Abstract
We present an intuitive and interactive web service for CARRIE (Computational Ascertainment of Regulatory Relationships Inferred from Expression). CARRIE is a computational method that analyzes microarray and promoter sequence data to infer a transcriptional regulatory network from the response to a specific stimulus. This service displays an interactive graph of the inferred network and provides easy access to the evidence for the involvement of each gene in the network. We provide functionality to include network data in KEGG XML (KGML) format in this graph. Our service also provides Gene Ontology annotation to aid the user in forming hypotheses about the role of each gene in the cellular response. The CARRIE web service is freely available at http://zlab.bu.edu/CARRIE-web.
INTRODUCTION
The computational mapping of biological regulatory networks is a crucial step in the evolution of individual-molecule bioinformatics to predictive systems biology. Furthermore, the determination of the flow of information through a cell in response to a large variety of external stimuli can be used to predict the effects of novel stimuli or to modulate a cell's response by altering the activity of specific members of a network. Deciphering the transcriptional portion of a cell's regulatory network is currently the most tractable part of the problem given the availability of high-throughput data on gene expression (1) and transcription factor binding (2). Several recent publications highlight the activities in this exciting area (3–5).
We have recently developed a computational method, CARRIE (Computational Ascertainment of Regulatory Relationships Inferred from Expression), for inferring a specific transcriptional network in response to a single stimulus or the deletion of a single transcription factor (TF). CARRIE combines two complementary approaches for detecting transcriptional regulation. First, microarray data are used to reveal the genes that respond to a given stimulus through changes in mRNA abundance. These genes are believed to form a co-regulated group. If there are TFs in the group, we propose that they regulate the observed expression changes of the other genes. Second, we identify TFs with binding sites that are statistically overrepresented in the promoter regions of the co-regulated group of genes. Even if their expression levels do not change upon stimulation, these TFs are also predicted to regulate the group of genes. The latter TF selection method is performed with the ROVER (Relative OVERabundance of cis-elements) component of CARRIE. ROVER determines the likelihood that a TF regulates a group of genes by calculating the statistical significance of any overabundance of binding sites for that TF in the promoters in question. We repeat the test of significance for all characterized TFs with binding site position specific scoring matrices (PSSMs), such as those available from TRANSFAC (6) or JASPAR (7), for the species being studied. Finally, a transcriptional network is built based on the most likely TFs and the genes they regulate. Details of CARRIE can be found in our recent publication (8). Here, we introduce an interactive web service for CARRIE with several new features and updates.
DESCRIPTION OF THE CARRIE WEB SERVICE
Using the CARRIE web service is a simple two-step process. In the first step, the user is asked to provide properly formatted microarray data and the statistical cutoffs for significant mRNA abundance changes and TF–promoter interactions. The user can either upload the promoter sequences of all genes on the microarray or use the ones we supply. We have prepared promoter sequences for 6221 yeast genes and the Affymetrix Mu11ksubB and HG-U95E arrays. The microarray data can be supplied preprocessed or unprocessed. In the former case, significant expression changes will be determined using probability of change and fold change measurements from the user's own analysis. In the latter case, genes with significant expression changes will be selected based upon the fold changes and P-values of such changes between the experimental and control conditions. The P-values are calculated using a permutation test. This test computes the significance of a simplified t-test statistic proposed by Golub and colleagues, using a null distribution of the statistic consisting of all possible permutations of the experimental condition labels (9,10). The user is also asked to provide two cutoff values for ROVER, with the first cutoff denoting the probability of binding sites occurring in random promoters and the second cutoff denoting the probability of a TF regulating a random gene.
In the second step, the user is presented with a list of TFs that ROVER predicts regulate the genes with significantly changed expression levels in the microarray data. The TFs are sorted by their P-values, computed using a binomial distribution. The P-value measures the overabundance of significant matches to a PSSM in the promoters of genes with significantly altered expression in contrast with the promoters of the genes with invariant expression (8). The TFs corresponding to genes that show significant expression changes in the microarray data are highlighted in red and pre-selected. The user can pick additional TFs, e.g. those TFs with P-values below a user-determined cutoff. Finally, the web service presents the inferred transcriptional regulatory network.
The network diagrams contain interactive elements that aid in their interpretation. Clicking any gene will bring up a LocusLink (11) webpage for that gene, if data are available. Passing the mouse pointer over any gene in the network graph will bring up a small window that details the microarray data and ROVER results that support the inclusion of that gene in the network. The Gene Ontology (GO) (12) annotation for each gene is also included, when available, using a local installation of the GO seqdblite database. The GO ‘Biological Process’, ‘Molecular Function’ and ‘Cellular Component’ categorizations can aid the user in interpreting the network graph, determining the role of each gene in the stimulus response and making hypotheses about other types of regulation involved in the response being studied.
Our first implementation of CARRIE (8) did not have a robust way of choosing appropriate cutoffs for the TF binding site abundance P-values. The web service has incorporated three improvements in this area. After the microarray and promoter analyses have been conducted the user is presented with a list of TFs, ranked by the likelihood of their involvement in the network according to ROVER. On this second page of the procedure, the user may apply any or all of the three methods for simplifying the selection of TFs.
Part of the problem stems from the fact that PSSM collections, such as the popular TRANSFAC database (6), may contain multiple PSSMs for a given TF. As the first method of addressing this issue, we allow the user to select several TFs from a ROVER output and view a pdf document of the hierarchical clustering of their PSSMs. This clustering is performed using a tool we have developed called Malign which finds the best alignment between two PSSMs, considering all relative shifts and both strands, but no internal gaps. End gaps are replaced with matrix columns containing equal preferences for the four nucleotides. Each PSSM of width K can be viewed as a probability vector over all 4K K-long oligomers: Malign scores each alignment with the Pearson correlation coefficient between these probability vectors. The method is similar in spirit but different in detail from CompareAce (13). After determining the similarity between all pairs of PSSMs, hierarchical clustering is performed using the hclust function from the R statistical package (http://www.r-project.org). This clustering allows the user to judge whether the high-scoring PSSMs are redundant or contain false positive TFs with binding sites that have high similarity to those of other TFs.
In the second approach, we use TRANSFAC annotation to recognize redundancies among PSSMs. Multiple PSSMs are annotated in TRANSFAC as belonging to a particular ‘Factor’, and we assign these PSSMs as a redundant group. Some PSSMs may belong to multiple Factors and we merge the PSSMs from each Factor into the same group. Finally, we adjust the PSSM list to include only the highest-scoring member of each non-redundant group. To use this option the user selects the ‘on’ option for ‘TRANSFAC matrix redundancy reduction’ and clicks ‘Apply’.
In the third approach, we can further reduce false positive PSSMs by applying the stepped multiple testing correction proposed by Benjamini and Yekutieli, which has been shown to be more powerful than more conservative corrections such as the well-known Bonferroni correction (10). This correction emphasizes the distinction between high- and low-scoring PSSMs. To use this option the user selects the ‘on’ option for ‘ROVER P-value multiple testing correction’ and clicks ‘Apply’. The P-values presented in the ROVER result are modified to reflect their corrected values.
Another new feature included in the CARRIE web service is the ability to incorporate additional pathway data into the inferred regulatory networks. The user can choose from a selection of verified KEGG pathways (13) or upload any additional pathway data formatted in the KEGG XML (KGML) format, assuming that the accession numbers in the pathway data match those in the user-submitted microarray data. This feature allows users to combine CARRIE's predictions with any other data sources on the relationships between the genes in their microarray study.
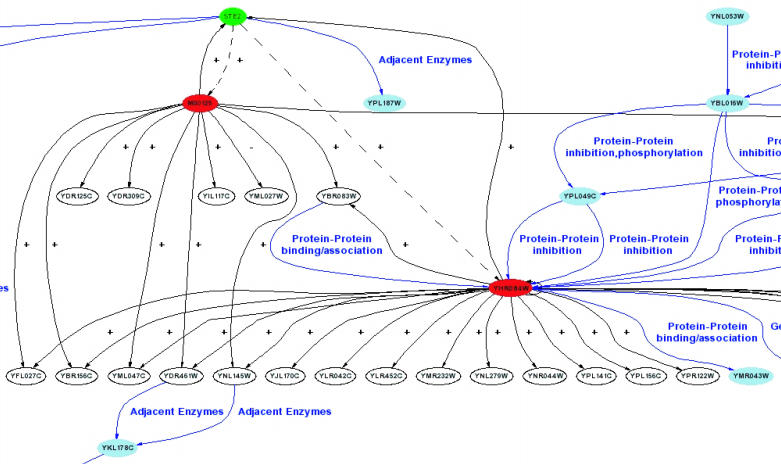
Figure 1 shows the response of Saccharomyces cerevisiae to alpha factor stimulation (14) inferred by the CARRIE web server using data from a microarray study (14) and the KEGG (13) MAP kinase cascade pathway (downloaded on January 27, 2004). If the user supplies the receptor for the experimental stimulus, which is alpha factor receptor STE2 in this case, it is shown in green. The TFs selected based upon the expression changes of their own genes or based upon ROVER's analysis are shown in red. Arrows are drawn from a TF to its regulated genes, as determined by ROVER, with a + depicting stimulation of transcription or a − for inhibition. These relationships are inferred from the directions of the expression changes of the TFs and the genes they regulate (8). Blue nodes, lines and labels are drawn to depict additional data imported from KEGG. We use GraphViz (http://www.research.att.com/sw/tools/graphviz/) and the PERL GraphViz API (http://theoryx5.uwinnipeg.ca/CPAN/data/GraphViz/GraphViz.html) to generate all network diagrams.
Figure 1.
A portion of a transcriptional regulatory network inferred by the CARRIE web service from a microarray study (14) of the transcriptional response to alpha factor stimulation in S.cerevisiae and the KEGG (13) MAP kinase pathway description file. Solid black lines indicate the transcriptional regulation of a gene (in white) from the microarray experiment by a transcription factor (in red). Dashed black lines indicate the inferred indirect regulation of transcription factors by the initial stimulus receptor (in green). Blue nodes and arrows indicate additional information gained by including KEGG data. Blue labels indicate the nature of the interactions of the corresponding blue arrows as annotated by KEGG.
CARRIE was introduced with an analysis of S.cerevisiae transcriptional regulation, but it is also applicable to higher eukaryotes. As a demonstration, CARRIE was applied to human gene expression data downloaded from the NCBI GEO database (15) (Accession no. GDS86) detailing the response of fibroblasts to serum stimulation after 15 min and after 2 h. Several TFs known to be involved in this response were ranked among the most significant of 341 human PSSMs in TRANSFAC (Professional version 7.2). Among them, MEF-2 (16) was ranked third and Pit1 (17) was fourth according to the 15-min data, and C/EBP (18) was ranked fourth according to the 2-h data. Other high-ranking TFs include Octamer Factor 1, STAT 3 and an interferon responsive element binding protein. The results of these analyses are available as Supplementary Data.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
M.C.F. is a Howard Hughes Medical Institute Predoctoral Fellow. This work has been supported in part by NSF grants DBI-0078194, MRI DBI-0116574 and IGERT-9870710 and NIH grants 1R01HG03110-01 and 1P20GM066401-01.
REFERENCES
- 1.Gollub J., Ball,C.A., Binkley,G., Demeter,J., Finkelstein,D.B., Hebert,J.M., Hernandez-Boussard,T., Jin,H., Kaloper,M., Matese,J.C. et al. (2003). The Stanford Microarray Database: data access and quality assessment tools. Nucleic Acids Res., 31, 94–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee T.I., Rinaldi,N.J., Robert,F., Odom,D.T., Bar-Joseph,Z., Gerber,G.K., Hannett,N.M., Harbison,C.T., Thompson,C.M., Simon,I. et al. (2002) Transcriptional regulatory networks in Saccharomyces cerevisiae. Science, 298, 799–804. [DOI] [PubMed] [Google Scholar]
- 3.Bar-Joseph Z., Gerber,G.K., Lee,T.I., Rinaldi,N.J., Yoo,J.Y., Robert,F., Gordon,D.B., Fraenkel,E., Jaakkola,T.S., Young,R.A. et al. (2003) Computational discovery of gene modules and regulatory networks. Nature Biotechnol., 21, 1337–1342. [DOI] [PubMed] [Google Scholar]
- 4.Segal E., Shapira,M., Regev,A., Pe'er,D., Botstein,D., Koller,D. and Friedman,N. (2003) Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nature Genet., 34, 166–176. [DOI] [PubMed] [Google Scholar]
- 5.Stuart J.M., Segal,E., Koller,D. and Kim,S.K. (2003) A gene-coexpression network for global discovery of conserved genetic modules. Science, 302, 249–255. [DOI] [PubMed] [Google Scholar]
- 6.Wingender E., Chen,X., Hehl,R., Karas,H., Liebich,I., Matys,V., Meinhardt,T., Pruss,M., Reuter,I. and Schacherer,F. (2000) TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res., 28, 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sandelin A., Alkema,W., Engstrom,P., Wasserman,W.W. and Lenhard,B. (2004) JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res., 32, D91–D94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haverty P.M., Hansen,U. and Weng,Z. (2004). Computational inference of transcriptional regulatory networks from expression profiling and transcription factor binding site identification. Nucleic Acids Res., 32, 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golub T.R., Slonim,D.K., Tamayo,P., Huard,C., Gaasenbeek,M., Mesirov,J.P., Coller,H., Loh,M.L., Downing,J.R., Caligiuri,M.A. et al. (1999). Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science, 286,531–537. [DOI] [PubMed] [Google Scholar]
- 10.Dudoit S., Shaffer,J.P. and Boldrick,J.C. (2002) Multiple hypothesis testing in microarray experiments. U.C. Berkeley Division of Biostatistics Working Paper Series.
- 11.Pruitt K.D. and Maglott,D.R. (2001) RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res., 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashburner M., Ball,C.A., Blake,J.A., Botstein,D., Butler,H., Cherry,J.M., Davis,A.P., Dolinski,K., Dwight,S.S., Eppig,J.T. et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes J.D., Estep,P.W., Tavazoie,S. and Church,G.M. (2000) Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol., 296, 1205–1214. [DOI] [PubMed] [Google Scholar]
- 14.Roberts C.J., Nelson,B., Marton,M.J., Stoughton,R., Meyer,M.R., Bennett,H.A., He,Y.D., Dai,H., Walker,W.L., Hughes,T.R. et al. (2000) Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science, 287, 873–880. [DOI] [PubMed] [Google Scholar]
- 15.Edgar R., Domrachev,M. and Lash,A.E. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res., 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato Y., Kravchenko,V.V., Tapping,R.I., Han,J., Ulevitch,R.J. and Lee,J.D. (1997) BMK1/ERK5 regulates serum-induced early gene expression through transcription factor MEF2C. EMBO J., 16, 7054–7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaiddon C., de Tapia,M. and Loeffler,J.P. (1999). The tissue-specific transcription factor Pit-1/GHF-1 binds to the c-fos serum response element and activates c-fos transcription. Mol. Endocrinol., 13, 742–751. [DOI] [PubMed] [Google Scholar]
- 18.Shimizu H. and Yamamoto,K. (1994). NF-kappa B and C/EBP transcription factor families synergistically function in mouse serum amyloid A gene expression induced by inflammatory cytokines. Gene, 149, 305–310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.