Abstract
The mutant RAC1P29S, a GTPase member of the RAS superfamily, was identified as the third most common recurrent mutation in melanomas. There is limited information regarding the functional role of RAC1P29S as a “driver” in melanogenesis and as a cause for drug resistance. This commentary describes the latest data and provides evidence that supports the notion that RAC1 is a good drug target to inhibit some melanomas that carry the mutation, but its role in conferring resistance to targeted BRAF or MEK inhibition is still in question.
RAC1 is a GTPase, member of small GTP-binding proteins of the RAS superfamily. The discovery of hotspot mutation in RAC1 revealed by next-generation sequencing of the coding regions of melanoma (1, 2) provided evidence for a new ‘player’ in melanoma genesis. The recurrent P29S mutation, i.e., substitution of proline at position 29 with serine, is the 3rd most frequent activating mutation in sun-exposed melanoma, occurring at about 4–7% of the patients. It is highly significant, because for the past several years the only two most well characterized hotspot mutations in melanoma were BRAFV600 (~40%) and NRASQ61 (~20%).
There are several novel aspects about RAC1P29S in melanoma. Unlike BRAFV600 and NRASQ61, it carries the characteristic signature for UVB induced DNA damage, a C to T transition in dipyrimidine, CCT>TCT, validating the critical role of sunlight in the onset of melanoma. Indeed, RAC1P29S is restricted to hair-bearing cutaneous melanoma, and most of the primary melanoma lesions in the Yale cohort are from sun-exposed areas, and most patients reported excess sun exposure. Unlike BRAFV600 and NRASQ61, the mutation is not present in benign nevi. In the original Yale cohort, it was predominant in males (95%), but additional studies showed only a slight increase in frequency in males over females (59%) (3).
Another important observation is that, like in BRAFV600 and NRASQ61, the mutation confers constitutive activity to RAC1. This is of critical importance because it constitutes an “oncogene” driver- type of mutation. The sequencing of over 600 sun-exposed melanoma (ours, the Broad Institute and the TCGA) showed that the only other activating mutations (in addition to the BRAF and NRAS mentioned above) is the MAP2K1P124S (~2.8%). This is not to diminish the importance of oncogenic mutations in c-KIT in mucosal and acral (10–20%), and in GNA11/GNAQ in uveal (~80%) melanomas, which are very rare in sun-exposed melanomas.
RAC1, like RAS, cycles between the GTP (active) and GDP (inactive) forms. Our functional studies showed that unlike the RAS mutations, RAC1P29S maintains its normal GTPase activity but displays an increased inherent GDP→GTP nucleotide exchange, which keeps it mostly in the active form (4). When mutant RAC1P29S is introduced into normal melanocytes, it increases membrane ruffling, promotes proliferation and migration (1). Biochemical studies showed that RAC1P29S maintains an increased binding to target proteins PAK1 and MAP3K11 (MLK3), likely to affect downstream molecular events. The general notion is that RAC1P29S has a particular role in early transformation, enhancing cell migration and proliferation, as supported by its early presence in primary melanoma (3).
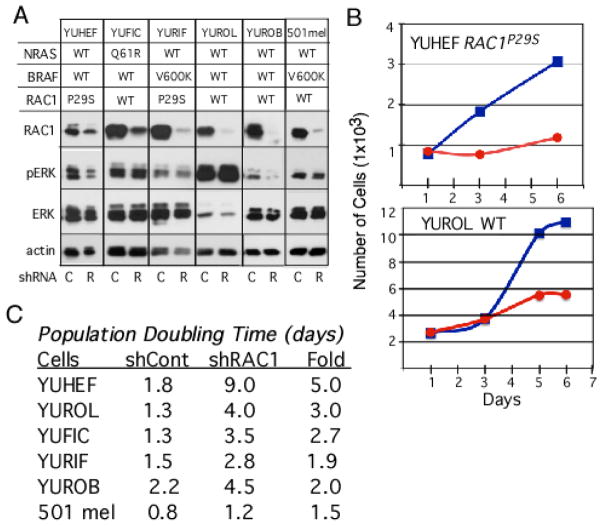
One of the main goals of cancer sequencing efforts is to identify new targets for therapy as well as new changes that confer drug-resistance and/or alters patients’ response to therapy, and RAC1P29S is no exception. We validated the role of RAC1 in melanoma cell proliferation by knockdown experiments with RAC1 specific shRNA (Figure 1, our unpublished results). The data show downregulation of RAC1 reduced the rate of cell proliferation of RAC1P29S mutant melanoma (such as YUHEF), as well as RAC1 wild type melanomas (such as YUROL and YUROB), with less of an effect on cells bearing the BRAFV600 mutation (Figure 1B, C). These data suggest that RAC1 is a good target for therapy, because it is active also in RAC1 WT melanomas by other molecular events. These can be changes in the activity of upstream regulators, especially GEFs and GAPs, i.e., enzymes that convert the inactive GDP to the active GTP state, and the GTP back to GDP-state, respectively (5). Unfortunately, the currently available drug that inhibits RAC1 (EHT1864) is not efficient at inhibiting RACP29S mutant or wild type melanoma cells (IC50 4–10 μM), although an inhibitor of its downstream target PAK (PF8055), is relatively potent (IC50 0.23–1 μM) (our unpublished results). Altogether, our experience show that RAC1, like RAS, is a target that is difficult to inhibit with small molecules. However, with the NCI new initiative to identify novel ways to inhibit RAS (5), there is still a possibility that new agents and methods will be developed.
Figure 1.
The consequences of RAC1 knockdown in melanoma cell signaling and proliferation. A. RAC1 and ERK (Extracellular regulated Kinase) in melanoma cells infected with control (C) or RAC1 specific (R) shRNA lentiviral particles. Cells were tested after selection in puromycin (1 μg/ml) for 10 days. Cell lysates were subjected to Western blot analysis by probing with antibodies against RAC1, phospho-ERK1/2 Thr202/Tyr204 mAb (pERK), ERK1/2 (ERK), and actin as a control. The data show that downreuglation of RAC can reduce ERK signaling as shown for YUHEF and YUROB, but not in the other cell lines. B. Cell proliferation in response to RAC1 knockdown. Cell proliferation monitored by counting the number of cells seeded in triplicate wells at 2–3 days intervals in medium containing puromycin; Blue and red curves indicate cells infected with control or RAC1 specific shRNA, respectively. C. Summary of melanoma cells response to RAC1 knockdown. Population-doubling time was calculated from the linear curves as shown in B.
The other major question is whether RAC1P29S confers resistance to drugs that are currently being used to target BRAFV600 mutant cancer cells, including melanomas. This possibility was recently suggested by Watson et al (6). The investigators describe resistance to BRAF inhibitors (vemurafenib and dabrafenib) and MEK inhibitors (trametinib and PD325901) of two patient-derived RAC1P29S mutant melanomas that also carry the BRAFV600K or the NRAS Q61K mutations, and by overexpressing RAC1P29S in melanoma cells. However, melanoma cells carry a large number of mutations and genomic alterations that are likely to affect their response to drugs, and two examples of melanoma are not sufficient to draw conclusion. We have recently performed drug response analyses on ~50 patient-derived early passage melanoma cells focusing on vemurafenib (originally known as PLX4032) and the MEK inhibitor selumetinib (AZD6244) currently used to treat melanoma patients. These melanoma cells are well characterized by whole exome-capture sequencing, so the presence of mutations, deletions/amplification and other genomic aberrations can be easily related to sensitivity and resistance to drugs. The results show that double-mutant BRAFV600E/RAC1P29S melanoma cells (YURIF) are in the mid-range of sensitivity to vemurafenib (IC50 0.25 μM) when compared to other BRAF mutant cells, where the range is 0.06–0.45 μM (7). On the other hand, these melanoma cells are highly sensitive to selumetinib, IC50 0.014 μM. In contrast, another RAC1P29S melanoma cell line (YUHEF, BRAF/NRAS WT, see Figure 1) is highly resistant to selumetinib, IC50 of over 10 μM. However, it is not yet clear whether the resistance to the MEK inhibitor is due to the presence of RAC1P29S, or to the loss of NF1 expression we observed in these cells. Loss of NF1 and the presence of RAC1P29S, do not confer resistance to the MEK inhibitors in another melanoma cell line that is BRAF/NRAS WT, and which displays IC50 0.025 μM. These data show that one cannot predict sensitivity or resistance to BRAF or MEK inhibitors by the presence of activated RAC1 or loss of NF1. More is needed to decipher the relevant molecular signature and markers that can guide us in pursue of precision medicine so patients are selected correctly to targeted therapy.
The unbiased sequencing of the whole-exome show that melanoma is one of the cancers with the highest number of mutations/tumor. Nevertheless, very few “oncogenes”, i.e., genes with activating mutations that can confer advantage to cancer cells and serve as targets, were identified so far (as mentioned above). In contrast, the number of “driver genes” in the tumor-suppressor category is much larger (such as NF1, TP53, CDKN2A, PTEN, PPP6C, RB1, ARID1A, ARID2, to mention the very top on the list). Unfortunately, it is more difficult to restore the function of tumor suppressors as an approach in cancer therapy. It is possible that their downstream target and/or the pathway that they affect can be a subject of further analyses, as for example the effect of PPP6C on Aurora kinase (8, 9). In additions, the mutations in melanoma likely contribute to the immunogenicity of this cancer and can be harnessed in immunotherapy (10, 11), raising the possibility that RAC1P29S is one such antigen that can be explored.
Altogether, we are in a new era in which next-generation sequencing of large number of tumors provided us already with a deep knowledge of the molecular changes in tumors. We are all aware that a single mutation, such as in BRAF, NRAS, RAC1, NF1 or any of the other ‘driver’ genes does not lead to cancer. There is a need to decipher the contribution of the additional changes, and combination of changes, so we can understand the cancer biology and responses to drugs. The development of bioinformatic algorithm that can take into account these changes is critical so we can move to the next step, i.e., apply “precision medicine” to the clinic to optimize the selection of individual patients to targeted and/or immunotherapy.
Acknowledgments
Supported by the Yale SPORE in Skin Cancer grant number P50 CA121974 (R. Halaban, PI).
References
- 1.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, Cheng E, Davis MJ, Goh G, Choi M, Ariyan S, Narayan D, Dutton-Regester K, Capatana A, Holman EC, Bosenberg M, Sznol M, Kluger HM, Brash DE, Stern DF, Materin MA, Lo RS, Mane S, Ma S, Kidd KK, Hayward NK, Lifton RP, Schlessinger J, Boggon TJ, Halaban R. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, Dicara D, Ramos AH, Lawrence MS, Cibulskis K, Sivachenko A, Voet D, Saksena G, Stransky N, Onofrio RC, Winckler W, Ardlie K, Wagle N, Wargo J, Chong K, Morton DL, Stemke-Hale K, Chen G, Noble M, Meyerson M, Ladbury JE, Davies MA, Gershenwald JE, Wagner SN, Hoon DS, Schadendorf D, Lander ES, Gabriel SB, Getz G, Garraway LA, Chin L. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mar VJ, Wong SQ, Logan A, Nguyen T, Cebon J, Kelly J, Wolfe R, Dobrovic A, McLean C, McArthur GA. Clinical and pathological associations of the activating RAC1 P29S mutation in primary cutaneous melanoma. Pigment Cell Melanoma Res. 2014 Jul 17; doi: 10.1111/pcmr.12295. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 4.Davis MJ, Ha BH, Holman EC, Halaban R, Schlessinger J, Boggon TJ. RAC1P29S is a spontaneously activating cancer-associated GTPase. Proc Natl Acad Sci U S A. 2013;110:912–917. doi: 10.1073/pnas.1220895110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25:272–281. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 6.Watson IR, Li L, Cabeceiras PK, Mahdavi M, Gutschner T, Genovese G, Wang G, Fang Z, Tepper JM, Stemke-Hale K, Tsai KY, Davies MA, Mills GB, Chin L. The RAC1 P29S hotspot mutation in melanoma confers resistance to pharmacological inhibition of RAF. Cancer Res. 2014;74:4845–4852. doi: 10.1158/0008-5472.CAN-14-1232-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halaban R, Zhang W, Bacchiocchi A, Cheng E, Parisi F, Ariyan S, Krauthammer M, McCusker JP, Kluger Y, Sznol M. PLX4032, a selective BRAF(V600E) kinase inhibitor, activates the ERK pathway and enhances cell migration and proliferation of BRAF(WT) melanoma cells. Pigment Cell Melanoma Res. 2010;23:190–200. doi: 10.1111/j.1755-148X.2010.00685.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gold HL, Wengrod J, de Miera EV, Wang D, Fleming N, Sikkema L, Kirchhoff T, Hochman T, Goldberg JD, Osman I, Gardner LB. PP6C hotspot mutations in melanoma display sensitivity to Aurora kinase inhibition. Mol Cancer Res. 2014;12:433–439. doi: 10.1158/1541-7786.MCR-13-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caputo E, Miceli R, Motti M, Tate R, Fratangelo F, Botti G, Mozzillo N, Carriero M, Cavalcanti E, Palmieri G, Ciliberto G, Pirozzi G, Ascierto P. AurkA inhibitors enhance the effects of B-RAF and MEK inhibitors in melanoma treatment. J Transl Med. 2014;12:216. doi: 10.1186/s12967-014-0216-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robbins PF, Lu YC, El-Gamil M, Li YF, Gross C, Gartner J, Lin JC, Teer JK, Cliften P, Tycksen E, Samuels Y, Rosenberg SA. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19:747–752. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tran E, Turcotte S, Gros A, Robbins PF, Lu YC, Dudley ME, Wunderlich JR, Somerville RP, Hogan K, Hinrichs CS, Parkhurst MR, Yang JC, Rosenberg SA. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344:641–645. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]