differing spatial resolution, as well as enabling searches that include anatomical structures and their substructures (described in detail below).
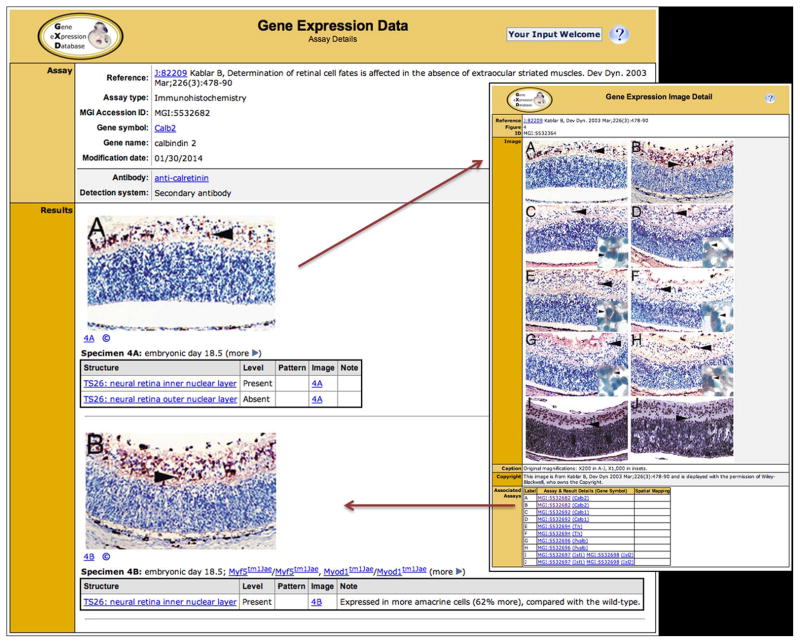
As illustrated in Figure 1, each database record describes the results obtained for each specimen, including the level and pattern of expression for each anatomical structure examined, as well as the molecular probe and the experimental conditions used. Images of the original expression data accompany the annotations whenever possible. By capturing these elemental data, different types of expression data can be represented and integrated in a robust manner. Genes and mutant alleles are recorded using official nomenclature, and all data are associated with a reference. Genes, probes, alleles, anatomical structures, and references are key points of data integration that tie genetic, genomic, expression, functional, and phenotypic data closely together, enabling search capabilities unique to GXD/MGI.
Figure 1.
Left: GXD assay details pages provide detailed annotations of experimental parameters and expression results. Shown is an example of an Assay Detail page for an immunohistochemistry experiment illustrating the detailed content of expression results in GXD. The Assay section reports the reference from which the data were derived, the assay type, the gene analyzed, and the antibody used, with links to more details about the antibody, reference, and gene. The Results section reports the tissue (Theiler stage and anatomical structure) analyzed, as well as the level and pattern of expression, as described by the authors. Images of the original expression data are displayed together with the corresponding annotations whenever possible. Major specimen details such as the age and mutant alleles are always displayed on this page. Other information, such as genetic background, sex, and specimen preparation method, is accessible via the ‘more’ toggle.
Right: Image detail pages are accessed by clicking on the image panes or pane labels shown in the assay details record. They show the entire figure, as published, providing the scientific context. As shown in the Associated Assays table of this example, several genes were studied in the same or similar tissue sections; the assay IDs and gene symbols link to the corresponding annotations for each image pane. While the image detail pages provide visual context, it is the detailed and standardized text annotations shown on the GXD assay detail pages (left) that make the expression data, including image data, accessible to searching.
GXD currently includes detailed expression records for > 13,800 genes. The data come from > 67,000 expression assays and include > 1.4 million expression result annotations and > 260,000 images of primary expression data. 82 % of the data are from RNA in situ hybridization studies and 10 % from RT-PCR experiments, reflecting the detailed spatial resolution and sensitivity required in developmental expression studies. In addition to data from different strains of wild-type mice, GXD currently includes expression data from > 1,900 mouse mutants. In the following we illustrate different ways to effectively search these data.
SEARCHING FOR EXPRESSION DATA
The GXD Home Page
One way to access the expression data for a given gene is to use the Quick Search, accessible from all MGI pages, in order to find the corresponding gene detail page and then follow the links in the “Expression” section (see Fig. 2). However, GXD’s search tools allow you to do much more than look up expression data gene by gene. To take full advantage of GXD, we recommend the GXD Home Page as a starting point: http://www.informatics.jax.org/expression.shtml. This page gives access to GXD’s search forms and provides more information about GXD, including user help, news announcements, and instructions for submitting data to GXD. We now describe the use of three of GXD’s search forms in more detail.
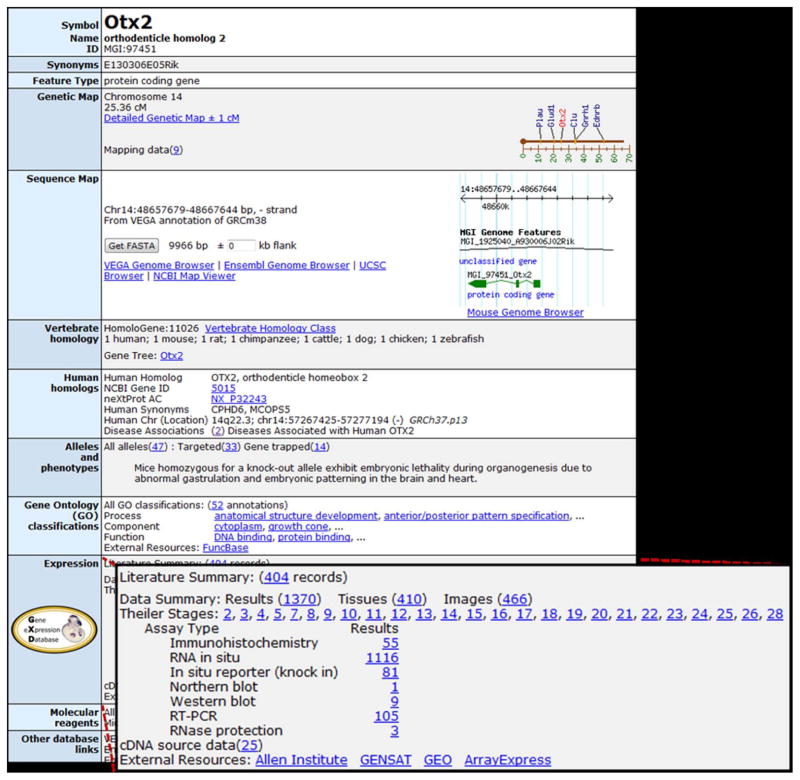
Figure 2.
Gene Detail pages summarize, and provide access to, all the information about a given gene in MGI, together with extensive links to external resources. The upper portion of the Otx2 gene detail page is shown. The expression section (expanded) indicates the types and amount of expression information available for the gene and provides links to the corresponding summary pages. Links to databases that store mouse expression data not available in GXD are provided as well: the Allen Institute (Lein et al., 2007), GENSAT (Heintz, 2004), GEO (Barret at al., 2013) and ArrayExpress (Petryszak et al., 2014).
Gene Expression Data Query
The Gene Expression Data Query form (http://www.informatics.jax.org/gxd) is the most versatile and powerful search form for detailed expression data in GXD. It offers two different query utilities via the Standard Search and Differential Expression Search tabs.
The Standard Search tab (shown in Fig. 3) allows investigators to ask both broad and very specific questions pertinent to their research interests and quickly obtain matching expression results. Researchers can find expression data for specific genes, or for sets of genes as defined by their biological function or by association with annotated mouse phenotypes or human diseases. They can search for expression data for specific anatomical structures and/or specific developmental stages. Searches can be limited to expression data from wild-type mice or to expression data from mice that have been mutated in specific genes. Assay type(s) can also be chosen. Further, using combinations of the query parameters described above makes it possible to formulate complex queries. For example, one could search for genes associated with DiGeorge Syndrome expressed in heart; for genes involved in left/right asymmetry that are expressed in the primitive streak; or for genes involved in signaling pathways that are expressed in the eye of Pax6 mutant mice.
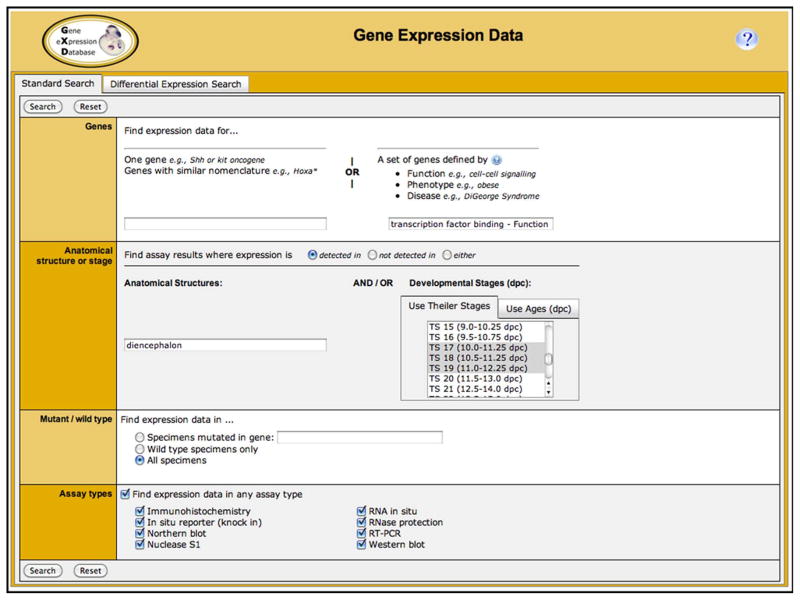
Figure 3.
The Gene Expression Data Query Form features two search tabs: Standard and Differential Expression. The Standard Search, shown here, enables queries for expression data using one or more parameters. The Genes section allows users to find expression data for a specific gene or for a set of genes based on their function [as defined by Gene Ontology terms (Gene Ontology Consortium, 2010)], their association with mouse phenotypes [as defined by Mammalian Phenotype Ontology terms (Smith et al., 2012)], or their association with human diseases [as defined by Online Mendelian Inheritance in Man (OMIM) terms (Amberger et al., 2011)]. In the anatomical/stage section, one can search for expression data in specific anatomical structures and/or developmental stages, and one can specify whether (1) all results should be returned or only those where expression was (2) detected (i.e. present) or (3) not detected (i.e. absent). Anatomical searches combine word searching and hierarchical searching. For example, a search for expression in “diencephalon” would return expression annotations for all anatomical structures that have “diencephalon” as part of their name as well as for all their anatomical substructures such as “thalamus”. In the mutant/wild type section one can limit the searches to expression data from wild-type mice or search for gene expression in specific mutants. The Assay types section allows selection of expression data types. Auto-fill utilities help to find appropriate search terms. The illustrated search asks for ‘transcription factor binding’ genes ‘detected’ in the ‘diencephalon’ at ‘Theiler stages 17, 18, or 19’. The corresponding search results page is shown in Fig. 4. The Differential Expression Search (not shown) allows searching for genes that are expressed in some anatomical structures but not others and/or at some developmental stages but not others.
The Differential Expression Search tab allows querying for genes that are expressed in one anatomical structure but not in another and/or at some developmental stages but not others. For example, one can search for genes that are expressed in the epithalamus but not in the hypothalamus; or for genes that are expressed at the morula stage (Theiler stage 3) but not at the blastocyst stages (Theiler stages 4 and 5). These searches will return a list of genes whose expression has been shown to be absent (not detected), as well as genes whose expression has not been analyzed or recorded in the database for the specified structures and stages. The two cases can be distinguished by filtering the results summaries for instances where expression was not detected (see below).
The Standard Search and Differential Expression Search return a results page with tabbed summaries for assay results, assays, genes, and images (see Fig. 4). This allows users to see the desired level of detail and focus on the data they are most interested in. The four tabs indicate the number of records returned, respectively. One can narrow down the returned results further by modifying the search or by filtering the expression results by Anatomical System, Assay Type, Detected/Not Detected, Theiler Stage, or Wild type/Mutant. Filters can be selected and de-selected to interactively revise and refine the data summaries. Most of the data columns of the summaries are sortable. Further, data from the Assay Results and Genes summaries can be downloaded in text or spreadsheet formats allowing upload into other applications.
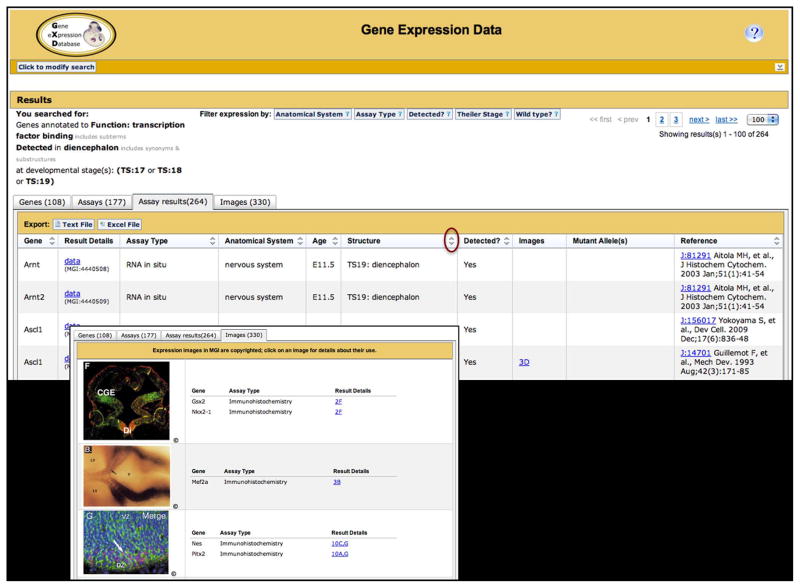
Figure 4.
GXD data summaries can be viewed at different levels of detail and interactively refined and sorted. Searches using the Gene Expression Data Query form return a page with four tabbed summaries for the assay results, assays, genes, and images that match the search parameters. The assay results tab (upper) is displayed by default. It lists the gene studied, the assay type used, the anatomical system, age and tissue examined, indicates whether expression was detected, provides a link to the corresponding images, lists the mutant alleles of the specimen (if applicable), and provides the reference from which the data were derived. Links in the Result Details and Images columns lead to detailed expression records, such as the one shown in Fig. 1. Arrows in column headers indicate that the column is sortable (one set is circled). The assay results tab (as well as the genes tab) allows for the export of results in text and spread sheet formats (buttons in table header). The images tab (lower) shows all the images that match the search criteria, together with the gene(s) examined in that image and the assay type used and provides a link to the corresponding part of the detailed expression record. The expression summaries can be refined by using the ‘click to modify search’ button or by employing the filter options provided on the summary page. The content of all four tabbed summaries will change accordingly.
Each row in the assays, assay results, and image summaries includes links to detailed expression records. Figure 1 shows, as an example, an entry for an immunohistochemistry experiment, illustrating the detail in which data in GXD are annotated. These Assay Detail pages display image panes together with their annotations. Each image pane is linked to the corresponding image detail page that shows the publication figure, which is often multi-paned, thus presenting the image pane in context. Image detail pages might also include links to external resources. For example, detail pages for the EurExpress and GenePaint images link back to the corresponding records at these sites where utilities such as serial-section browsers and high-resolution images are available. Some image detail pages also have links to the Edinburgh Mouse Atlas and Gene Expression Database (EMAGE). GXD makes all its RNA in situ and immunohistochemistry images, together with their annotations, available to EMAGE (Richardson et al., 2014), so that the expression patterns can be mapped and queried spatially. If in situ images have been spatially mapped, GXD provides links from image detail pages to the corresponding mapped images in EMAGE. However, only a subset of the wild-type expression images can be spatially mapped (the 3D atlas is based on wild-type embryos). The standardized text-based description of expression patterns employed by GXD is essential as it allows the representation and integration of all types of expression data from wild-type and mutant mice, as well as the further integration with other data that relate to anatomy, such as mouse phenotype and human disease data.
In short, the Gene Expression Data Query form enables researchers to quickly find specific sets of expression data. Query summaries can be interactively refined and lead to the detailed expression records.
Mouse Developmental Anatomy Browser
As discussed above, GXD and EMAP have developed an extensive ontology for mouse developmental anatomy to describe the time and space of gene expression in a standardized way. This enables intuitive and comprehensive expression searches at variable anatomical resolution. The ontology is structured hierarchically. Each term can have multiple parents, allowing the anatomy to be represented and searched from different perspectives. For example, “brain” is represented as part of the “nervous system” and as part of the “head”. Searches for expression in the “nervous system” or in the “head” will both return expression data for the brain.
The newest version of Mouse Developmental Anatomy comprises all 28 Theiler stages, including the embryonic (TS 1-26), newborn (TS 27), and postnatal mouse (TS 28). There is one “abstract” (non-stage specific) representation of the mouse anatomy that lists the anatomical structures for all Theiler stages, as well as the stage range during which each structure is present. We refer to these terms as EMAPA terms (A for abstract). In addition, there are 28 stage-specific representations (derived from the abstract version). Currently, the ontology includes more than 6500 EMAPA terms and more than 25,000 stage-specific terms (referred to as EMAPS terms). Expression data in GXD are annotated to the stage-specific anatomical terms.
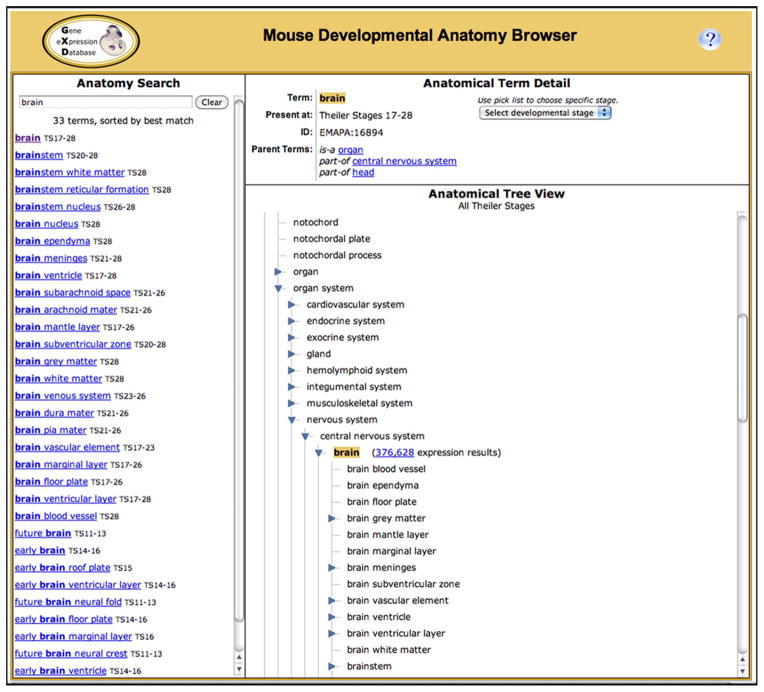
The Mouse Developmental Anatomy Browser (http://www.informatics.jax.org/vocab/gxd/anatomy) allows users to navigate through this anatomical ontology, view a specific anatomical structure, and to obtain the expression data associated with that structure and its substructures (see Fig. 5). The browser consists of three interactive sections: a search section for finding and selecting anatomical terms; a detail section that provides additional information for a selected term and lets users toggle between the abstract and stage-specific versions of the anatomical ontology; and a tree view section that allows users to view the terms in their hierarchical context, to expand and collapse branches of the hierarchy, and to retrieve the expression data for specific anatomical structures. The expression result summaries accessed from the Mouse Developmental Anatomy Browser have the same features and utilities as those obtained upon using the Gene Expression Data Query Form. They can be sorted and filtered and link to the detailed expression records, as described above.
Figure 5.
The Mouse Developmental Anatomy Browser allows users to search for anatomical terms, to explore the anatomical hierarchies and locate specific anatomical structures in context, and to retrieve the expression data associated with these structures and their substructures. The anatomy search is facilitated by an auto-fill utility. As soon as a term is selected from the pick list, all matching anatomical structures are displayed in the search column, together with the developmental stage range during which these structures are present in the embryo. The best match is listed first and selected by default. Other matching terms can be selected by clicking. Upon selection, the Anatomical Tree View and the Anatomical Term Detail section are updated and the selected anatomical structure is highlighted. Using the Tree View, users can explore the ontology further by expanding and collapsing branches. Clicking on a term in the tree view will select (and highlight) that term. The number of expression results associated with each term is listed; following that link will lead to an expression summary page similar to the one shown in Fig. 4. The initial tree view shows the abstract version of the anatomy ontology. Accordingly, the associated expression results will include the annotations for all developmental stages at which the selected anatomical structure is present. The developmental stage pick list in the Anatomical Term Detail section allows users to toggle between stage-independent terms and tree views and stage-specific terms and tree views. Stage-specific terms will link to the expression results for the anatomical structure at that specific stage.
Terms from the mouse developmental ontology are also being used to label the EMAP 3D atlas and to describe expression patterns in EMAGE (Armit et al., 2012). Efforts to establish cross-references to anatomical ontologies from other model organism and human databases are underway (Mungall et al., 2012; Dahdul et al., 2012; Hayamizu et al., 2012; Van Slyke et al., 2014; Segerdell et al., 2013, Costa et al., 2013). This will foster the comparative analysis of expression patterns between model organisms used in developmental research.
Gene Expression Literature Query
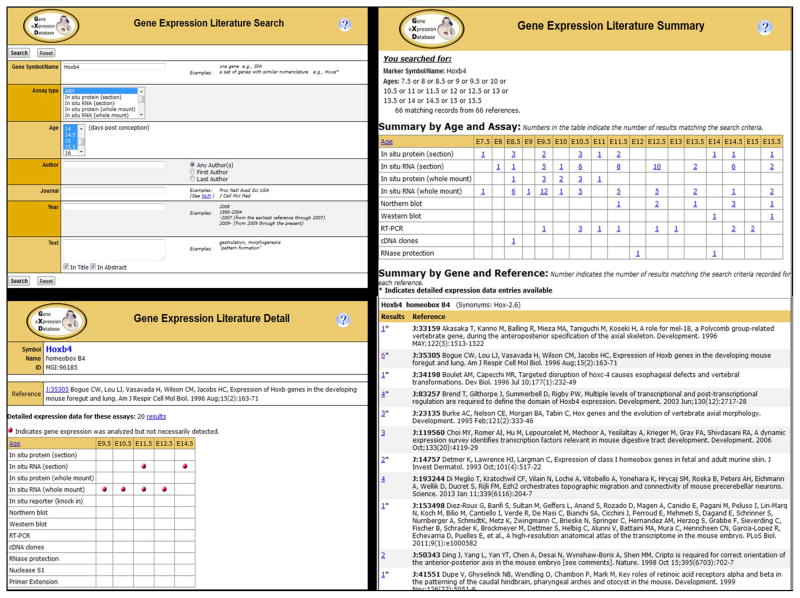
GXD provides researchers with an effective way to search the mouse embryonic expression literature. Our curators survey journals to find all publications that contain the types of data that GXD collects. As a first annotation step, they index all these publications with regard to the genes that have been studied, the expression assay types used, and the ages analyzed. These annotations are then combined with bibliographic information from PubMed to generate the Gene Expression Literature Index. The index is complete and up-to-date from 1990 onwards for all major journals (~150). Currently, the index covers > 21,800 references reporting expression data for > 15,100 genes. An average of 1,200 papers are added to the index per year. All this information is available via the Gene Expression Literature Query form (http://www.informatics.jax.org/gxdlit). The query form allows researchers to quickly find publications that report specific sets of expression data (see Fig. 6). These searches are more effective and complete than PubMed searches because the index uses standard nomenclature for genes, assay types, and ages and because the annotations are based on the entire article, including supplemental data.
Figure 6.
Querying the embryonic mouse expression literature. The Gene Expression Literature Search (top left) allows querying of the embryonic expression literature for genes and ages analyzed and expression assay types used, as well as querying for bibliographic information or specific words in the title or abstract. A portion of the summary return for the query formulated in the figure is displayed at right. The table at the top, “Index Results by Age and Assay”, shows the number of matching records grouped by the age of the specimen and the assay type used. The lower portion of the page, “Index Results by Gene and Reference”, lists the citations for the references where the matching index results were reported, as well as the number of matching results contained therein. Entries marked with an * indicate they have been annotated in detail in GXD. Links on this summary page access detail pages (lower left). These pages display the expression information about the gene contained in the reference and provide links to gene and reference detail pages, as well as to the detailed expression data from the paper if they have already been annotated in GXD.
ACQUIRING AND CURATING DATA
Data are acquired from the literature, via electronic data submissions, and through collaborations with projects that generate pertinent expression data at a large-scale. All data are reviewed by curators and annotated in standardized ways by making extensive use of controlled vocabularies and ontologies as illustrated in Fig. 1. This is a prerequisite for the data integration and search capabilities that GXD provides.
GXD is the only effort that curates mouse developmental expression data from the literature in a systematic way. The first step in this process is populating the Gene Expression Literature Index as described above. The next step is to annotate the details of the expression data from these articles, including supplemental data, as illustrated in Fig. 1. Papers are prioritized for detailed annotation based on the information in the Gene Expression Literature Index. Prioritization criteria include: genes for which there is no detailed data in the database; genes associated with human diseases; genes which, based on the number of publications, are underrepresented in the detailed portion of the database; publications that include a large amount of expression data; and publications that characterize the developmental expression pattern of a given gene in detail (often the first publications for that gene).
Large-scale electronic data sets are reviewed by a combination of computational and manual checks to make sure that the probe-to-gene assignments are up-to-date and that data entries are complete. Nomenclature issues, data ambiguities, and questions that might arise during the mapping of submitted data to ontologies are resolved together with the data provider before the data are loaded into GXD.
All data, including those from large-scale data sets, gain significant value when integrated with the other data in GXD and the larger MGI resource. All data can be explored together and searches can be done that are unavailable elsewhere. Further, GXD ensures that data and data connections are maintained and kept up-to-date after large-scale projects have ended and gene models or the gene names used in the literature have changed.
Authors base their conclusions on much more primary data than will appear in a publication, they are specialists in their fields, and they have detailed knowledge about specific experimental parameters and potential pitfalls that is not available to curators. For these reasons, GXD curators rely on the text of the manuscript, or of data submissions, to derive annotations of expression patterns. These annotations are standardized by using terms from the anatomical ontology (which is expanded as required). However, curators do not interpret images themselves, thereby trying to derive additional expression results that have not been asserted in the paper. For the reasons discussed above, interpretation of images by curators to infer expression or absence of expression would be problematic and error-prone. Instead, GXD displays standardized text annotations together with the corresponding images so users can see the original expression data (see Fig. 1). We have obtained permissions to include images from all major developmental journals, as well as from many others. Currently, over 70% of the result annotations are accompanied by images. Also in cases where GXD does not have the permission to include images, we annotate the data based on the information in the manuscript and provide a reference to the corresponding figure panes.
It is thus important to note that the detail of expression annotations in GXD is determined by the details provided in the text of published articles and electronic data submissions. Detailed descriptions of expression results will lead to more fine-grained and complete annotations. Further, a clear correspondence in the publication between stated expression results, probes, specimens, and images facilitates data annotation. Concise, unambiguous descriptions of expression patterns result in robust annotations. Issues of completeness, consistency, data identity, and clarity apply not only to GXD but to all databases that curate data from the published scientific literature. This includes other developmental organism databases such as GEISHA (Antin et al., 2014), Xenbase (James-Zorn et al., 2013), ZFIN (Howe et al., 2013), FlyBase (St. Pierre et al., 2014), and Wormbase (Harris at al., 2014). Publications that present data, results, and interpretations clearly and identify genes, strains, reagents, and methods adequately can significantly facilitate data curation and dissemination, thus greatly benefiting the researchers who generated the data initially as well as improving data access to the research community at large.
THE CASE FOR ELECTRONIC DATA SUBMISSION
Both journal publications and databases are essential for the research enterprise, and they fulfill complementary roles. Journal publications excel in the narrative descriptions of novel discoveries and discussions of data implications and future applications. However, they usually include only part of the data pertinent to a study due to space constraints or because authors report only the results most relevant to the papers’ narrative. Expression (or non-expression) in anatomical systems peripheral to the narrative is often not reported. Further, journal publications cannot and do not provide a framework for data integration and regular updates, with the result that the data cannot be searched adequately. Databases, on the other hand, do not have space constraints or an incentive to focus on specific scientific narratives; and they excel at the handling, maintenance, and integration of data, and in making them accessible to searches. Currently, data generated from conventional (non large-scale) laboratories are primarily reported in free-text descriptions in journal publications. Unless database curators extract data from these publications and bring them into formats proper for integration and searching, these data will remain poorly integrated into our scientific knowledge base. This is clearly not a good solution for making research data accessible.
An obvious and effective solution to address this problem is to combine journal publications with electronic data submissions. Following the example of sequence and array expression data, the types of developmental expression data discussed here could be submitted to pertinent public databases in conjunction with publications. Submitters would receive accession numbers that can be cited in the publications. Without causing an undue burden on submitters, they could, for example, provide an expanded legend for each image that includes more complete descriptions of the expression patterns seen. Thus, electronic submission can include more primary data and results can be described in greater detail and in more standardized ways. While such submissions would still require review and annotation by database curators, it would significantly facilitate the acquisition, integration, and dissemination of data. Having the data widely accessible to many types of searches would benefit everyone: the journal (because all the data would be tied to the publication reference), the submitter (because their data would be more accessible), and the scientific community as a whole (because of the increased database content and utility).
GXD accepts electronic data submissions for the types of data it collects; see the “Send us your data” tab at the bottom of the GXD Home Page for instructions (http://www.informatics.jax.org/mgihome/GXD/GEN/gxd_submission_guidelines.shtml). Other developmental databases, such as FlyBase, GEISHA, ZFIN and Xenbase, are welcoming electronic data submissions as well. The value of all these community resources is proportional to the amount and the quality of data they contain, and it is time for the research community to realize the potential of electronic data submissions.
FUTURE DIRECTIONS
GXD will continue to acquire data from the literature and electronic data submissions and to improve its search and display capabilities. Later this year, we plan to add interactive matrix views of expression data. Tissue-by-developmental stage matrices will provide high-level overviews of the spatio-temporal expression patterns of genes. Tissue-by-gene matrices will enable a comparison of expression patterns. Both types of matrices can be expanded (and collapsed) along the tissue axis, based on the hierarchical organization of the anatomy. Thus, these matrix views will provide users with intuitive high-level summaries of expression results from where they can drill down to more detail. GXD and the larger MGI resource are also planning to implement additional links to other developmental organism databases. In the shorter term, gene-based links will enable users to look up and compare expression data (and other types of data) for orthologous genes. As described above, we are also working on establishing cross-references between anatomical ontologies to enable, in the longer term, an anatomy-based comparison of developmental gene expression patterns.
USER SUPPORT AND OUTREACH
User Support and GXD curatorial staff actively seek to provide presentations, demonstrations and training sessions on GXD and the larger MGI resource at many meetings. Upon request user support personnel also provide on-site visits and training workshops, as well as remote interactive sessions. GXD is continually looking for ways to improve the user experience and, thus, direct user interactions, surveys and collaborations provide valuable feedback on the project from biologists, biomedical researchers, ontology developers and computational biologists who utilize our data. Further, the GXD Advisory Board provides critical input and guidance for the project.
DATABASE ACCESS AND CONTACT INFORMATION
The GXD home page can be accessed directly at http://www.informatics.jax.org/expression.shtml; via the MGI home page (http://www.informatics.jax.org/) by following the topic “Gene Expression Database (GXD)”; or from any other page within MGI by clicking the “Expression” tab of the navigation bar. For web-based access to GXD and MGI, we recommend Firefox, Chrome, or Safari. Online help is available via the FAQs on the GXD home page and by clicking on the question marks in the upper right corner of most GXD pages. User Support personnel can be contacted via email to mgi-help@jax.org or via the “Contact Us” link in the navigation bar. This article focuses on web-based access to the GXD database proper. Additional computational tools to explore GXD’s expression data are available, such as the GXD BioMart (accessible via the GXD home page) and MouseMine (www.mousemine.org). MouseMine, in particular, offers advanced iterative search and filtering capabilities.
Acknowledgments
We would like to thank our colleagues from the GXD project, as well as our colleagues from other MGI projects for their contributions to GXD and the larger MGI resource. In particularly, we would like to thank Janan Eppig and Joanne Berghout for their critical reading of the manuscript and the following individuals for their contributions to the most recent GXD release: Richard Baldarelli, Jonathan Beal, Olin Blodgett, Lori Corbani, Sharon Giannatto, Terry Hayamizu, Jill Lewis, Ingeborg McCright, Dave Miers, David Shaw, and Jingxia Xu.
FUNDING
Grant Sponsor: Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) of the National Institutes of Health (NIH)
Grant Number: HD062499
References
- Amberger J, Bocchini C, Hamosh A. A new face and new challenges for Online Mendelian Inheritance in Man (OMIM(R)) Hum Mutat. 2011;32:564–567. doi: 10.1002/humu.21466. [DOI] [PubMed] [Google Scholar]
- Antin PB, Yatskievych TA, Davey S, Darnell DK. GEISHA: an evolving gene expression resource for the chicken embryo. Nucleic Acids Res. 2014;42:D933–937. doi: 10.1093/nar/gkt962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armit C, Venkataraman S, Richardson L, Stevenson P, Moss J, Graham L, Ross A, Yang Y, Burton N, Rao J, Hill B, Rannie D, Wicks M, Davidson D, Baldock R. eMouseAtlas, EMAGE, and the spatial dimension of the transcriptome. Mamm Genome. 2012;23:514–524. doi: 10.1007/s00335-012-9407-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard JBL, Kaufman MH, Dubreuil C, Brune RM, Burger A, Baldock RA, Davidson DR. An internet-accessible database of mouse developmental anatomy based on a systematic nomenclature. Mech Dev. 1998;74:111–120. doi: 10.1016/s0925-4773(98)00069-0. [DOI] [PubMed] [Google Scholar]
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake JA, Bult CJ, Eppig JT, Kadin JA, Richardson JE The Mouse Genome Database Group. The Mouse Genome Database: integration of and access to knowledge about the laboratory mouse. Nucleic Acids Res. 2014;42:D810–D817. doi: 10.1093/nar/gkt1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Reeve S, Grumbling G, Osumi-Sutherland D. The Drosophila anatomy ontology. J Biomed Semantics. 2013;4:32. doi: 10.1186/2041-1480-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahdul WM, Balhoff JP, Blackburn DC, Diehl AD, Haendel MA, Hall BK, Lapp H, Lundberg JG, Mungall CJ, Ringwald M, Segerdell E, Van Slyke CE, Vickaryous MK, Westerfield M, Mabee PM. A unified anatomy ontology of the vertebrate skeletal system. PLoS One. 2012;7:e51070. doi: 10.1371/journal.pone.0051070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, Rozado D, Magen A, Canidio E, Pagani M, Peluso I, Lin-Marq N, Koch M, Bilio M, Cantiello I, Verde R, De Masi C, Bianchi SA, Cicchini J, Perroud E, Mehmeti S, Dagand E, Schrinner S, Nürnberger A, Schmidt K, Metz K, Zwingmann C, Brieske N, Springer C, Hernandez AM, Herzog S, Grabbe F, Sieverding C, Fischer B, Schrader K, Brockmeyer M, Dettmer S, Helbig C, Alunni V, Battaini MA, Mura C, Henrichsen CN, Garcia-Lopez R, Echevarria D, Puelles E, Garcia-Calero E, Kruse S, Uhr M, Kauck C, Feng G, Milyaev N, Ong CK, Kumar L, Lam M, Semple CA, Gyenesei A, Mundlos S, Radelof U, Lehrach H, Sarmientos P, Reymond A, Davidson DR, Dollé P, Antonarakis SE, Yaspo ML, Martinez S, Baldock RA, Eichele G, Ballabio A. A High-Resolution Anatomical Atlas of the Transcriptome in the Mouse Embryo. PLoS Biol. 2011;9:e1000582. doi: 10.1371/journal.pbio.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger JH, Smith CM, Hayamizu TF, McCright IJ, Eppig JT, Kadin JA, Richardson JE, Ringwald M. The mouse Gene Expression Database (GXD): 2011 update. Nucleic Acids Res. 2011;39:D835–D841. doi: 10.1093/nar/gkq1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium. The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38:D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding SD, Armit C, Armstrong J, Brennan J, Cheng Y, Haggarty B, Houghton D, Lloyd-MacGilp S, Pi X, Roochun Y, Sharghi M, Tindal C, McMahon AP, Gottesman B, Little MH, Georgas K, Aronow BJ, Potter SS, Brunskill EW, Southard-Smith EM, Mendelsohn C, Baldock RA, Davies JA, Davidson D. The GUDMAP database--an online resource for genitourinary research. Development. 2011;138:2845–2853. doi: 10.1242/dev.063594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TW, Baran J, Bieri T, Cabunoc A, Chan J, Chen WJ, Davis P, Done J, Grove C, Howe K, Kishore R, Lee R, Li Y, Muller HM, Nakamura C, Ozersky P, Paulini M, Raciti D, Schindelman G, Tuli MA, Van Auken K, Wang D, Wang X, Williams G, Wong JD, Yook K, Schedl T, Hodgkin J, Berriman M, Kersey P, Spieth J, Stein L, Sternberg PW. WormBase 2014: new views of curated biology. Nucleic Acids Res. 2014;42:D789–793. doi: 10.1093/nar/gkt1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayamizu TF, de Coronado S, Fragoso G, Sioutos N, Kadin JA, Ringwald M. The mouse-human anatomy ontology mapping project. Database (Oxford) 2012;2012:bar066. doi: 10.1093/database/bar066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayamizu TF, Wicks MN, Davidson DR, Burger A, Ringwald M, Baldock RA. EMAP/EMAPA ontology of mouse developmental anatomy: 2013 update. J Biomed Semantics. 2013;4:15. doi: 10.1186/2041-1480-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N. Gene expression nervous system atlas (GENSAT) Nature Neurosci. 2004;7:483. doi: 10.1038/nn0504-483. [DOI] [PubMed] [Google Scholar]
- Howe DG, Bradford YM, Conlin T, Eagle AE, Fashena D, Frazer K, Knight J, Mani P, Martin R, Moxon SA, Paddock H, Pich C, Ramachandran S, Ruef BJ, Ruzicka L, Schaper K, Shao X, Singer A, Sprunger B, Van Slyke CE, Westerfield M. ZFIN, the Zebrafish Model Organism Database: increased support for mutants and transgenics. Nucleic Acids Res. 2013;41:D854–860. doi: 10.1093/nar/gks938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James-Zorn C, Ponferrada VG, Jarabek CJ, Burns KA, Segerdell EJ, Lee J, Snyder K, Bhattacharyya B, Karpinka JB, Fortriede J, Bowes JB, Zorn AM, Vize PD. Xenbase: expansion and updates of the Xenopus model organism database. Nucleic Acids Res. 2013;41:D865–870. doi: 10.1093/nar/gks1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Yuan XF, Zhang B, Zwingman TA, Jones AR. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Magdaleno S, Jensen P, Brumwell CL, Seal A, Lehman K, Asbury A, Cheung T, Cornelius T, Batten DM, Eden C, Norland SM, Rice DS, Dosooye N, Shakya S, Mehta P, Curran T. BGEM: an in situ hybridization database of gene expression in the embryonic and adult mouse nervous system. PLoS Biol. 2006;4:e86. doi: 10.1371/journal.pbio.0040086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungall CJ, Torniai C, Gkoutos GV, Lewis SE, Haendel MA. Uberon, an integrative multi-species anatomy ontology. Genome Biol. 2012;13:R5. doi: 10.1186/gb-2012-13-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryszak R, Burdett T, Fiorelli B, Fonseca NA, Gonzalez-Porta M, Hastings E, Huber W, Jupp S, Keays M, Kryvych N, McMurry J, Marioni JC, Malone J, Megy K, Rustici G, Tang AY, Taubert J, Williams E, Mannion O, Parkinson HE, Brazma A. Expression Atlas update--a database of gene and transcript expression from microarray- and sequencing-based functional genomics experiments. Nucleic Acids Res. 2014;42:D926–932. doi: 10.1093/nar/gkt1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson L, Venkataraman S, Stevenson P, Yang Y, Moss J, Graham L, Burton N, Hill B, Rao J, Baldock RA, Armit C. EMAGE mouse embryo spatial gene expression database: 2014 update. Nucleic Acids Res. 2014;42:D835–844. doi: 10.1093/nar/gkt1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringwald M, Baldock R, Bard J, Kaufman M, Eppig JT, Richardson JE, Nadeau JH, Davidson D. A database for mouse development. Science. 1994;265:2033–2034. doi: 10.1126/science.8091224. [DOI] [PubMed] [Google Scholar]
- Ringwald M, Mangan ME, Eppig JT, Kadin JA, Richardson JE. GXD: a gene expression database for the laboratory mouse. The Gene Expression Database Group. Nucleic Acids Res. 1999;27:106–112. doi: 10.1093/nar/27.1.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segerdell E, Ponferrada VG, James-Zorn C, Burns KA, Fortriede JD, Dahdul WM, Vize PD, Zorn AM. Enhanced XAO: the ontology of Xenopus anatomy and development underpins more accurate annotation of gene expression and queries on Xenbase. J Biomed Semantics. 2013;4:31. doi: 10.1186/2041-1480-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Eppig JT. The Mammalian Phenotype Ontology as a unifying standard for experimental and high-throughput phenotyping data. Mamm Genome. 2012;23:653–668. doi: 10.1007/s00335-012-9421-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CM, Finger JH, Hayamizu TF, McCright IJ, Xu J, Berghout J, Campbell J, Corbani LE, Forthofer KL, Frost PJ, Miers D, Shaw DR, Stone KR, Eppig JT, Kadin JA, Richardson JE, Ringwald M. The mouse Gene Expression Database (GXD): 2014 update. Nucleic Acids Res. 2014;42:D818–D824. doi: 10.1093/nar/gkt954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Pierre SE, Ponting L, Stefancsik R, McQuilton P FlyBase Consortium. FlyBase 102--advanced approaches to interrogating FlyBase. Nucleic Acids Res. 2014;42:D780–788. doi: 10.1093/nar/gkt1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Slyke CE, Bradford YM, Westerfield M, Haendel MA. The zebrafish anatomy and stage ontologies: representing the anatomy and development of Danio rerio. J Biomed Semantics. 2014;5:12. doi: 10.1186/2041-1480-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Thaller C, Eichele G. GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004;32:D552–D556. doi: 10.1093/nar/gkh029. [DOI] [PMC free article] [PubMed] [Google Scholar]