Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) infections are a serious global problem, with considerable impact on patients and substantial health care costs. This systematic review provides an overview on the clonal diversity of MRSA, as well as the prevalence of Panton-Valentine leukocidin (PVL)-positive MRSA in Africa. A search on the molecular characterization of MRSA in Africa was conducted by two authors using predefined terms. We screened for articles published in English and French through to October 2014 from five electronic databases. A total of 57 eligible studies were identified. Thirty-four reports from 15 countries provided adequate genotyping data. CC5 is the predominant clonal complex in the healthcare setting in Africa. The hospital-associated MRSA ST239/ST241-III [3A] was identified in nine African countries. This clone was also described with SCCmec type IV [2B] in Algeria and Nigeria, and type V [5C] in Niger. In Africa, the European ST80-IV [2B] clone was limited to Algeria, Egypt and Tunisia. The clonal types ST22-IV [2B], ST36-II [2A], and ST612-IV [2B] were only reported in South Africa. No clear distinctions were observed between MRSA responsible for hospital and community infections. The community clones ST8-IV [2B] and ST88-IV [2B] were reported both in the hospital and community settings in Angola, Cameroon, Gabon, Ghana, Madagascar, Nigeria, and São Tomé and Príncipe. The proportion of PVL-positive MRSA carriage and/or infections ranged from 0.3 to 100% in humans. A number of pandemic clones were identified in Africa. Moreover, some MRSA clones are limited to specific countries or regions. We strongly advocate for more surveillance studies on MRSA in Africa.
Keywords: Staphylococcus aureus, MRSA, molecular epidemiology, Africa, systematic review
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is a major public health concern and is responsible for both hospital- and community-associated infections worldwide (De Kraker et al., 2011; CDC, 2013; Falagas et al., 2013; Garza-González and Dowzicky, 2013; Lee et al., 2013; Chen and Huang, 2014). It is estimated that MRSA infections within the health care setting alone affected more than 150,000 patients annually in the European Union, with an additional cost of 380 million Euros (Köck et al., 2010). In the United States of America, 80,461 invasive MRSA infections and 11,285 related deaths occurred in 2011, and an estimated annual burden of between $1.4 billion and 13.8 billion was attributed to community-acquired MRSA (CDC, 2013; Lee et al., 2013). Besides, MRSA has been established as a pathogen for domestic animals and linked with livestock-associated infections (Verkade and Kluytmans, 2013).
Methicillin resistance is usually due to the mecA gene, borne on the staphylococcal cassette chromosome mec (SCCmec) that codes for a 78-kDa penicillin binding protein (PBP2a), with decreased affinity to methicillin and all beta-lactam antibiotics (Chambers, 1997). To date, eleven SCCmec types have been identified (IWG-SCC, 2009). Some cassettes, for example, SCCmec II (53 kb) and SCCmec III (67 kb), are large and possess mobile genetic elements (MGE), such as integrated plasmids (pUB110, pI258, and pT181) and transposons (e.g., Tn554) (Ito et al., 2001), and are frequently associated with hospital-acquired MRSA (Ma et al., 2002; Ito et al., 2004). In contrast, SCCmec IV (21–24 kb) and V (27 kb) are shorter elements, generally susceptible to non-beta-lactam antibiotics, and linked with community MRSA (Chambers and Deleo, 2010). However, the spread of various MRSA clones between the hospital and community settings has made the dichotomous ranking difficult (Deurenberg and Stobberingh, 2008). Recently, a variant mecA gene (named mecC) which is situated on an SCCmec XI element has been described (Shore et al., 2011). It has a higher relative affinity for oxacillin as compared with cefoxitin (Kim et al., 2012), and exhibits only 69% sequence similarity at the nucleotide level and 63% amino-acid identity to mecA/PBP2a (Paterson et al., 2014b). Furthermore, based on whole genome sequencing, mutations of the endogenous penicillin-binding proteins (PBP) 1, 2, and 3 in mecA and mecC negative strains have been postulated as a possible alternative mechanism for beta-lactam resistance in MRSA (Ba et al., 2014).
There is great interest in tracking, identifying and understanding the diversity of MRSA in various settings. Currently, the most widely used molecular techniques include Staphylococcus protein A gene typing (spa) and multilocus sequence typing (MLST). Studies (particularly using MLST) have provided evidence that a small set of lineages, clonal complex (CC)5, CC8, CC22, CC30, and CC45, are associated with most of the MRSA infections in hospitals (Stefani et al., 2012). Besides, a number of different geographically distinct lineages, CC1, CC8, CC30, and CC80, have also been associated with community MRSA infections (Chatterjee and Otto, 2013), while CC8 and CC30 have been identified as pandemic lineages both in the hospital and community setting (Chatterjee and Otto, 2013). Furthermore, regional clones have been described in Australia (sequence type [ST] 93) (Coombs et al., 2009), India (ST772) (D'Souza et al., 2010; Shambat et al., 2012), South Korea (ST72) (Kim et al., 2007), Taiwan and China (ST59) (Chen and Huang, 2014).
The distribution of MRSA clones in Africa is not well-described. Understanding the molecular epidemiology of MRSA in Africa is important as a recent review indicated that since the year 2000, the prevalence of MRSA appears to be increasing in many African countries and pose a visible threat to the continent (Falagas et al., 2013). Furthermore, there is evidence of the replacement of existing MRSA clones with different and new clonal types in a number of countries (Conceição et al., 2007; Aires-de-Sousa et al., 2008; Albrecht et al., 2011; Espadinha et al., 2013; Lim et al., 2013; Nimmo et al., 2013) but information on this trend is lacking in Africa. The occurrence and changes in clonal identities, and their geographic spread is important to understand the spread and evolution of MRSA.
The Panton-Valentine Leukocidin (PVL) is a two-component pore-forming toxin with cytolytic activity on defined cells of the immune system (neutrophils, macrophages and monocytes) (Löffler et al., 2010; Yoong and Torres, 2013). It is encoded by the lukS-PV and lukF-PV genes (Boakes et al., 2011), and PVL-producing S. aureus exhibit a propensity for causing mainly severe and often recurrent skin and soft tissue infections (Shallcross et al., 2013). In addition, PVL-positive MRSA are associated with community onset-pneumonia (Vandenesch et al., 2003). Although the PVL genes are mainly carried by community-associated MRSA (CA-MRSA) (Vandenesch et al., 2003), data from West and Central Africa showed that at least 40% of clinical methicillin-susceptible S. aureus (MSSA) isolates in this region are PVL-positive (Breurec et al., 2011a; Schaumburg et al., 2011; Shittu et al., 2011; Egyir et al., 2014a). Therefore, the acquisition of the mecA gene by PVL-positive MSSA and the possible dissemination of PVL-positive CA-MRSA could present a significant challenge in disease management and infection control in resource-limited countries in Africa.
This systematic review examined published literature on the molecular epidemiology of MRSA in Africa. By summarizing currently available data on the continent, our objective was to describe the distribution of MRSA clones, the prevalence of PVL-positive MRSA, and to highlight the need to develop more comprehensive surveillance and reporting systems for multidrug-resistant organisms such as MRSA in Africa.
Methods
This systematic review was conducted in accordance with the preferred reporting items for systematic reviews and meta-analyses (PRISMA) guidelines (Moher et al., 2009).
Literature search strategy
The relevant English and French articles available in five electronic databases (MEDLINE, EBSCOhost, ISI Web of knowledge, Scopus, and African Journals Online) were retrieved by two authors using predefined search terms (Table S1). The literature search was conducted until 31 October 2014.
Eligible article identification
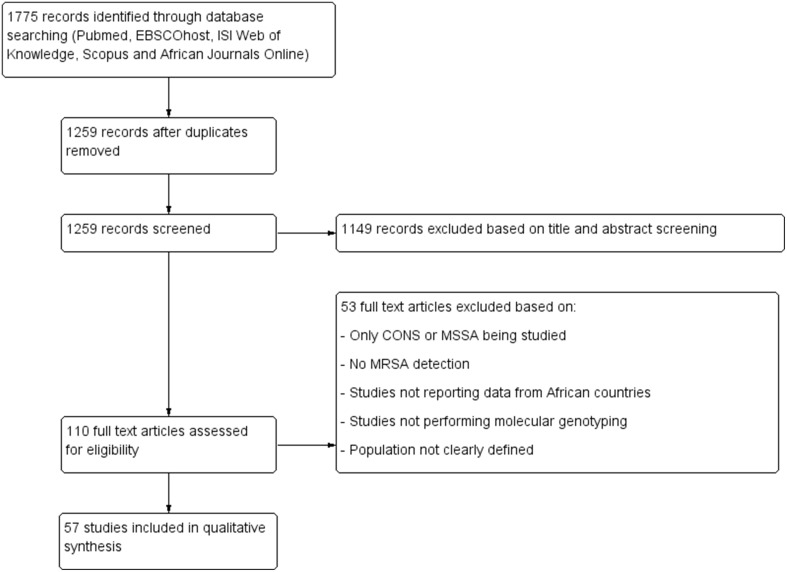
Figure 1 summarizes the study selection process. All duplicate articles were removed and data on MSSA as well as in-vitro studies were also excluded. The eligibility of published reports in this review was based primarily on polymerase chain reaction (PCR) detection of the mecA gene, and the use of at least one molecular tool for genotyping of MRSA strains (Table 1). In addition, worldwide surveys that covered African countries were also included. An MRSA clone was defined based on the combination of MLST sequence type (ST) and SCCmec typing data as previously reported (Okuma et al., 2002). The nomenclature of the SCCmec types was as proposed by the International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC, 2009). SCCmec elements that could not be classified were indicated as non-typeable (SCCmec-NT). In this study, we categorized MRSA into various CCs according to the current eBURST scheme, Version 3 (accessed 30 October 2014) (eBURST, www.mlst.net, V3)1.
Figure 1.
Preferred reporting item for systematic reviews. CONS, coagulase negative staphylococci; MSSA, methicillin susceptible S. aureus; MRSA, methicillin resistant S. aureus.
Table 1.
Characteristics of eligible articles that studied Methicillin resistant Staphylococcus aureus.
| Country | Study period | Study population (sample type) | No. of S. aureus isolates | S. aureus molecular identification | No. of MRSA¶ | Setting (no.) | Genotyping tools | PVL | Detection of genes | References | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SCCmec | coa | agr | spa typing | MLST | PFGE | Antibiotic resistance | Toxin/Virulence | |||||||||
| STUDIES CONDUCTED IN HUMANS | ||||||||||||||||
| Algeria | 2003–2004 | Clinical samples from hospitals and community | 614 | – | 204 | HA (40)/CA (21) | ✓ | – | ✓ | – | ✓ | ✓ | ✓ | – | – | Ramdani-bouguessa et al., 2006 |
| 2004–2007 | Human infections (in- and-out patients) | 65 | – | 23 | NR | ✓ | – | – | ✓ | ✓ | – | ✓ | – | – | Bekkhoucha et al., 2009 | |
| 2005–2007 | From military hospital (Pus, venous catheter, tracheal aspirates, lumbar puncture fluid, blood culture and urine) | NR | – | 64 | HA (50)/CA (14) | ✓ | – | – | – | – | – | ✓ | – | ✓ | Ouchenane et al., 2011, 2013 | |
| 2006–2007 | Healthy and hospitalized individuals | 221α 52β | gyrA PCR | 99α 23β | HA (65)/CA (84) | ✓ | – | ✓ | ✓ | ✓ | NR | ✓ | – | ✓ | Antri et al., 2011 | |
| 2010–2011 | Children and neonates (SSTI, bacteraemia, otitis and bone/joint infections) | 129 | – | 25 | HA (15)/CA (10) | ✓ | – | – | – | ✓ | ✓ | ✓ | – | – | Djoudi et al., 2013 | |
| Angola | 2012 | Nasal swabs from inpatients and HCW | 117 | – | 68 | NR | ✓ | – | ✓ | ✓ | ✓ | ✓ | ✓ | – | ✓ | Conceição et al., 2014 |
| Egypt | 2007–2008 | Pus, sputum, wounds, abscess, blood, urine, and discharge | NR | – | 21 | CA (4) | ✓ | ✓ | ✓ | ✓ | ✓ | NR | ✓ | – | ✓ | Enany et al., 2010 |
| NR | SSTI and nasal swabs | 38 | – | 18 | CA (18) | ✓ | – | – | – | – | – | ✓ | – | – | Sobhy et al., 2012 | |
| NR | Septic wounds, UTI and RTI (nasal swabs) | 10 | – | 7 | – | – | ✓ | – | ✓ | – | – | – | – | – | El-Jakee et al., 2011* | |
| Gabon | 2008–2010 | asymptomatic carriers (nares, axillae, inguinal swabs) and patients (abscess, wound, blood and others) | 217 | nuc and 16S rRNA PCR | 12 | HA (6)/ CA (6) | ✓ | – | ✓ | ✓ | ✓ | – | ✓ | – | ✓ | Schaumburg et al., 2011; Ateba Ngoa et al., 2012 |
| 2010–2013 | swabs from S. aureus carrier mothers (nasal and mammillary) and their infants (Nasal and pharyngeal) | 460 | – | 9 | NR | ✓ | – | – | ✓ | ✓ | – | ✓ | – | ✓ | Schaumburg et al., 2014 | |
| NR | Blood culture of one patient | 1 | – | 1 | NR | – | – | – | ✓ | ✓ | – | ✓ | – | ✓ | Huson et al., 2014 | |
| Ghana | 2011–2012 | In-patients and hospital staff | 105 | spa gene PCR | 6 | NR | ✓ | – | – | ✓ | ✓ | – | ✓ | – | – | Egyir et al., 2013 |
| 2010–2012 | SSTI and blood samples from six hospitals | 308 | – | 9 | NR | ✓ | – | – | ✓ | ✓ | – | ✓ | – | – | Egyir et al., 2014a | |
| 2011–2012 | Nasal swabs from apparently healthy carriers | 124 | – | 2 | HA (2) | ✓ | – | – | ✓ | ✓ | – | ✓ | – | – | Egyir et al., 2014b | |
| Kenya | 2005–2007 | In and out-patients with SSTI boil, abscess, cellulitis and ulcer | 84 | – | 69 | NR | ✓ | – | – | – | – | – | ✓ | – | – | Maina et al., 2013 |
| 2011 | Nasal and axillary skin swabs from hospitalized patients | 85 | – | 6 | NR | ✓ | – | – | ✓ | ✓ | – | ✓ | ✓ | ✓ | Aiken et al., 2014 | |
| Libya | 2009–2010 | Nasal swabs from in-patient children, their mothers, out-patient children and HCW | 758 | – | 70 | HA (12) CA (6) | – | – | – | – | – | ✓ | ✓ | – | – | Al-haddad et al., 2014 |
| Mali | 2005 | Asymptomatic nasal carriers | 88 | – | 1 | CA (1) | – | – | – | – | ✓ | – | – | – | – | Ruimy et al., 2008 |
| Mozambique | 2010–2011 | Post-operative, burn wound infections, skin and soft tissue abscesses | 99 | – | 9 | HA (8), CA (1) | – | – | – | ✓ | – | – | ✓ | ✓ | – | Van der Meeren et al., 2014 |
| Nigeria | 1998–2002 | Wounds, aspirate, amniotic fluid | 276 | – | 4 | NR | ✓ | – | – | – | ✓ | ✓ | – | – | – | Adesida et al., 2005 |
| 2002–2004 | Wound samples, blood cultures, urine, otitis media and ocular related infections | 200 | – | 3 | NR | ✓ | – | – | – | – | ✓ | – | – | – | Shittu and Lin, 2006b | |
| 2007–2012 | Clinical specimen | 150 | – | 12 | NR | ✓ | – | – | – | ✓ | – | ✓ | – | ✓ | Okon et al., 2009, 2014 | |
| 2007 | Surgical and pediatric patients wound samples, corneal, conjunctival, auricular, genital and nasal swabs | 346 | – | 70 | HA (42), CA (28) | ✓ | – | ✓ | ✓ | ✓ | – | ✓ | – | ✓ | Ghebremedhin et al., 2009 | |
| 2008–2010 | HIV-positive and healthy individuals (nasal swabs) | 202 | – | 26 | NR | – | ✓ | – | ✓ | ✓ | – | ✓ | – | – | Olalekan et al., 2012 | |
| 2009 | Wound infections, semen, UTI, chronic ulcer, conjunctivitis, throat infections | 68 | – | 11 | NR | ✓ | – | – | ✓ | ✓ | – | ✓ | ✓ | – | Shittu et al., 2011 | |
| 2009–2011 | Patients and carriers | 62 | – | 22 | NR | ✓ | – | – | – | ✓ | – | ✓ | – | – | Raji et al., 2013 | |
| 2010 | Clinical samples from patients with burns, septicaemia, wound infections, osteomyelitis, bronchitis and GIT | 51 | tuf gene PCR | 15 | NR | ✓ | – | – | ✓ | ✓ | – | ✓ | ✓ | – | Shittu et al., 2012 | |
| NR | Urine, blood and aspirates, wound, eye and ear, urethral and endocervical swab | 116 | – | 48 | HA (40), CA (8) | ✓ | – | – | – | – | – | ✓ | – | ✓ | Terry Alli et al., 2012 | |
| South Africa | 2001–2003 | Wound samples, sputum, otitis media and blood culture | 227 | nuc gene PCR | 61 | NR | ✓ | ✓ | – | – | – | ✓ | – | ✓ | – | Shittu and Lin, 2006a; Shittu et al., 2007 |
| 2001–2003 | Isolates from 16 laboratories in KZN | 241 | – | 24 | NR | ✓ | – | – | – | ✓ | – | – | – | – | Essa et al., 2009 | |
| 2001–2003 | Wounds, sputum, otitis media, urine and blood culture | NR | – | 61 | NR | ✓ | – | – | ✓ | ✓ | ✓ | ✓ | – | – | Shittu et al., 2009 | |
| 2005–2006 | Bacteraemia, SSTI, urine, catheter tip, cerebrospinal and drainage fluids | NR | – | 320 | HA | ✓ | – | – | ✓ | ✓ | ✓ | ✓ | – | ✓ | Moodley et al., 2010 | |
| 2006–2007 | Clinical samples | NR | – | 97 | HA (79), CA (4) | ✓ | – | – | ✓ | – | – | ✓ | – | – | Makgotlho et al., 2009 | |
| 2007–2008 2007–2011 | Pus and pus swabs, urine, blood, RTS and CVCT | NR | – | 100 | CA (10) | ✓ | – | – | ✓ | ✓ | ✓ | ✓ | – | ✓ | Jansen van Rensburg et al., 2011, 2012 | |
| 2009–2010 | A wide range of clinical specimens mostly SSTI | 367 | – | 56 | NR | ✓ | – | ✓ | ✓ | ✓ | ✓ | ✓ | – | – | Oosthuysen et al., 2014 | |
| São Tome and Príncipe | 2010–2012 | Patients and healthy carriers | 52 | – | 14 | NR | ✓ | – | ✓ | ✓ | ✓ | ✓ | ✓ | – | ✓ | Conceição et al., 2013 |
| Tanzania | 2008 | Wound, nasal swab and pus | 160 | – | 24 | HA | – | – | – | ✓ | ✓ | – | ✓ | – | – | Moremi et al., 2012 |
| 2010 | Apparently healthy children under 5 years (nasal swabs) | 114 | nuc gene PCR | 12 | CA | – | – | – | – | – | ✓ | – | – | – | Moyo et al., 2014 | |
| Tunisia | 1998–2007 | Clinical specimens from neutropenic patients | 72 | nuc gene PCR | 13 | HA (13) | ✓ | – | – | – | – | ✓ | – | – | – | Bouchami et al., 2009 |
| 2002 | Patients who developed MRSA infections | NR | – | 6 | HA (6) | – | – | – | – | – | ✓ | – | – | – | Ben Ayed et al., 2010 | |
| 2003–2004 | Pus, blood, pleural fluid, venous catheter | NR | – | 72 | NR | ✓ | – | ✓ | – | – | – | ✓ | – | ✓ | Ben Nejma et al., 2006 | |
| 2003–2004 | Pathological samples from different wards | 147 | – | 19 | NR | – | – | – | – | – | ✓ | – | – | – | Ben Saida et al., 2005 | |
| 2003–2005 | Pus and associated with cutaneous infections | NR | – | 64 | CA (64) | ✓ | – | ✓ | ✓ | ✓ | ✓ | ✓ | – | ✓ | Ben Nejma et al., 2013 | |
| 2004 | Cutaneous pus, blood cultures, urine and puncture fluids | NR | – | 34 | HA (32), CA (2) | ✓ | – | – | – | – | – | – | – | – | Ben Jomaa-Jemili et al., 2006 | |
| 2004–2005 | Cutaneous pus, RTS, urine, blood culture, | 475 | – | 57 | NR | – | – | ✓ | – | – | – | – | – | – | Ben Ayed et al., 2006 | |
| 2004–2008 | Samples from hospitals and community | NR | – | 69 | HA (41), CA (28) | ✓ | – | ✓ | ✓ | ✓ | – | ✓ | – | – | Ben Jomàa-Jemili et al., 2013 | |
| 2006–2008 | Children with CA invasive infections bacteraemia and osteomyelitis | 36 | – | 8 | CA (8) | ✓ | – | ✓ | ✓ | ✓ | ✓ | ✓ | – | – | Kechrid et al., 2011 | |
| 2007 | Pus and skin infections | NR | – | 11 | CA (11) | ✓ | – | ✓ | ✓ | ✓ | ✓ | ✓ | – | – | Ben Nejma et al., 2009 | |
| 2008 | Pus and blood culture (case report) | 2 | – | 2 | NR | ✓ | – | ✓ | ✓ | ✓ | – | ✓ | – | ✓ | Zribi et al., 2011 | |
| 2008–2009 | Humans in contact with animals | 55 | – | 1 | CA (1) | ✓ | – | ✓ | ✓ | ✓ | – | ✓ | ✓ | ✓ | Ben Slama et al., 2011 | |
| Uganda | 2009–2010 | Swabs from patients, HCW and from hospital environment (sinks, door handles, surgical trays, bed and table surfaces) | 41 | – | 41 | NR | ✓ | – | – | – | – | – | ✓ | ✓ | ✓ | Kateete et al., 2011 |
| 2011–2012 | SSI | 64 | nuc gene PCR | 24 | NR | ✓ | – | – | ✓ | – | – | ✓ | – | – | Seni et al., 2013 | |
| Multicenter# | 2007–2008 | SSTI, bacteraemia/septicaemia, urine, wounds osteomyelitis and myositis | NR | – | 86 | CA (9), HA (77) | ✓ | – | ✓ | ✓ | ✓ | – | ✓ | – | ✓ | Breurec et al., 2011b |
| Multicentre✖ | 2004–2005 | Uncomplicated skin infections | 292 | – | 105 | HA (3) | ✓ | – | – | – | ✓ | ✓ | ✓ | – | – | Goering et al., 2008 |
| STUDIES CONDUCTED IN ANIMALS | ||||||||||||||||
| Egypt | NR | Cows and buffaloes milk, cattle septic wounds | 9 | – | 5 | NR | – | ✓ | – | ✓ | – | – | – | – | – | El-Jakee et al., 2011* |
| Senegal | 2009–2011 | Pigs (nasal swabs) | 73 | – | 6 | NR | ✓ | – | – | ✓ | ✓ | – | ✓ | – | ✓ | Fall et al., 2012 |
| Tunisia | 2010 | Healthy sheep (nasal swabs) | 73 | – | 5 | CA (6) | ✓ | – | ✓ | ✓ | ✓ | ✓ | – | – | ✓ | Gharsa et al., 2012 |
agr, Accessory gene regulator; CA, Community-acquired methicillin-resistant S. aureus; coa, Coagulase gene; CVCT, Central venous catheter tips; GIT, Genital tract infections; gyrA, DNA gyrase gene; HA, Hospital-acquired methicillin resistant S. aureus; HCW, Health care workers; HIV, Human immunodeficiency virus; KZN, KwaZulu-Natal province; MLST, Multilocus locus sequence typing; MRSA, Methicillin-resistant Staphylococcus aureus; No., Number of isolates; NR, Not reported; nuc, Thermonuclease gene; PCR, Polymerase chain reaction; PFGE, Pulsed-field gel electrophoresis; PVL, Panton-Valentine-Leukocidin genes; SCCmec, Staphylococcal chromosomal cassette mec element; RTI, Respiratory tract infections; rRNA, Ribosomal ribonucleic acid; RTS, Respiratory tract specimens; spa, Staphylococcus aureus protein A gene; SSTI, Skin and soft tissue infections; SSI, Surgical site infections; tuf, Elongation factor tu; UTI, Urinary tract infections.
MRSA as confirmed by mecA PCR;
✓, Test was conducted; −, Test was not conducted;
Hospitalized individuals;
Nasal carrier study;
African multicenter study which included Cameroon, Madagascar, Morocco, Niger and Senegal;
An international multicenter study which included only South Africa;
Study was conducted in both animal and human host.
Data extraction and synthesis
The relevant data were extracted from each of the articles as stated in Table 1. Separate articles that analyzed the same S. aureus isolates but answered different questions were considered as a single study.
eBURST analysis
The relationship between the MRSA STs described in this review and other lineages reported world-wide was analyzed using the eBURST scheme. The allelic profiles were downloaded from the MLST website (http://saureus.mlst.net/) which included the African MRSA STs as well as 223 representative and randomly selected STs (from each CC) based on the differences in their allelic profiles. The minimum spanning tree was constructed by the goeBURST algorithm using the Phyloviz software v1.1 (http://www.phyloviz.net/).
Results
Literature search
The systematic search of the five electronic databases yielded 1775 articles (Figure 1). No additional studies were identified from AJOL. After the removal of duplicate studies and assessment of titles and abstracts, 110 full-text articles were screened, of which 57 studies were considered eligible for the qualitative analysis according to our inclusion criteria.
Characteristics of the studies included in the systematic review
Most of the data analyzed were obtained from single center studies conducted mainly in five countries; Tunisia (n = 13), Nigeria (n = 9), South Africa (n = 7), Algeria (n = 5), and Egypt (n = 3) (Table 1). Multicenter studies were only reported in two articles (Goering et al., 2008; Breurec et al., 2011b), including a survey which comprised five African countries, Cameroon, Madagascar, Morocco, Niger and Senegal (Breurec et al., 2011b), and an inter-continental multicenter study, which included South Africa (Goering et al., 2008). Only three studies investigated the detection of MRSA in animals (Table 1).
In most of the reports included in this study, S. aureus was identified by phenotypic and culture characteristics, while molecular identification (16S rRNA, detection of the thermonuclease and the elongation factor tu - nuc, tuf - genes) was performed in only 12.3% (7/57). The screening for antibiotic resistance and toxin/virulence genes were carried out in seven and 22 studies, respectively (Table 1). Furthermore, all the eligible studies analyzed MRSA using at least one genotyping technique, and 59.6% (34/57) provided adequate genotyping data on MRSA clones from 15 African countries (Tables 1, 2). Studies included in this systematic review did not investigate on the mecC gene.
Table 2.
Methicillin resistant Staphylococcus aureus clones reported in 34 eligible studies.
| Country | Clonal type ST-SCCmec | Clonal complex | spa type | PVL status | agr | References |
|---|---|---|---|---|---|---|
| Algeria | ST80-IV [2B] | 80 | ND | + | III | Ramdani-bouguessa et al., 2006 |
| ST37-IVa [2B] | 30 | ND | + | III | ||
| ST239-III [3A] | 5 | ND | − | I | ||
| ST239-IVa [2B] | 5 | ND | − | I | ||
| ST241-III [3A] | 5 | ND | − | I | ||
| ST637-III [3A] | 5 | ND | − | I | ||
| ST5-IV, IVa [2B] | 5 | ND | +, − | II | ||
| ST635-IV [2B] | 80 | ND | − | III | ||
| ST636-NT | 22 | ND | − | I | ||
| ST80-IV [2B] | 80 | t044 | + | ND | Bekkhoucha et al., 2009 | |
| ST239-III [3A] | 5 | t037 | − | ND | ||
| ST5-IVa [2B] | 5 | t311, t450 | − | ND | ||
| ST88-NT | 88 | t188, t267 | − | ND | ||
| ST80-IV [2B] | 80 | t044, t4143 | + | III | Antri et al., 2011 | |
| ST241-III [3A] | 5 | ND | − | III | ||
| ST8-V [5C] | 5 | ND | − | I | ||
| ST80-IVc [2B] | 80 | ND | +, − | ND | Djoudi et al., 2013 | |
| ST39-II [2A] | 30 | ND | − | ND | ||
| Angola | ST5-IVa [2B] | 5 | t105, t311, t11657 | − | II | Conceição et al., 2014 |
| ST8-IVd, V [2B] | 5 | t104, t1774 | − | I | ||
| ST72-V [5C] | 5 | t3092 | − | I | ||
| ST88-IVa [2B] | 88 | t186, t325, t786, | − | III | ||
| t1951, t3869 | ||||||
| ST5-V [5C] | 5 | t6065 | − | II | ||
| ST2629-V [5C] | 5 | t6065 | − | II | ||
| ST789-V [5C] | 7 | t091 | − | II | ||
| Cameroon | ST5-V [5C] | 5 | t311 | + | II | Breurec et al., 2011b |
| ST88-IV [2B] | 88 | t186 | − | III | ||
| ST8-IV [2B] | 5 | t024, t121, t451 | + | I | ||
| ST1289-V [5C] | 88 | t1339 | + | III | ||
| Egypt | ST80-IVc [2B] | 80 | t042, t044, t070, t983 | + | III | Enany et al., 2010 |
| ST30-IVa [2B] | 30 | t251, t318 | + | III | ||
| ST1010-Xa | 121 | t159, t312 | + | IV | ||
| Gabon | ST8-IV [2B] | 5 | t121 | + | I | Schaumburg et al., 2011; Ateba Ngoa et al., 2012 |
| ST88-IV [2B] | 88 | t186 | − | III | ||
| ST5-IV [2B] | 5 | t653 | − | II | ||
| ST5-IV [2B] | 5 | t653 | − | ND | Schaumburg et al., 2014 | |
| ST8-NT | 5 | t112, t121 | + | ND | ||
| ST45-V [5C] | 45 | t437, t8860 | − | ND | ||
| ST88-IV [2B] | 88 | t4195 | − | ND | ||
| Ghana | ST72-V [5C] | 5 | t537 | − | ND | Egyir et al., 2013 |
| ST8-V [5C] | 5 | t064 | − | ND | ||
| ST88-IV [2B] | 88 | t325, t1951, t2649 | − | ND | ||
| ST72-V [5C] | 5 | t537 | − | ND | Egyir et al., 2014b | |
| ST8-IV [2B] | 5 | t121 | + | ND | ||
| ST239-III [3A] | 5 | t037 | − | ND | ||
| ST250-I [1B] | 5 | t928 | − | ND | ||
| ST2021-V [5C] | 5 | t024 | − | ND | ||
| ST88-IV [2B] | 88 | t186 | − | ND | ||
| ST789-IV [2B] | 7 | t547 | + | ND | ||
| ST508-V [5C] | 45 | t5132 | − | ND | Egyir et al., 2014a | |
| Kenya | ST239-III [3A] | 5 | t037 | − | ND | Aiken et al., 2014 |
| Madagascar | ST8-IV [2B] | 5 | t121 | + | I | Breurec et al., 2011b |
| ST30-V [5C] | 30 | t4686 | − | III | ||
| ST88-IV [2B] | 88 | t186 | − | III | ||
| Morocco | ST239, ST241-III [3A] | 5 | t037, t138 | − | I | |
| ST5-IV [2B] | 5 | t311 | + | II | ||
| Niger | ST239, ST241-III [3A] | 5 | t138 | − | I | |
| ST239, ST241-V [5C] | 5 | t037 | − | I | ||
| ST88-IV [2B] | 88 | t186 | − | III | ||
| Nigeria | ST8-IV [2B] | 5 | ND | − | ND | Adesida et al., 2005 |
| ST88-IV [2B] | 88 | t186 | + | III | Ghebremedhin et al., 2009 | |
| ST241-IV [2B] | 5 | t037 | − | I | ||
| ST250-I [1B] | 5 | t194, t292 | − | I | ||
| ST241-III [3A] | 5 | t037 | − | ND | Shittu et al., 2011 | |
| ST8-V [5C] | 5 | t064 | − | ND | ||
| ST8-V [5C] | 5 | t451 | − | ND | ||
| ST94-IV [2B] | 5 | t008 | − | ND | ||
| ST5-V [5C] | 5 | t002 | − | ND | ||
| ST241-III [3A] | 5 | t037 | − | ND | Shittu et al., 2012 | |
| ST88-IV [2B] | 88 | t729, t1603 | − | ND | ||
| ST37-III [3A] | 30 | t074 | − | ND | ||
| ST39-II [2A] | 30 | t007 | − | ND | ||
| ST8-V [5C], IV [2B], ST8-NT | 5 | t064 | − | ND | ||
| ST152-NT | 152 | t4690 | + | ND | ||
| ST1-V [5C] | 5 | ND | + | ND | Raji et al., 2013 | |
| ST239-III[3A]mercury | 5 | ND | − | ND | ||
| ST5-II [2A] | 5 | ND | − | ND | ||
| ST8-V [5C] | 5 | ND | − | ND | ||
| ST247-I [1B] | 5 | ND | − | ND | ||
| ST772-V [5C] | 5 | ND | + | ND | ||
| ST88-IV [2B] | 88 | ND | − | ND | ||
| ST241-III [3A] | 5 | ND | − | ND | Okon et al., 2009 | |
| Senegal | ST5-IV [2B]* | 5 | t311 | + | ND | Fall et al., 2012 |
| ST88-IV [2B]* | 88 | t3489 | − | ND | ||
| ST239, ST241-III [3A] | 5 | t037, t138 | − | I | Breurec et al., 2011b | |
| ST5-II [2A] | 5 | t311 | + | II | ||
| ST5-IV [2B] | 5 | t311 | + | II | ||
| ST88-IV [2B] | 88 | t168 | − | III | ||
| South Africa | ST5-IV [2B] | 5 | ND | ND | ND | Essa et al., 2009 |
| ST8-IV [2B] | 5 | ND | ND | ND | ||
| ST8-II [2A] | 5 | ND | ND | ND | ||
| ST239-III [3A] | 5 | ND | ND | ND | ||
| ST45-IV [2B] | 45 | ND | ND | ND | ||
| ST612-IV [2B] | 5 | ND | − | ND | Goering et al., 2008 | |
| ST36-II [2A] | 30 | ND | − | ND | ||
| ST1173-IV [2B] | 5 | t064 | − | ND | Shittu et al., 2009 | |
| ST1338-IV [2B] | 5 | t064 | − | ND | ||
| ST239-III [3A] | 5 | t037 | − | ND | ||
| ST5-III [3A] | 5 | t045 | − | ND | ||
| ST239-III [3A] | 5 | t037 | − | ND | Moodley et al., 2010 | |
| ST612-IV [2B] | 5 | t064 | − | ND | ||
| ST5-I [1B] | 5 | t045 | − | ND | ||
| ST22-IV [2B] | 22 | t032 | − | ND | ||
| ST22-IV [2B] | 22 | t891 | + | ND | ||
| ST36-II [2A] | 30 | t012 | − | ND | ||
| ST239-III [3A] | 5 | t037 | − | ND | Jansen van Rensburg et al., 2011 | |
| ST5-I [1B] | 5 | t045 | ND | ND | ||
| ST650-IV [2B] | 5 | t002 | ND | ND | ||
| ST612-IV [2B] | 5 | t064, t1443, t2196 | ND | ND | ||
| ST72-NT | 5 | t3092 | ND | ND | ||
| ST22-IV [2B] | 22 | t032 | ND | ND | ||
| ST36-II [2A] | 30 | t012, t021 | ND | ND | ||
| ST5-I [1B] | 5 | t002 | − | II | Oosthuysen et al., 2014 | |
| ST8-V [5C] | 5 | t064 | − | I | ||
| ST612-IV [2B] | 5 | t064 | + | I | ||
| ST239-III [3A] | 5 | t021 | − | I | ||
| ST22-V [5C] | 22 | t891 | + | I | ||
| ST22-IV [2B] | 22 | t891 | − | I | ||
| ST36-II [2A] | 30 | t021 | − | III | ||
| São Tome | ST5-IVa [2B] | 5 | t105 | − | II | Conceição et al., 2013 |
| and Príncipe | ST105-II [2A] | 5 | t002 | − | II | |
| ST8-V [5C] | 5 | t451 | − | I | ||
| ST8-IV [2B] | 5 | t451 | − | I | ||
| ST88-IVa [2B] | 88 | t186, t786 | − | III | ||
| Tunisia | ST80-IV [2B] | 80 | t044 | + | III | Ben Nejma et al., 2009 |
| ST80-IV [2B] | 80 | t044 | + | III | Ben Nejma et al., 2013 | |
| ST728-IVc [2B] | 80 | t042, t044 | + | III | Kechrid et al., 2011 | |
| ST8-IVc [2B] | 5 | t062 | + | II | ||
| ST80-IVc [2B] | 80 | t203 | + | III | Ben Slama et al., 2011 | |
| ST1-NT | 5 | t035 | − | III | Ben Jomàa-Jemili et al., 2013 | |
| ST247-I [1B] | 5 | t040 | − | I | ||
| ST239-III [3A] | 5 | t003 | − | I | ||
| ST241-III [3A] | 5 | t125 | − | I | ||
| ST97-NT | 5 | t003 | − | I | ||
| ST1819-I [1B] | 5 | NS | − | I | ||
| ST80-IVc [2B] | 80 | t070 | + | III | ||
| ST2563-IVc [2B] | 80 | t070 | + | III | ||
| ST1440-IVc [2B] | 80 | t070 | + | III | ||
| ST80-IVc [2B] | 80 | t1021 | − | II | ||
| ST80-IVc [2B] | 80 | ND | − | III | ||
| ST22-NT | 22 | t998 | − | II | ||
| ST45-NT | 45 | ND | − | I | ||
| ST153-NT | 80 | NST | + | III | ||
| ST153-NT | 80 | t044 | ND | III | Gharsa et al., 2012 |
agr, Accessory gene regulator; CC, Clonal complex; NT, Non typeable; ND, Not determined; NST, New spa type; PVL, Panton-Valentine Leukocidin genes; SCCmec, Staphylococcal chromosomal cassette mec element; ST, Sequence type; spa, Staphylococcus aureus protein A gene; Xa, Unknown SCCmec type other than I, II, III, IV or V;
Clones isolated from pigs;
+, PVL positive; −, PVL negative.
Community- and hospital- acquired MRSA
Overall, 51% (29/57) of the eligible studies provided the potential source (hospital- or community-associated) of the MRSA strains. Only 17.5% (10/57) reported MRSA from community settings (Table 1). USA300 (ST8-IV [2B]) and other related sequence types were noted both in health care and community settings in nine African countries (Tables 1, 2). The “Brazilian/Hungarian clone” (ST239-III [3A]) was associated with hospital-acquired infections in nine countries (Tables 1, 2). Furthermore, the “West Australia MRSA-2” (ST88-IV [2B]) was reported in community- and hospital-acquired infections in several African countries (Table 2).
Detection of panton-valentine leukocidin (PVL) genes
The screening for PVL-associated genes (lukF-PV and lukS-PV) was carried out in 44 studies, and the detection of PVL genes was only reported in 32 studies (Table 1). In animals, PVL-positive MRSA (ST5) was described in nasal samples of pigs from Senegal (Fall et al., 2012). In humans, the proportion of PVL-positive MRSA carriage and/or infections ranged from 0.3 to 100%. Studies from Algeria and Tunisia reported higher PVL prevalence while investigations from South Africa reported the lowest prevalence (Table 3). Overall, PVL-positive MRSA were more frequently reported with skin and soft tissue infections, and community-associated clones (Tables 1, 2). There was no report on the role of PVL in necrotizing pneumonia caused by MRSA in Africa.
Table 3.
Panton-Valentine Leukocidin prevalence as reported by the eligible studies with sample size of 30 or above.
| Country | PVL positive (no. positive/total tested) | Prevalence (%) | References |
|---|---|---|---|
| Algeria | 46/61 | 75 | Ramdani-bouguessa et al., 2006 |
| 19/64 | 30 | Ouchenane et al., 2011 | |
| 94/122 | 77 | Antri et al., 2011 | |
| Kenya | 14/69 | 20 | Maina et al., 2013 |
| Libya | 10/35 | 29 | Al-haddad et al., 2014 |
| Nigeria | 33/70 | 47 | Ghebremedhin et al., 2009 |
| South Africa | 1/320 | 0.3 | Moodley et al., 2010 |
| 4/97 | 4 | Makgotlho et al., 2009 | |
| 5/56 | 9 | Oosthuysen et al., 2014 | |
| Tunisia | 68/72 | 94 | Ben Nejma et al., 2006 |
| 64/64 | 100 | Ben Nejma et al., 2013 | |
| 43/69 | 62 | Ben Jomàa-Jemili et al., 2013 | |
| Uganda | 30/41 | 73 | Kateete et al., 2011 |
| Multicenter* | 20/86 | 23 | Breurec et al., 2011b |
Multicenter study which included Cameroon, Madagascar, Morocco, Niger and Senegal.
PVL, Panton-Valentine Leukocidin; no., Number.
MRSA clones reported in africa using the current eBURST scheme
Figures 2, 3 summarize the MRSA clones identified in Africa based on MLST CCs. By the current eBURST scheme, six main CCs were identified: CC5, CC22, CC30, CC45, CC80, and CC88. In addition, a number of diverse spa types were identified among the MRSA clones in Africa (Table 2), but the distribution of spa types t042 and t044 (associated with CC80-IV [2B]) appear to be limited to three North African (Algeria, Egypt and Tunisia) countries (Table 2).
Figure 2.
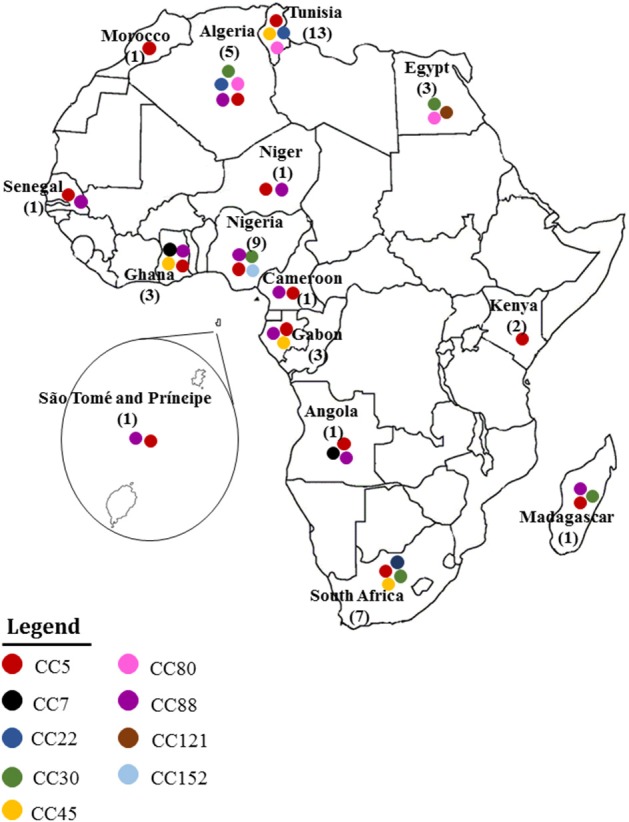
MRSA clones reported in Africa. Each clonal complex is annotated with a colored circle. The number of studies conducted in each country is also indicated.
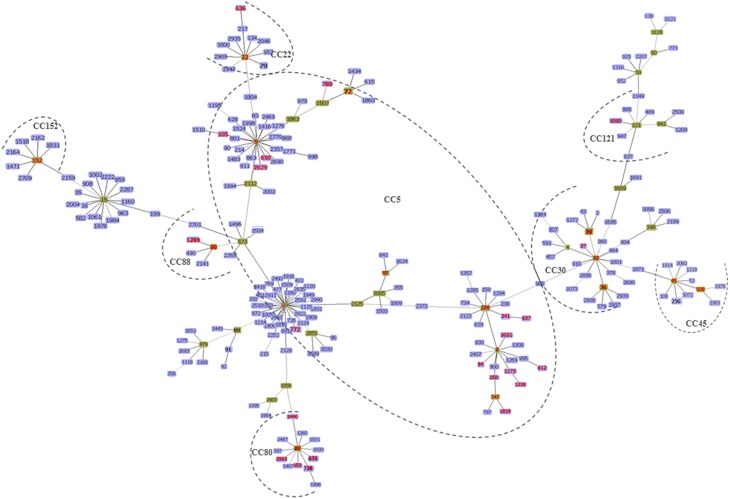
Figure 3.
The minimum spanning tree was constructed by the goeBURST algorithm using the Phyloviz software v1.1 (http://www.phyloviz.net/). The allelic profiles were downloaded from the MLST website (http://saureus.mlst.net/) which included the MRSA sequence types (STs) described in this review as well as 223 randomly selected STs (from each CC) based on the differences in their allelic profiles. The Group founder is colored in green and the related STs are in blue. The six main CCs described in this review are indicated by the dotted lines and the STs reported in Africa are indicated by the red color.
Clonal complex 5
This clonal complex is considered the largest group based on the eBURST scheme (Figure 3). It was subdivided into three main clusters and designated as CC5-ST1, ST5, and ST8.
MRSA CC5 with sequence type 1
This group was reported in Nigeria (Raji et al., 2013) and Tunisia (Ben Jomàa-Jemili et al., 2013). The clonal type included the PVL-positive ST1-V [5C] isolated from patients in a tertiary hospital in Nigeria (Raji et al., 2013), and the PVL-negative ST1 with a non-typeable SCCmec element (spa type t035 and agr type III) identified in Tunisia (Ben Jomàa-Jemili et al., 2013). In addition, an ST1 related sequence type (ST772-V [5C]), “the Bengal-Bay clone” has been described in Nigeria (Raji et al., 2013).
MRSA CC5 with sequence type 5
This clone was documented in 14 studies and consisted of diverse SCCmec elements (Table 2). The ST5-I [1B]/III [3A] were identified from clinical samples in health care institutions in South Africa (Shittu et al., 2009; Moodley et al., 2010; Jansen van Rensburg et al., 2011; Oosthuysen et al., 2014). ST5-II [2A] has been described in Nigeria (Raji et al., 2013), and Senegal (Breurec et al., 2011b). ST5-IV [2B]-PVL-positive was the dominant clone in hospitalized patients with skin and soft tissue infections in Dakar, Senegal (Breurec et al., 2011b). In addition, ST5-IV [2B] was detected from nasal samples of pigs in the same geographical area (Fall et al., 2012). ST5-IV [2B] has also been identified in Algeria (Ramdani-bouguessa et al., 2006), Gabon (Schaumburg et al., 2011; Ateba Ngoa et al., 2012), Morocco (Breurec et al., 2011b), and South Africa (Essa et al., 2009), while the SCCmec IVa [2B] variant was recovered from hospitalized patients in Algeria (Ramdani-bouguessa et al., 2006; Bekkhoucha et al., 2009), Angola (Conceição et al., 2014), and São Tomé and Príncipe (Conceição et al., 2013). Moreover, ST5-IVa [2B] was reported from nasal samples of apparently healthy-hospital workers in Angola (Conceição et al., 2014). Other ST5 and related clones identified are ST5-V [5C] in Angola (Conceição et al., 2014), Cameroon (Breurec et al., 2011b), and Nigeria (Shittu et al., 2011), ST72-SCCmec-NT in South Africa (Jansen van Rensburg et al., 2011), ST72-V [5C] in Angola and Ghana (Egyir et al., 2013; Egyir et al., 2014b; Conceição et al., 2014), and ST105-II [2A] from a patient in São Tomé and Príncipe (Conceição et al., 2013). Furthermore, ST650-IV [2B] was detected from clinical samples in health care institutions in South Africa (Jansen van Rensburg et al., 2011). Finally, ST2629-V [5C] was described in Angola (Conceição et al., 2014).
MRSA CC5 with sequence type 8
MRSA assigned to this clone are widespread and diverse across Africa as evidenced in 27 studies (Table 2). The first known early or ancestral clone, ST250-I [1B], was mainly associated with hospital-acquired infections in Ibadan, South-West Nigeria (Ghebremedhin et al., 2009), and recently observed in Ghana (Egyir et al., 2014b). ST8-II [2A] was only described in the KwaZulu-Natal region of South Africa (Essa et al., 2009), while a number of investigations reported ST8-IV [2B] in Angola (Conceição et al., 2014), Cameroon (Breurec et al., 2011b), Gabon (Schaumburg et al., 2011; Ateba Ngoa et al., 2012), Ghana (Egyir et al., 2014b), Madagascar (Breurec et al., 2011b), Nigeria (Adesida et al., 2005; Shittu et al., 2012), São Tomé and Príncipe (Conceição et al., 2013) and South Africa (Essa et al., 2009). The MRSA isolates from Angola possessed the SCCmec type IVd element (Conceição et al., 2014). ST612-IV [2B], a double locus variant (dlv) of ST8-IV [2B], and only recently reported as PVL-positive (Oosthuysen et al., 2014), is widespread across South Africa (Goering et al., 2008; Moodley et al., 2010; Jansen van Rensburg et al., 2011; Oosthuysen et al., 2014), alongside other variants such as ST1173/ST1338-IV [2B] (Shittu et al., 2009). The ST8-IV [2B] clone in South Africa was identified from a variety of clinical infections, in particular, bacteraemia, skin and soft tissue and wound infections (Shittu et al., 2009; Moodley et al., 2010; Jansen van Rensburg et al., 2011; Oosthuysen et al., 2014). An ST8-IVc [2B] strain (PVL-positive; spa type t062) was identified from a 4 day old male child with community-acquired invasive infection in Tunisia (Kechrid et al., 2011). Furthermore, ST8-V [5C] was described in Algeria (Antri et al., 2011), Angola (Conceição et al., 2014), Ghana (Egyir et al., 2013), Nigeria (Shittu et al., 2011, 2012; Raji et al., 2013), São Tomé and Príncipe (Conceição et al., 2013), and South Africa (Oosthuysen et al., 2014). Other STs observed within the CC5-ST8 cluster include ST8-SCCmec-NT in Gabon (Schaumburg et al., 2014) and Nigeria (Shittu et al., 2012), ST94-IV [2B] described in Nigeria (Shittu et al., 2011) and ST97-SCCmec-NT in Tunisia (Ben Jomàa-Jemili et al., 2013). In addition, ST247-I [1B] was reported only in Tunisia (Ben Jomàa-Jemili et al., 2013) and Nigeria (Raji et al., 2013), ST637-III [3A] in Algeria (Ramdani-bouguessa et al., 2006), ST1819-I [1B] in Tunisia (Ben Jomàa-Jemili et al., 2013), and ST2021-V [5C] in Ghana (Egyir et al., 2014b).
The “Brazilian/Hungarian clone” (ST239-III [3A]) is an hybrid of CC30 and CC8 based on a single large chromosomal replacement (Robinson and Enright, 2004), and ST241-III [3A] is a single locus variant (slv) of ST239-III [3A]. These two STs were identified concurrently in Algeria (Ramdani-bouguessa et al., 2006), Morocco, Niger and Senegal (Breurec et al., 2011b), and Tunisia (Ben Jomàa-Jemili et al., 2013). ST239-III [3A] has also been described in Ghana (Egyir et al., 2014b) and Kenya (Aiken et al., 2014), and consistently since 2001 in South Africa (Essa et al., 2009; Shittu et al., 2009; Moodley et al., 2010; Jansen van Rensburg et al., 2011; Oosthuysen et al., 2014). A recent study detected ST239 with the SCCmec type IIImercury [3A] in a tertiary health care facility in South-West Nigeria (Raji et al., 2013). ST241-III [3A] is the dominant clone in North-East Nigeria (Okon et al., 2009; Shittu et al., 2011, 2012). Interestingly, three SCCmec variants, ST239-IVa [2B], ST239/ST241-V [5C], and ST241-IV [2B], and associated with hospital-acquired infections were reported in Algeria (Ramdani-bouguessa et al., 2006), Niger (Breurec et al., 2011b), and Nigeria (Ghebremedhin et al., 2009).
Clonal complex 22
In Africa, ST22 was identified only in Algeria (Ramdani-bouguessa et al., 2006), South Africa (Moodley et al., 2010; Jansen van Rensburg et al., 2011; Oosthuysen et al., 2014), and Tunisia (Ben Jomàa-Jemili et al., 2013). ST22-IV [2B] was related with hospital-associated infections in the Western Cape and KwaZulu-Natal provinces of South Africa. A variant of ST22 (ST22-V [5C]-PVL-positive) was also reported in an hospital in Western Cape, South Africa (Oosthuysen et al., 2014). The ST22 identified in Tunisia possessed a non-typeable SCCmec element (Ben Jomàa-Jemili et al., 2013). Besides, an ST636-SCCmec-NT isolate has also been reported in Algeria (Ramdani-bouguessa et al., 2006).
Clonal complex 30
ST30-IVa [2B]-PVL-positive, also known as “South-West Pacific clone,” has been reported in Egypt (Enany et al., 2010), and a multicenter African study identified ST30-V [5C] only in Antananarivo, Madagascar (Breurec et al., 2011b). The hospital associated ST36-II [2A] (UK-EMRSA-16), was described only in South Africa (Goering et al., 2008; Moodley et al., 2010; Jansen van Rensburg et al., 2011; Oosthuysen et al., 2014), while ST39-II [2A] a dlv was identified in an hospital in Algiers, Algeria (Djoudi et al., 2013), and Ile-Ife, South-West Nigeria (Shittu et al., 2012). MRSA assigned with these groups (ST36-II [2A] and ST39-II [2A]) were PVL-negative. Furthermore, two SCCmec variants, ST37-IVa [2B] and ST37-III [3A], were reported in Algeria (Ramdani-bouguessa et al., 2006) and Nigeria (Shittu et al., 2012), respectively.
Clonal complex 45
ST45-IV [2B], the “Berlin clone,” was detected in an hospital in the KwaZulu-Natal (South Africa) during a multicenter surveillance study (Essa et al., 2009) and ST45-V [5C] was reported in mother-infant pairs in Gabon (Schaumburg et al., 2014). An MRSA with a non-typeable SCCmec associated with community-acquired infections has been identified in Tunisia (Ben Jomàa-Jemili et al., 2013). Finally, ST508-V [5C], a slv to ST45, and also associated with community-acquired infections was described in Ghana (Egyir et al., 2014a).
Clonal complex 80
The CC80 was limited to three North African countries: Algeria, Egypt, and Tunisia (Table 2). The European clone, ST80-IV [2B]-PVL-positive, was first described in Algeria from both hospitalized and outpatients (Ramdani-bouguessa et al., 2006), and has continued to be the leading clone in the country (Ramdani-bouguessa et al., 2006; Bekkhoucha et al., 2009; Antri et al., 2011; Djoudi et al., 2013). ST80-IVc [2B] has been identified in Egypt (Enany et al., 2010), and Tunisia (Ben Slama et al., 2011; Ben Jomàa-Jemili et al., 2013). In addition, sequence types related to ST80 have been recovered from human clinical samples (ST153-SCCmec-NT, ST728-IVc [2B], ST1440-IVc [2B], and ST2563-IVc [2B]) (Kechrid et al., 2011; Ben Jomàa-Jemili et al., 2013), and nasal specimen of healthy sheep (ST153-SCCmec-NT) (Gharsa et al., 2012) in Tunisia. The afore-mentioned sequence types, ST728, ST1440 and ST2563 belonged to accessory gene regulator (agr) type III and were PVL-positive. Moreover, a PVL-negative ST80-IVc [2B] with agr type II has also been detected in Tunisia (Ben Jomàa-Jemili et al., 2013), and a PVL-negative ST635-IV [2B] in Algeria (Ramdani-bouguessa et al., 2006).
Clonal complex 88
The “West Australia MRSA-2 clone” (WA-MRSA-2), ST88-IV [2B], was reported in both hospital and community settings in eight African countries; Angola (Conceição et al., 2014), Cameroon (Breurec et al., 2011b), Gabon (Schaumburg et al., 2011; Ateba Ngoa et al., 2012), Ghana (Egyir et al., 2013, 2014b), Madagascar (Breurec et al., 2011b), Niger (Breurec et al., 2011b), Nigeria (Ghebremedhin et al., 2009; Shittu et al., 2012; Raji et al., 2013) and Senegal (Breurec et al., 2011b). The MRSA isolates from Angola possessed an SCCmec IVa [2B] element (Conceição et al., 2014). PVL-positive ST88-IV [2B] were detected in Nigeria (Ghebremedhin et al., 2009), and an SCCmec subtype ST88-IVa [2B] was identified among three health care workers and a patient in São Tomé and Príncipe (Conceição et al., 2013). The ST88-IV [2B] with spa type t3489 was also recovered from nasal samples of swine in Senegal (Fall et al., 2012). In addition, an SCCmec non-typeable ST88 was described from an out-patient in Algeria (Bekkhoucha et al., 2009), and a strain related to WA-MRSA-2 (ST1289-IV [2B]) was identified in Yaoundé, Cameroon (Breurec et al., 2011b).
Other clonal complexes
CC7, CC121, and CC152 have been reported in Africa. The PVL-negative ST789 (assigned with CC7) was identified in Angola (with SCCmec IV [2B]) (Conceição et al., 2014). However, in Ghana, ST789 was PVL-positive and carried an SCCmec IV element [2B] (Egyir et al., 2014b). An ST1010-PVL-positive (CC121) with non-typeable SCCmec element has only been described in Egypt (Enany et al., 2010). Furthermore, PVL-positive MRSA assigned to CC152 (ST152-SCCmec-NT) was reported in Nigeria (Shittu et al., 2012).
Discussion
MRSA has been reported in Africa, at least since 1978 (Scragg et al., 1978). This systematic review showed that adequate data on the molecular epidemiology of MRSA are limited, with reports from only 15 of the 54 African countries. No spa type was dominant, however, t042 and t044 were the major spa types identified in three North African countries (Table 2). Moreover, we did not observe a clear distinction between hospital- and community-associated MRSA clones in Africa which is in agreement with other investigations worldwide (Fossum Moen et al., 2013; Pasquale et al., 2013; Sherwood et al., 2013; Tavares et al., 2013). In this systematic review, the use of the current eBURST scheme grouped several African MRSA CCs (CC1, CC5, CC8, and CC7) into a single cluster, (CC5). This raises some concern on a suitable method for discrimination and grouping of S. aureus isolates. To overcome the above mentioned issue, whole genome sequencing approach might be the alternative (Dabul and Camargo, 2014).
Although a combination of factors could be responsible for the dissemination of clones between continents, increased movement of human population within or between countries might be one of the potential factors (Rogers et al., 2011). International travel could play a significant role in the transmission of MRSA, particularly the replacement of existing MRSA with fitter and more transmissible clones (Zhou et al., 2014). We observed that the predominant hospital-associated epidemic clones, EMRSA-15 [ST22-IV [2B]) and (EMRSA-16 [ST36-II [2A]), in the United Kingdom (UK) (Johnson et al., 2005) were reported only in South Africa (Goering et al., 2008; Moodley et al., 2010; Jansen van Rensburg et al., 2011; Oosthuysen et al., 2014). Moreover, ST80-IV [2B] (the European clone) has consistently been recognized as the predominant PVL-positive MRSA clone in North Africa (Ramdani-bouguessa et al., 2006; Bekkhoucha et al., 2009; Ben Nejma et al., 2009, 2013; Enany et al., 2010; Antri et al., 2011; Ben Slama et al., 2011; Ben Jomàa-Jemili et al., 2013; Djoudi et al., 2013). A recent report based on whole genome analysis provided strong evidence that the European ST80-IV [2B] was derived from a PVL-positive MSSA ancestor in sub-Saharan Africa that acquired the SCCmec IV element, and clonal spread was enhanced by increased transnational movement (Stegger et al., 2014). However, the factors responsible for the limited spread of the ST80-IV [2B] only in North Africa observed so far are unclear.
The SCCmec IV (and its subtypes) and SCCmec V were identified in several MRSA clones, and ST5 and ST8 clearly showed more diversity in terms of SCCmec types compared with other STs in Africa. The success of these SCCmec types (IV and V) could be due to their small sizes and low fitness costs (Enright et al., 2002; Okuma et al., 2002; Monecke et al., 2011). It is also noteworthy that the SCCmec types IVa and IVc were identified in genetically unrelated clones, e.g., ST5-IVa [2B] (CC5) in Algeria (Ramdani-bouguessa et al., 2006; Bekkhoucha et al., 2009), São Tomé and Príncipe (Conceição et al., 2013), ST8-IVc [2B] in Tunisia (CC5) (Kechrid et al., 2011), and ST37-IVa [2B] (CC30) in Algeria (Ramdani-bouguessa et al., 2006). This might suggest horizontal gene transfer or independent acquisition (Mašlaòová et al., 2013). Another interesting observation was the detection of the SCCmec type IVa and V in the hospital-associated ST239/ST241-III [3A] in Algeria (Ramdani-bouguessa et al., 2006), Nigeria (Ghebremedhin et al., 2009), and Niger (Breurec et al., 2011b). Since ancient MSSA strains for this ST have not been reported (Enright et al., 2002), our observation suggests that acquisition of these SCCmec types by MSSA is less likely, and points to the possible replacement of SCCmec type III with IV and V on the ST239/241 genome (Li et al., 2013).
Data on the epidemiology of MRSA in animals are limited in Africa (EL Seedy et al., 2012; Fall et al., 2012; Gharsa et al., 2012). Moreover, the genetic relatedness between human and animal MRSA has not been investigated (Table 2). It should be noted, however, that human-associated ST5-IV [2B], ST88-IV [2B], and ST153-SCCmec-NT have been reported from animals in Tunisia (Gharsa et al., 2012) and Senegal (Fall et al., 2012). Recently, human-associated S. aureus lineages were described in captive Chimpanzees in Gabon, Madagascar, Uganda and Zambia (Schaumburg et al., 2012, 2013; Nagel et al., 2013). Notably, a likely case of S. aureus transmission from a veterinarian to a chimpanzee from the same sanctuary was demonstrated (Schaumburg et al., 2012). Zoonotic transmission may constitute a major concern in Africa, where there is often substantial exposure to domesticated animals (Fall et al., 2012; Gharsa et al., 2012). Furthermore, animal-adapted clones might undergo further host-adaptive evolutionary changes, which could result in an epidemic spread of new and more virulent strains in the human population (Spoor et al., 2013). Other risk factors for animal to human MRSA transmission, which include contaminated environment (Verkade and Kluytmans, 2013) and meat products (Hamid and Youssef, 2013), have not been investigated in Africa. Livestock-associated MRSA are widespread in Europe, but the transmission of these strains to humans is either rare or limited to people with direct contact with MRSA infected/carrier animals (Verkade and Kluytmans, 2013). Using whole genome sequencing, evidence of zoonotic transmission of MRSA harboring mecC was reported in Denmark (Harrison et al., 2013). The mecC-positive MRSA, initially known as a livestock MRSA belonging to the CC130, is recognized in both animals and humans in Europe (Paterson et al., 2014a). In addition, this clone has been implicated in severe infections in humans (Paterson et al., 2014b), resulting in one death (García-Garrote et al., 2014). The clinical importance of mecC-positive MRSA is not yet clear in Africa as data is unavailable. Therefore, we suggest that surveillance for MRSA should include detection of the mecC gene where mecA is not detected in resistant isolates.
This systematic review did not seek to provide comprehensive information on the burden of PVL-positive MRSA in Africa. However, it provided some interesting observations on their epidemiology in Africa such as the identification of PVL-positive isolates assigned with CC7 (originally classified with CC152) in Ghana (Egyir et al., 2014a), CC88-IV [2B] in Cameroon (Breurec et al., 2011b) and Nigeria (Ghebremedhin et al., 2009), ST612-IV [2B] in South Africa (Oosthuysen et al., 2014), and CC152 in Nigeria (Shittu et al., 2012). Until now, CC152 was only described in the Balkan region (Francois et al., 2008). The mode of acquisition of the mecA gene by ST152 is still unknown, but it might be explained by either its introduction through international travel or the acquisition of the methicillin resistance gene by PVL-positive MSSA, which is prevalent in West and Central Africa (Ruimy et al., 2008; Okon et al., 2009; Breurec et al., 2011a; Schaumburg et al., 2011; Shittu et al., 2011, 2012; Egyir et al., 2014a). These observations highlight the need for further surveillance data (including information on community-acquired necrotizing pneumonia) to understand the epidemiology of PVL-associated S. aureus in both hospital and community settings on the African continent.
Conclusion
A number of pandemic MRSA clones were identified in Africa. In contrast, some MRSA clones are limited to specific countries or regions. Although the eBURST snapshot provided a description of the relationship between the MRSA clones reported in Africa and other lineages submitted into the MLST database from other continents, the objective of this review was not to understand the origin of MRSA clones in Africa, as this will require in depth analysis like whole genome sequencing. However, it did show that CC5 is the largest group and predominant in Africa. Nevertheless, the limited data available on MRSA in Africa draw attention to the need for increased surveillance of MRSA and molecular epidemiological studies. We strongly recommend improved co-operation between clinicians and microbiologists in Africa. This synergy could provide an understanding on the local epidemiology of MRSA. In addition, we strongly advocate the establishment of effective diagnostic microbiology facilities that will incorporate high-throughput technologies for monitoring the clonal expansion and dissemination of MRSA. In the meantime, increased networking through collaboration with S. aureus reference centers could provide support for genotyping services to African countries with limited resources. Finally, population-based surveillances for MRSA are needed to evaluate the situation of community associated MRSA as well as studies on MRSA from animal hosts. To understand the origin of the newly emerged clones, MSSA genotyping needs to be incorporated with MRSA surveillance studies.
Author contributions
MK, AS, and SMA initiated the project. SMA extracted the data and reviewed the articles with MK. SMA, AS, MN, and MK wrote the manuscript. All the authors reviewed the final version of the manuscript prior to submission for publication
Financial support
This systematic review was supported by the Organization for Women in Science for the Developing World; Clinical Infectious Diseases Research Initiative, University of Cape Town, South Africa; Bill and Melinda Gates Foundation Global Health Grant (OPP107641), United States of America; Deutscher Akademischer Austausch Dienst, Germany; Carnegie Corporation of New York, United States of America. Any opinions, findings and conclusions, or recommendations expressed in this review are those of the authors, and therefore do not represent the official position of the funders.
Conflict of interest statement
The authors have no conflict of interest to declare related to the content of this paper. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The first and corresponding authors had full access to the study data. All authors had final responsibility for the decision to submit the article for publication. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
SMA is supported by the Organization for Women in Science for the developing World (OWSD) and the Drakenstein Child Health Study, Cape Town, South Africa; AS was supported by Deutscher Akademischer Austausch Dienst (DAAD award) Staff Exchange Programme (2013); MK was a recipient of Carnegie Corporation of New York (USA) fellowship and is currently a Wellcome Trust (UK) Fellow.
Footnotes
1eBURST [Online]. Available online at: http://www.mlst.net version 3.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00348/abstract
References
- Adesida S., Boelens H., Babajide B., Kehinde A., Snijders S., van Leeuwen W., et al. (2005). Major epidemic clones of Staphylococcus aureus in Nigeria. Microb. Drug. Resist. 11, 115–121. 10.1089/mdr.2005.11.115 [DOI] [PubMed] [Google Scholar]
- Aiken A. M., Mutuku I. M., Sabat A. J., Akkerboom V., Mwangi J., Scott J. A. G., et al. (2014). Carriage of Staphylococcus aureus in Thika Level 5 Hospital, Kenya: a cross-sectional study. Resist. Infect. Control. 3:22. 10.1186/2047-2994-3-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aires-de-Sousa M., Correia B., de Lencastre H. (2008). Changing patterns in frequency of recovery of five methicillin-resistant Staphylococcus aureus clones in Portuguese hospitals: surveillance over a 16-year period. J. Clin. Microbiol. 46, 2912–2917. 10.1128/JCM.00692-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht N., Jatzwauk L., Slickers P., Ehricht R., Monecke S. (2011). Clonal replacement of epidemic methicillin-resistant Staphylococcus aureus strains in a German university hospital over a period of eleven years. PLoS ONE 6:e28189. 10.1371/journal.pone.0028189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-haddad O. H., Zorgani A., Ghenghesh K. S. (2014). Nasal carriage of multi-drug resistant Panton-Valentine leucocidin-positive methicillin-resistant Staphylococcus aureus in children in Tripoli-Libya. Am. J. Trop. Med. Hyg. 90, 724–727. 10.4269/ajtmh.13-0715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antri K., Rouzic N., Dauwalder O., Boubekri I., Bes M., Lina G., et al. (2011). High prevalence of methicillin-resistant Staphylococcus aureus clone ST80-IV in hospital and community settings in Algiers. Clin. Microbiol. Infect. 17, 526–532. 10.1111/j.1469-0691.2010.03273.x [DOI] [PubMed] [Google Scholar]
- Ateba Ngoa U., Schaumburg F., Adegnika A. A., Kösters K., Möller T., Fernandes J. F., et al. (2012). Epidemiology and population structure of Staphylococcus aureus in various population groups from a rural and semi urban area in Gabon, Central Africa. Acta Tropica 124, 42–47. 10.1016/j.actatropica.2012.06.005 [DOI] [PubMed] [Google Scholar]
- Ba X., Harrison E. M., Edwards G. F., Holden M. T. G., Larsen A. R., Petersen A., et al. (2014). Novel mutations in penicillin-binding protein genes in clinical Staphylococcus aureus isolates that are methicillin resistant on susceptibility testing, but lack the mec gene. J. Antimicrob. Chemother. 69, 594–597. 10.1093/jac/dkt418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekkhoucha S. N., Cady A., Gautier P., Itim F., Donnio P. Y. (2009). A portrait of Staphylococcus aureus from the other side of the Mediterranean Sea: molecular characteristics of isolates from Western Algeria. Eur. J. Clin. Microbiol. Infect. Dis. 28, 553–555. 10.1007/s10096-008-0660-x [DOI] [PubMed] [Google Scholar]
- Ben Ayed S., Boutiba-Ben Boubaker I., Boukadida J., Hammami S., Ben Redjeb S. (2010). Hospital acquired outbreak of methicillin-resistant Staphylococcus aureus infection initiated by a health care worker. Tunis. Med. 88, 199–202. [PubMed] [Google Scholar]
- Ben Ayed S., Boutiba-Ben Boubaker I., Samir E., Ben Redjeb S. (2006). Prevalence of agr specificity groups among methicilin resistant Staphylococcus aureus circulating at Charles Nicolle hospital of Tunis. Pathol. Biol. 54, 435–438. 10.1016/j.patbio.2006.07.010 [DOI] [PubMed] [Google Scholar]
- Ben Jomaa-Jemili M., Boutiba-Ben Boubaker I., Ben Redjeb S. (2006). Identification of staphylococcal cassette chromosome mec encoding methicillin resistance in Staphylococcus aureus isolates at Charles Nicolle Hospital of Tunis. Pathol. Biol. 54, 453–455. 10.1016/j.patbio.2006.07.013 [DOI] [PubMed] [Google Scholar]
- Ben Jomàa-Jemili M., Ito T., Zhang M., Jin J., Li S., Ilhem B.-B. B., et al. (2013). Molecular characterization of methicillin-resistant Panton-valentine leukocidin positive Staphylococcus aureus clones disseminating in Tunisian hospitals and in the community. BMC Microbiol. 13:2. 10.1186/1471-2180-13-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Nejma M. B. E. N., Mastouri M., Bel B., Jrad H., Makhlouf M., Nour M. (2009). Community-acquired methicillin resistant isolated in dermatology department Staphylococcus aureus. Ann. Microbiol. 59, 823–826 10.1007/BF03179229 [DOI] [Google Scholar]
- Ben Nejma M., Mastouri M., Bel Hadj Jrad B., Nour M. (2013). Characterization of ST80 Panton-Valentine leukocidin-positive community-acquired methicillin-resistant Staphylococcus aureus clone in Tunisia. Diagn. Microbiol. Infect. Dis. 77, 20–24. 10.1016/j.diagmicrobio.2008.02.010 [DOI] [PubMed] [Google Scholar]
- Ben Nejma M., Mastouri M., Frih S., Sakly N., Ben Salem Y., Nour M. (2006). Molecular characterization of methicillin-resistant Staphylococcus aureus isolated in Tunisia. Diagn. Microbiol. Infect. Dis. 55, 21–26. 10.1016/j.diagmicrobio.2005.10.017 [DOI] [PubMed] [Google Scholar]
- Ben Saida N., Ben Abdallah H., Hannachi N., Boukadida J. (2005). Multiclonality of methicillin-resistant Staphylococcus aureus in a university hospital. Méd. Mal. Infect. 35, 363–366. 10.1016/j.medmal.2005.02.014 [DOI] [PubMed] [Google Scholar]
- Ben Slama K., Gharsa H., Klibi N., Jouini, A, Lozano C., Gómez-Sanz E., et al. (2011). Nasal carriage of Staphylococcus aureus in healthy humans with different levels of contact with animals in Tunisia: genetic lineages, methicillin resistance, and virulence factors. Eur. J. Clin. Microbiol. Infect. Dis. 30, 499–508. 10.1007/s10096-010-1109-6 [DOI] [PubMed] [Google Scholar]
- Boakes E., Kearns A. M., Ganner M., Perry C., Hill R. L., Ellington M. J. (2011). Distinct bacteriophages encoding Panton-Valentine leukocidin (PVL) among international methicillin-resistant Staphylococcus aureus clones harboring PVL. J. Clin. Microbiol. 49, 684–692. 10.1128/JCM.01917-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchami O., Achour W., Ben Hassen A. (2009). Typing of staphylococcal cassette chromosome mec encoding methicillin resistance in Staphylococcus aureus strains isolated at the bone marrow transplant centre of Tunisia. Curr. Microbiol. 59, 380–385. 10.1007/s00284-009-9448-1 [DOI] [PubMed] [Google Scholar]
- Breurec S., Fall C., Pouillot R., Boisier P., Brisse S., Diene-Sarr F., et al. (2011a). Epidemiology of methicillin-susceptible Staphylococcus aureus lineages in five major African towns: high prevalence of Panton-Valentine leukocidin genes. Clin. Microbiol. Infect. 17, 633–639. 10.1111/j.1469-0691.2010.03320.x [DOI] [PubMed] [Google Scholar]
- Breurec S., Zriouil S., Fall C., Boisier P., Brisse S., Djibo S., et al. (2011b). Epidemiology of methicillin-resistant Staphylococcus aureus lineages in five major African towns: emergence and spread of atypical clones. Clin. Microbiol. Infect. 17, 160–165. 10.1111/j.1469-0691.2010.03219.x [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and prevention. (2013). Antibiotic Resistance Threats in the United States, 508 Available online at: http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf
- Chambers H. F. (1997). Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin. Microbiol. Rev. 10, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers H. F., Deleo F. R. (2010). Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 7, 629–641. 10.1038/nrmicro2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S. S., Otto M. (2013). Improved understanding of factors driving epidemic waves. Clin. Epidemiol. 5, 205–217. 10.2147/CLEP.S37071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. J., Huang Y.-C. (2014). New epidemiology of Staphylococcus aureus infection in Asia. Clin. Microbiol. Infect. 20, 605–623. 10.1111/1469-0691.12705 [DOI] [PubMed] [Google Scholar]
- Conceição T., Aires-de-Sousa M., Füzi M., Tóth A., Pászti J., Ungvári E., et al. (2007). Replacement of methicillin-resistant Staphylococcus aureus clones in Hungary over time: a 10-year surveillance study. Clin. Microbiol. Infect. 13, 971–979. 10.1111/j.1469-0691.2007.01794.x [DOI] [PubMed] [Google Scholar]
- Conceição T., Coelho C., Santos-Silva I., de Lencastre H., Aires-de-Sousa M. (2014). Epidemiology of methicillin-resistant and -susceptible Staphylococcus aureus in Luanda, Angola: first description of the spread of the MRSA ST5-IVa clone in the African continent. Microb. Drug Resist. 20, 441–449. 10.1089/mdr.2014.0007 [DOI] [PubMed] [Google Scholar]
- Conceição T., Silva I. S., de Lencastre H., Aires-de-Sousa M. (2013). Staphylococcus aureus nasal carriage among patients and health care workers in São Tomé and Príncipe. Microb. Drug Resist. 20, 57–66. 10.1089/mdr.2013.0136 [DOI] [PubMed] [Google Scholar]
- Coombs G., Nimmo G., Pearson J., Christiansen K., Bell JM, Collignon PJ, et al. (2009). Prevalence of MRSA strains among Staphylococcus aureus isolated from outpatients. Commun. Dis. Intell. Q. Rep. 33, 10–20. [DOI] [PubMed] [Google Scholar]
- Dabul A. N. G., Camargo I. L. B. C. (2014). Clonal complexes of Staphylococcus aureus: all mixed and together. FEMS Microbiol. Lett. 35, 7–8. 10.1111/1574-6968.12358 [DOI] [PubMed] [Google Scholar]
- De Kraker M. E. A, Davey P. G., Grundmann H. (2011). Mortality and hospital stay associated with resistant Staphylococcus aureus and Escherichia coli bacteremia; estimating the burden of antibiotic resistance in Europe. PLoS Med. 8:e1001104. 10.1371/journal.pmed.1001104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deurenberg R. H., Stobberingh E. E. (2008). The evolution of Staphylococcus aureus. Infect. Genet. Evol. 8, 747–763. 10.1016/j.meegid.2008.07.007 [DOI] [PubMed] [Google Scholar]
- Djoudi F., Bonura C., Benallaoua S., Touati A., Touati D., Aleo A., et al. (2013). Panton-Valentine leukocidin positive sequence type 80 methicillin-resistant Staphylococcus aureus carrying a staphylococcal cassette chromosome mec type IVc is dominant in neonates and children in an Algiers hospital. New Microbiol. 36, 49–55. [PubMed] [Google Scholar]
- D'Souza N., Rodrigues C., Mehta A. (2010). Molecular characterization of methicillin-resistant Staphylococcus aureus with emergence of epidemic clones of sequence type (ST)22 and ST772 in Mumbai, India. J. Clin. Microbiol. 48, 1806–1811. 10.1128/JCM.01867-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egyir B., Guardabassi L., Esson J., Nielsen S. S., Newman M. J., Addo K. K., et al. (2014a). Insights into nasal carriage of Staphylococcus aureus in an urban and a rural community in Ghana. PLoS ONE 9:e96119. 10.1371/journal.pone.0096119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egyir B., Guardabassi L., Nielsen S. S., Larsen J., Addo K. K., Newman M. J., et al. (2013). Prevalence of nasal carriage and diversity of Staphylococcus aureus among inpatients and hospital staff at Korle Bu Teaching Hospital, Ghana. J. Glob. Antimicrob. Resist. 1, 189–193 10.1016/j.jgar.2013.05.006 [DOI] [PubMed] [Google Scholar]
- Egyir B., Guardabassi L., Sørum M., Nielsen S. S., Kolekang A., Frimpong E., et al. (2014b). Molecular epidemiology and antimicrobial susceptibility of clinical Staphylococcus aureus from healthcare institutions in Ghana. PLoS ONE 9:e89716. 10.1371/journal.pone.0089716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Jakee J. K., Atta N., Samy A., Bakry M. E. A. E., Kandil M., Gad El-Said W. (2011). Antimicrobial resistance in clinical isolates of Staphylococcus aureus from bovine and human sources in Egypt. Glob. Vet. 7, 581–586. [Google Scholar]
- EL Seedy F. R., El Hakim A. S., Syame S. F., Osman N. M. (2012). Advanced techniques used for isolation and characterization of Staphylococcus aureus isolated from mastitic buffaloes. Glob. Vet. 8, 144–152. [Google Scholar]
- Enany S., Yaoita E., Yoshida Y., Enany M., Yamamoto T. (2010). Molecular characterization of Panton-Valentine leukocidin-positive community-acquired methicillin-resistant Staphylococcus aureus isolates in Egypt. Microbiol. Res. 165, 152–162. 10.1016/j.micres.2009.03.005 [DOI] [PubMed] [Google Scholar]
- Enright M. C., Robinson D. A., Randle G., Feil E. J., Grundmann H., Spratt B. G. (2002). The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. U.S.A. 99, 7687–7692. 10.1073/pnas.122108599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espadinha D., Faria N. A., Miragaia M., Lito L. M., Melo-Cristino J., de Lencastre H. (2013). Extensive dissemination of methicillin-resistant Staphylococcus aureus (MRSA) between the hospital and the community in a country with a high prevalence of nosocomial MRSA. PLoS ONE 8:e59960. 10.1371/journal.pone.0059960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essa Z. I., Connolly C., Essack S. Y. (2009). Staphylococcus aureus from public hospitals in KwaZulu-Natal, South Africa—infection detection and strain-typing. South African J. Epidemiol. Infect. 24, 4–7. [Google Scholar]
- Falagas M. E., Karageorgopoulos D. E., Leptidis J., Korbila I. P. (2013). MRSA in Africa: filling the global map of antimicrobial resistance. PLoS ONE 8:e68024. 10.1371/journal.pone.0068024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fall C., Seck A., Richard V., Ndour M., Sembene M., Laurent F., et al. (2012). Epidemiology of Staphylococcus aureus in pigs and farmers in the largest farm in Dakar, Senegal. Foodborne Pathog. Dis. 9, 962–965. 10.1089/fpd.2012.1197 [DOI] [PubMed] [Google Scholar]
- Fossum Moen A. E., Tannaes T. M., Leegaard T. M. (2013). USA300 methicillin-resistant Staphylococcus aureus in Norway. APMIS 121, 1091–1096. 10.1111/apm.12077 [DOI] [PubMed] [Google Scholar]
- Francois P., Harbarth S., Huyghe A., Renzi G. (2008). Methicillin-resistant Staphylococcus aureus, Geneva, Switzerland, 1993-2005. Emerg. Infect. Dis. 14, 304–307. 10.3201/eid1402.070229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Garrote F., Cercenado E., Marín M., Bal M., Trincado P., Corredoira J., et al. (2014). Methicillin-resistant Staphylococcus aureus carrying the mecC gene: emergence in Spain and report of a fatal case of bacteraemia. J. Antimicrob. Chemother. 69, 45–50. 10.1093/jac/dkt327 [DOI] [PubMed] [Google Scholar]
- Garza-González E., Dowzicky M. J. (2013). Changes in Staphylococcus aureus susceptibility across Latin America between 2004 and 2010. Braz. J. Infect. Dis. 17, 13–19. 10.1016/j.bjid.2012.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharsa H., Ben Slama K., Lozano C., Gómez-Sanz E., Klibi N., Ben Sallem R., et al. (2012). Prevalence, antibiotic resistance, virulence traits and genetic lineages of Staphylococcus aureus in healthy sheep in Tunisia. Vet. Microbiol. 156, 367–373. 10.1016/j.vetmic.2011.11.009 [DOI] [PubMed] [Google Scholar]
- Ghebremedhin B., Olugbosi M. O., Raji A. M., Layer F., Bakare R. A., König B., et al. (2009). Emergence of a community-associated methicillin-resistant Staphylococcus aureus strain with a unique resistance profile in Southwest Nigeria. J. Clin. Microbiol. 47, 2975–2980. 10.1128/JCM.00648-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goering R. V, Shawar R. M., Scangarella N. E., O'Hara F. P., Amrine-Madsen H., West J. M., et al. (2008). Molecular epidemiology of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates from global clinical trials. J. Clin. Microbiol. 46, 2842–2847. 10.1128/JCM.00521-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid D., Youssef A. (2013). Clinical Features of Methicillin-resistant Staphylococcus aureus (MRSA) Infection in Rabbits and its Zoonotic potentials. Pakistan J. Nutr. 12, 244–249 10.3923/pjn.2013.244.249 [DOI] [Google Scholar]
- Harrison E. M., Paterson G. K., Holden M. T. G., Larsen J., Stegger M., Larsen A. R., et al. (2013). Whole genome sequencing identifies zoonotic transmission of MRSA isolates with the novel mecA homologue mecC. EMBO Mol. Med. 5, 509–515. 10.1002/emmm.201202413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson M. A. M., Kalkman R., Remppis J., Beyeme J. O., Kraef C., Schaumburg F., et al. (2014). Methicillin-resistant Staphylococcus aureus as a cause of invasive infections in Central Africa: a case report and review of the literature. Infection 42, 451–457. 10.1007/s15010-014-0589-1 [DOI] [PubMed] [Google Scholar]
- Ito T., Katayama Y., Asada K., Mori N. (2001). Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45, 1323–1336. 10.1128/AAC.45.5.1323-1336.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Ma X. X., Takeuchi F., Okuma K. (2004). Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 48, 2637–2651. 10.1128/AAC.48.7.2637-2651.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IWG-SCC. (2009). Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother. 53, 4961–4967. 10.1128/AAC.00579-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen van Rensburg M. J., Eliya Madikane V., Whitelaw A., Chachage M., Haffejee S., Gay Elisha B. (2011). The dominant methicillin-resistant Staphylococcus aureus clone from hospitals in Cape Town has an unusual genotype: ST612. Clin. Microbiol. Infect. 17, 785–792. 10.1111/j.1469-0691.2010.03373.x [DOI] [PubMed] [Google Scholar]
- Jansen van Rensburg M. J., Whitelaw A. C., Elisha B. G. (2012). Genetic basis of rifampicin resistance in methicillin-resistant Staphylococcus aureus suggests clonal expansion in hospitals in Cape Town, South Africa. BMC Microbiol. 12:46. 10.1186/1471-2180-12-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. P., Pearson A., Duckworth G. (2005). Surveillance and epidemiology of MRSA bacteraemia in the UK. J. Antimicrob. Chemother. 56, 455–462. 10.1093/jac/dki266 [DOI] [PubMed] [Google Scholar]
- Kateete D. P., Namazzi S., Okee M., Okeng A., Baluku H., Musisi N. L., et al. (2011). High prevalence of methicillin resistant Staphylococcus aureus in the surgical units of Mulago hospital in Kampala, Uganda. BMC Res. Notes 4:326. 10.1186/1756-0500-4-326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kechrid A., Pérez-Vázquez M., Smaoui H., Hariga D., Rodríguez-Baños M., Vindel A., et al. (2011). Molecular analysis of community-acquired methicillin-susceptible and resistant Staphylococcus aureus isolates recovered from bacteraemic and osteomyelitis infections in children from Tunisia. Clin. Microbiol. Infect. 17, 1020–1026. 10.1111/j.1469-0691.2010.03367.x [DOI] [PubMed] [Google Scholar]
- Kim C., Milheiriço C., Gardete S., Holmes M. A., Holden M. T. G., de Lencastre H., et al. (2012). Properties of a novel PBP2A protein homolog from Staphylococcus aureus strain LGA251 and its contribution to the β-lactam-resistant phenotype. J. Biol. Chem. 287, 36854–36863. 10.1074/jbc.M112.395962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. S., Song J. S., Lee H. J., Choe P. G., Park K. H., Cho J. H., et al. (2007). A survey of community-associated methicillin-resistant Staphylococcus aureus in Korea. J. Antimicrob. Chemother. 60, 1108–1114. 10.1093/jac/dkm309 [DOI] [PubMed] [Google Scholar]
- Köck R., Becker K., Cookson B., van Gemert-Pijnen J. E., Harbarth S., Kluytmans J., et al. (2010). Methicillin-resistant Staphylococcus aureus (MRSA): burden of disease and control challenges in Europe. Euro. Surveill. 15, 19688. [DOI] [PubMed] [Google Scholar]
- Lee B. Y., Singh A., David M. Z., Bartsch S. M., Slayton R. B., Huang S. S., et al. (2013). The economic burden of community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA). Clin. Microbiol. Infect. 19, 528–536. 10.1111/j.1469-0691.2012.03914.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Song Y., Zhu Y., Du X., Li M. (2013). Current status of Staphylococcus aureus infection in a central teaching hospital in Shanghai, China. BMC Microbiol. 13:153. 10.1186/1471-2180-13-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K. T., Hanifah Y. A., Mohd Yusof M. Y., Ito T., Thong K. L. (2013). Comparison of methicillin-resistant Staphylococcus aureus strains isolated in 2003 and 2008 with an emergence of multidrug resistant ST22: SCCmec IV clone in a tertiary hospital, Malaysia. J. Microbiol. Immunol. Infect. 46, 224–233. 10.1016/j.jmii.2013.02.001 [DOI] [PubMed] [Google Scholar]
- Löffler B., Hussain M., Grundmeier M., Brück M., Holzinger D., Varga G., et al. (2010). Staphylococcus aureus Panton-Valentine leukocidin is a very potent cytotoxic factor for human neutrophils. PLoS Pathog. 6:e1000715. 10.1371/journal.ppat.1000715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X. X., Ito T., Tiensasitorn C., Chongtrakool P., Boyle-Vavra S., Daum R. S., et al. (2002). Novel type of staphylococcal cassette chromosome mec identified in Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46, 1147–1152. 10.1128/AAC.46.4.1147-1152.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maina E. K., Kiiyukia C., Wamae C. N., Waiyaki P. G., Kariuki S. (2013). Characterization of methicillin-resistant Staphylococcus aureus from skin and soft tissue infections in patients in Nairobi, Kenya. Int. J. Infect. Dis. 17, e115–e119. 10.1016/j.ijid.2012.09.006 [DOI] [PubMed] [Google Scholar]
- Makgotlho P. E., Kock M. M., Hoosen A., Lekalakala R., Omar S., Dove M., et al. (2009). Molecular identification and genotyping of MRSA isolates. FEMS Immunol. Med. Microbiol. 57, 104–115. 10.1111/j.1574-695X.2009.00585.x [DOI] [PubMed] [Google Scholar]
- Mašlaòová I., Doškaø J., Varga M., Kuntová L., Mužík J., Malúšková D., et al. (2013). Bacteriophages of Staphylococcus aureus efficiently package various bacterial genes and mobile genetic elements including SCCmec with different frequencies. Environ. Microbiol. Rep. 5, 66–73. 10.1111/j.1758-2229.2012.00378.x [DOI] [PubMed] [Google Scholar]
- Moher D., Liberati A., Tetzlaff J., Altman D. G. (2009). Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 6:e1000097. 10.1371/journal.pmed.1000097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monecke S., Coombs G., Shore A. C., Coleman D. C., Akpaka P., Borg M., et al. (2011). A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS ONE 6:e17936. 10.1371/journal.pone.0017936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moodley A., Oosthuysen W. F., Dusé A. G., Marais E. (2010). Molecular characterization of clinical methicillin-resistant Staphylococcus aureus isolates in South Africa. J. Clin. Microbiol. 48, 4608–4611. 10.1128/JCM.01704-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moremi N., Mshana S. E., Kamugisha E., Kataraihya J. B., Tappe D., Vogel U., et al. (2012). Predominance of methicillin resistant Staphylococcus aureus -ST88 and new ST1797 causing wound infection and abscesses. J. Infect. Dev. Ctries. 6, 620–625. 10.3855/jidc.2093 [DOI] [PubMed] [Google Scholar]
- Moyo S. J., Aboud S., Blomberg B., Mkopi N., Kasubi M., Manji K., et al. (2014). High Nasal carriage of methicillin-resistant Staphylococcus aureus among healthy tanzanian under-5 children. Microb. Drug Resist. 20, 82–88. 10.1089/mdr.2013.0016 [DOI] [PubMed] [Google Scholar]
- Nagel M., Dischinger J., Türck M., Verrier D., Oedenkoven M., Ngoubangoye B., et al. (2013). Human-associated Staphylococcus aureus strains within great ape populations in Central Africa (Gabon). Clin. Microbiol. Infect. 19, 1072–1077. 10.1111/1469-0691.12119 [DOI] [PubMed] [Google Scholar]
- Nimmo G. R., Bergh H., Nakos J., Whiley D., Marquess J., Huygens F., et al. (2013). Replacement of healthcare-associated MRSA by community-associated MRSA in Queensland: confirmation by genotyping. J. Infect. 67, 439–447. 10.1016/j.jinf.2013.07.020 [DOI] [PubMed] [Google Scholar]
- Okon K. O., Basset P., Uba A., Lin J., Oyawoye B., Shittu A. O., et al. (2009). Cooccurrence of predominant Panton-Valentine leukocidin-positive sequence type (ST) 152 and multidrug-resistant ST 241 Staphylococcus aureus clones in Nigerian hospitals. J. Clin. Microbiol. 47, 3000–3333. 10.1128/JCM.01119-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okon K. O., Shittu A. O., Kudi A. A., Umar H., Becker K., Schaumburg F. (2014). Population dynamics of Staphylococcus aureus from Northeastern Nigeria in 2007 and 2012. Epidemiol. Infect. 142, 1737–1740. 10.1017/S0950268813003117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuma K., Iwakawa K., Turnidge J. D., Warren B., Bell J. M., Brien F. G. O., et al. (2002). Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40, 4289–4294. 10.1128/JCM.40.11.4289-4294.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olalekan A. O., Schaumburg F., Nurjadi D., Dike A. E., Ojurongbe O., Kolawole D. O., et al. (2012). Clonal expansion accounts for an excess of antimicrobial resistance in Staphylococcus aureus colonising HIV-positive individuals in Lagos, Nigeria. Int. J. Antimicrob. Agents 40, 268–272. 10.1016/j.ijantimicag.2012.05.016 [DOI] [PubMed] [Google Scholar]
- Oosthuysen W. F., Orth H., Lombard C. J., Sinha B., Wasserman E. (2014). Population structure analyses of Staphylococcus aureus at Tygerberg Hospital, South Africa, reveals a diverse population, high prevalence of Panton-Valentine leukocidin genes and unique local MRSA clones. Clin. Microbiol. Infect. 20, 652–659. 10.1111/1469-0691.12452 [DOI] [PubMed] [Google Scholar]
- Ouchenane Z., Agabou A., Smati F., Rolain J.-M., Raoult D. (2013). Staphylococcal cassette chromosome mec characterization of methicillin-resistant Staphylococcus aureus strains isolated at the military hospital of Constantine/Algeria. Pathol. Biol. 61, 280–281. 10.1016/j.patbio.2013.05.006 [DOI] [PubMed] [Google Scholar]
- Ouchenane Z., Smati F., Rolain J.-M., Raoult D. (2011). Molecular characterization of methicillin-resistant Staphylococcus aureus isolates in Algeria. Pathol. Biol. 59, e129–e132. 10.1016/j.patbio.2009.11.004 [DOI] [PubMed] [Google Scholar]
- Pasquale T. R., Jabrocki B., Salstrom S. J., Wiemken T. L., Peyrani P., Haque N. Z., et al. (2013). Emergence of methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of late-onset nosocomial pneumonia in intensive care patients in the USA. Int. J. Infect. Dis. 17, e398–e403. 10.1016/j.ijid.2012.12.013 [DOI] [PubMed] [Google Scholar]
- Paterson G. K., Harrison E. M., Holmes M. A. (2014a). The emergence of mecC methicillin-resistant Staphylococcus aureus. Trends Microbiol. 22, 42–47. 10.1016/j.tim.2013.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson G. K., Morgan F. J. E., Harrison E. M., Cartwright E. J. P., Török M. E., Zadoks R. N., et al. (2014b). Prevalence and characterization of human mecC methicillin-resistant Staphylococcus aureus isolates in England. J. Antimicrob. Chemother. 69, 907–910. 10.1093/jac/dkt462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raji A., Ojemhen O., Umejiburu U., Ogunleye A., Blanc D. S., Basset P. (2013). High genetic diversity of Staphylococcus aureus in a tertiary care hospital in Southwest Nigeria. Diagn. Microbiol. Infect. Dis. 77, 367–369. 10.1016/j.diagmicrobio.2013.08.030 [DOI] [PubMed] [Google Scholar]
- Ramdani-bouguessa N., Bes M., Meugnier H., Forey F., Reverdy M., Lina G., et al. (2006). Detection of methicillin-resistant Staphylococcus aureus strains resistant to multiple antibiotics and carrying the panton-valentine leukocidin genes in an Algiers Hospital. Antimicrob. Agents Chemother. 50, 1083–1085. 10.1128/AAC.50.3.1083-1085.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D. A., Enright M. C. (2004). Evolution of Staphylococcus aureus by large chromosomal replacements. J. Bacteriol. 186, 1060–1064. 10.1128/JB.186.4.1060-1064.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers B. A., Aminzadeh Z., Hayashi Y., Paterson D. L. (2011). Country-to-country transfer of patients and the risk of multi-resistant bacterial infection. Clin. Infect. Dis. 53, 49–56. 10.1093/cid/cir273 [DOI] [PubMed] [Google Scholar]
- Ruimy R., Maiga A., Armand-Lefevre L., Maiga I., Diallo A., Koumaré A. K., et al. (2008). The carriage population of Staphylococcus aureus from Mali is composed of a combination of pandemic clones and the divergent Panton-Valentine leukocidin-positive genotype ST152. J. Bacteriol. 190, 3962–3968. 10.1128/JB.01947-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaumburg F., Alabi A. S., Mombo-Ngoma G., Kaba H., Zoleko R. M., Diop D. A, et al. (2014). Transmission of Staphylococcus aureus between mothers and infants in an African setting. Clin. Microbiol. Infect. 20, 390–396. 10.1111/1469-0691.12417 [DOI] [PubMed] [Google Scholar]
- Schaumburg F., Mugisha L., Kappeller P., Fichtel C., Köck R., Köndgen S., et al. (2013). Evaluation of non-invasive biological samples to monitor Staphylococcus aureus colonization in great apes and lemurs. PLoS ONE 8:e78046. 10.1371/journal.pone.0078046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaumburg F., Mugisha L., Peck B., Becker K., Gillespie T. R., Peters G., et al. (2012). Drug-resistant human Staphylococcus aureus in sanctuary apes pose a threat to endangered wild ape populations. Am. J. Primatol. 74, 1071–1075. 10.1002/ajp.22067 [DOI] [PubMed] [Google Scholar]
- Schaumburg F., Ngoa U. A., Kösters K., Köck R., Adegnika A. A, Kremsner P. G., et al. (2011). Virulence factors and genotypes of Staphylococcus aureus from infection and carriage in Gabon. Clin. Microbiol. Infect. 17, 1507–1513. 10.1111/j.1469-0691.2011.03534.x [DOI] [PubMed] [Google Scholar]
- Scragg J. N., Appelbaum P. C., Govender D. A. (1978). The spectrum of infection and sensitivity of organisms isolated from African and Indian children in a Durban hospital. Trans. R. Soc. Trop. Med. Hyg. 72, 325–328. 10.1016/0035-9203(78)90118-9 [DOI] [PubMed] [Google Scholar]
- Seni J., Bwanga F., Najjuka C. F., Makobore P., Okee M., Mshana S. E., et al. (2013). Molecular characterization of Staphylococcus aureus from patients with surgical site infections at Mulago Hospital in Kampala, Uganda. PLoS ONE 8:e66153. 10.1371/journal.pone.0066153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shallcross L. J., Fragaszy E., Johnson A. M., Hayward A. C. (2013). The role of the Panton-Valentine leucocidin toxin in staphylococcal disease: a systematic review and meta-analysis. Lancet Infect. Dis. 13, 43–54. 10.1016/S1473-3099(12)70238-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shambat S., Nadig S., Prabhakara S., Bes M., Etienne J., Arakere G. (2012). Clonal complexes and virulence factors of Staphylococcus aureus from several cities in India. BMC Microbiol. 12:64. 10.1186/1471-2180-12-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood J., Park M., Robben P., Whitman T., Ellis M. W. (2013). USA300 methicillin-resistant Staphylococcus aureus emerging as a cause of bloodstream infections at military medical centers. Infect. Control Hosp. Epidemiol. 34, 393–399. 10.1086/669866 [DOI] [PubMed] [Google Scholar]
- Shittu A. O., Lin J. (2006a). Antimicrobial susceptibility patterns and characterization of clinical isolates of Staphylococcus aureus in KwaZulu-Natal province, South Africa. BMC Infect. Dis. 6:125. 10.1186/1471-2334-6-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shittu A. O., Lin J. (2006b). Antimicrobial susceptibility patterns of Staphylococcus aureus and characterization of MRSA in Southwestern Nigeria. Wounds 18, 77–84. [Google Scholar]
- Shittu A. O., Lin J., Morrison D. (2007). Molecular identification and characterization of mannitol-negative methicillin-resistant Staphylococcus aureus. Diagn. Microbiol. Infect. Dis. 57, 93–95. 10.1016/j.diagmicrobio.2006.05.004 [DOI] [PubMed] [Google Scholar]
- Shittu A. O., Nübel U., Udo E., Lin J., Gaogakwe S. (2009). Characterization of meticillin-resistant Staphylococcus aureus isolates from hospitals in KwaZulu-Natal province, Republic of South Africa. J. Med. Microbiol. 58, 1219–1226. 10.1099/jmm.0.011452-0 [DOI] [PubMed] [Google Scholar]
- Shittu A. O., Okon K., Adesida S., Oyedara O., Witte W., Strommenger B., et al. (2011). Antibiotic resistance and molecular epidemiology of Staphylococcus aureus in Nigeria. BMC Microbiol. 11:92. 10.1186/1471-2180-11-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shittu A. O., Oyedara O., Abegunrin F., Okon K., Raji A., Taiwo S., et al. (2012). Characterization of methicillin-susceptible and -resistant staphylococci in the clinical setting: a multicentre study in Nigeria. BMC Infect. Dis. 12:286. 10.1186/1471-2334-12-286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore A. C., Deasy E. C., Slickers P., Brennan G., O'Connell B., Monecke S., et al. (2011). Detection of staphylococcal cassette chromosome mec type XI carrying highly divergent mecA, mecI, mecR1, blaZ, and ccr genes in human clinical isolates of clonal complex 130 methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 55, 3765–3773. 10.1128/AAC.00187-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhy N., Aly F., Abd El Kader O., Ghazal A., Elbaradei A. (2012). Community-acquired methicillin-resistant Staphylococcus aureus from skin and soft tissue infections (in a sample of Egyptian population): analysis of mec gene and staphylococcal cassette chromosome. Braz. J. Infect. Dis. 16, 426–431. 10.1016/j.bjid.2012.08.004 [DOI] [PubMed] [Google Scholar]
- Spoor L. E., McAdam P. R., Weinert L. A. (2013). Livestock origin for a human pandemic clone of Community- Associated Methicillin-Resistant Staphylococcus aureus. MBio 4, e00356–e00313. 10.1128/mBio.00356-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani S., Chung D. R., Lindsay J. A., Friedrich A. W., Kearns A. M., Westh H., et al. (2012). Meticillin-resistant Staphylococcus aureus (MRSA): global epidemiology and harmonisation of typing methods. Int. J. Antimicrob. Agents 39, 273–282. 10.1016/j.ijantimicag.2011.09.030 [DOI] [PubMed] [Google Scholar]
- Stegger M., Wirth T., Andersen P. S., Skov R. L., Grassi A., De Simões M., et al. (2014). Origin and evolution of european methicillin-resistant Staphylococcus aureus. MBio 5, e01044–e01014. 10.1128/mBio.01044-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares A., Miragaia M., Rolo J., Coelho C., de Lencastre H. (2013). High prevalence of hospital-associated methicillin-resistant Staphylococcus aureus in the community in Portugal: evidence for the blurring of community-hospital boundaries. Eur. J. Clin. Microbiol. Infec. Dis. 32, 1269–1283. 10.1007/s10096-013-1872-2 [DOI] [PubMed] [Google Scholar]
- Terry Alli O. A, Ogbolu D. O., Mustapha J. O., Akinbami R., Ajayi A. O. (2012). The non-association of Panton-Valentine leukocidin and mecA genes in the genome of Staphylococcus aureus from hospitals in South Western Nigeria. Indian J. Med. Microbiol. 30, 159–164. 10.4103/0255-0857.96675 [DOI] [PubMed] [Google Scholar]
- Vandenesch F., Naimi T., Enright M. C., Lina G., Nimmo G. R., Heffernan H., et al. (2003). Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9, 978–984. 10.3201/eid0908.030089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Meeren B. T., Millard P. S., Scacchetti M., Hermans M. H., Hilbink M., Concelho T. B., et al. (2014). Emergence of methicillin resistance and Panton-Valentine leukocidin positivity in hospital- and community-acquired Staphylococcus aureus infections in Beira, Mozambique. Trop. Med. Int. Health 19, 169–176. 10.1111/tmi.12221 [DOI] [PubMed] [Google Scholar]
- Verkade E., Kluytmans J. (2013). Livestock-associated Staphylococcus aureus CC398: animal reservoirs and human infections. Infect. Genet. Evol. 21, 523–530. 10.1016/j.meegid.2013.02.013 [DOI] [PubMed] [Google Scholar]
- Yoong P., Torres V. J. (2013). The effects of Staphylococcus aureus leukotoxins on the host: cell lysis and beyond. Curr. Opin. Microbiol. 16, 63–69. 10.1016/j.mib.2013.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y. P., Wilder-Smith A., Hsu L.-Y. (2014). The role of international travel in the spread of methicillin-resistant Staphylococcus aureus. J. Trav. Med. 21, 272–281. 10.1111/jtm.12133 [DOI] [PubMed] [Google Scholar]
- Zribi M., Etienne J., El Euch D., Zribi H., Bes M., Meugnier H., et al. (2011). Detection of the first strain of glycopeptide intermediary Staphylococcus aureus in Tunis Rabta hospital. Path. Biol. 59, 334–335. 10.1016/j.patbio.2009.10.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.