Abstract
NKp44 (NCR2) is a distinct member of Natural Cytotoxicity Receptors (NCRs) family that can induce cytokine production and cytolytic activity in human NK cells. Heparan sulfate proteoglycans (HSPGs) are differentially expressed in various normal and cancerous tissues. HSPGs were reported to serve as ligands/co-ligands for NKp44 and other NCRs. However, HSPG expression is not restricted to either group and can be found also in NK cells. Our current study reveals that NKp44 function can be modulated through interactions with HSPGs on NK cells themselves in -cis rather than on target cells in -trans. The intimate interaction of NKp44 and the NK cell-associated HSPG syndecan-4 (SDC4) in –cis can directly regulate membrane distribution of NKp44 and constitutively dampens the triggering of the receptor. We further demonstrate, that the disruption of NKp44 and SDC4 interaction releases the receptor to engage with its ligands in –trans and therefore enhances NKp44 activation potential and NK cell functional response.
Keywords: NKp44, NCR2, Heparan Sulfate, Heparan Sulfate Proteoglycans (HSPGs), Syndecan 4 (SDC4), Immune regulation, NK cell activation, Natural Cytotoxicity Receptors (NCRs)
Introduction
Natural killer (NK) cells are innate immune cells that are capable of directly attacking tumor, virus-infected and stressed cells [1, 2]. Human NK cell activation is mediated by activating forms of killer cell Ig-like receptors (KIR [KIR2DS, KIR3DS, and KIR2DL4]), 2B4, NKG2D, NKp80, and natural cytotoxicity receptors (NCRs) -1, -2, and -3 called NKp46, NKp44, and NKp30, respectively. Selective engagement of primary activating receptors such as NCRs can trigger both focused target cell lysis through release of perforin and granzymes from cytolytic granules and the secretion of numerous cytokines, especially interferon (IFN)-γ. [3–5]. NKp44 is a structurally distinct member of the NCR family and a functionally unique NK cell receptor with expression primarily restricted to NK cells in higher primates [6] and in contrast to NKp46 and NKp30, NKp44 is predominantly expressed on activated NK cells [7].
NKp44 is a structurally unique receptor that contains: (a) an extracellular domain consisting of a single V-type Ig-like domain, forms a highly positive-charged groove that is directly involved in ligand binding [8–11], (b) a cytoplasmic domain possessing a single immunoreceptor tyrosine-based inhibitory motif (ITIM), which functionality depends on what ligand binds to NKp44 ([12, 13], and (c) a transmembrane domain containing a charged lysine residue, which can associate with a dimer of the immunoreceptor tyrosine-based activating motif (ITAM)-containing adaptor DAP12 [12, 14–16]. Therefore, functionally, NKp44 can trigger either inhibitory or activating responses by NK cells in a ligand-dependent manner.
Several NKp44 ligands have previously been characterized. NKp44 was reported to interact with viral hemagglutinin and hemagglutinin-neuraminidase of Orthomyxoviridae and the envelope glycoprotein (E-protein) of Flaviviridae [17–19]. In addition, two novel cellular ligands for NKp44 were recently identified: PCNA [13] and a splice variant of MLL5 [20]. Furthermore, NKp44 can also directly bind heparan sulfate/heparin (HS) and these interactions can modulate function of NKp44 to impact NK cell activation [9]. Although NKp44, NKp46 and NKp30 can bind HS with differential individual specificities, all the NCRs preferably recognize HS glycosaminoglycans (HSGAGs) that are sulfated above average [3, 5, 9, 10, 21–23].
Most of the HS expressed on the surface of mammalian cells is derived from the Syndecan (SDC) HS proteoglycans (HSPGs) and SDC4 in particular [24, 25]. We and others have found high expression of SDC4 also in NK cells [25, 26]. Therefore, we propose that HSPGs can affect receptor engagement on NK cell surface (acting in –cis), rather than on the ligand-expressing target cell (acting in –trans).
Results
Characterization of HS binding site mutation and its effect on NKp44-mediated NK cell’s functional response
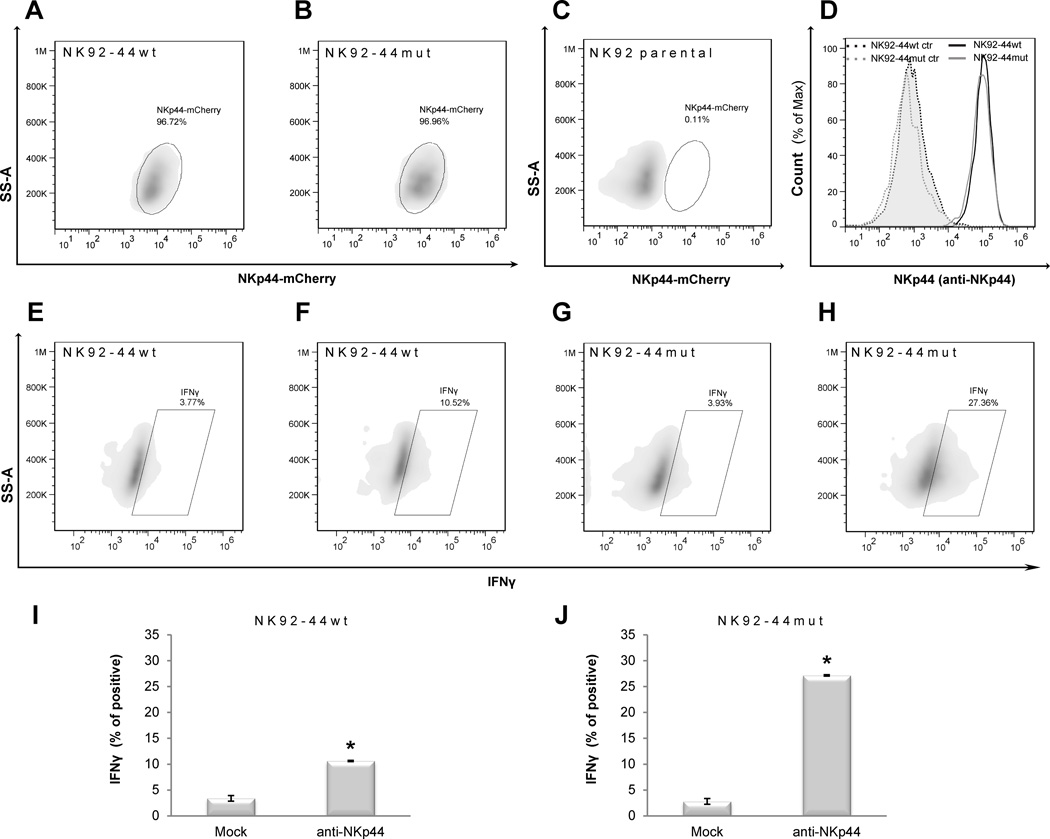
To assess the contribution of NK-expressed HSPGs to the membrane distribution and function of NK-expressed NCRs, we chose NKp44 since its HS binding site was already fully characterized: HS binding site of NKp44 is composed of a basic patch of 8 amino acids (R47, K53, R55, H88, R92, R95, K103 and R106) [9, 11]. For this study we used cDNA constructs encoding for wild type NKp44 (NKp44wt) and mutated NKp44 (NKp44mut) in which R47, K53 and R55 in the highly positive charged ligand binding groove were replaced by Q to considerably reduce binding to HS and HSPG [9]. In both constructs, NKp44 was tagged with intracellular mCherry at the C-terminus and expressed in NK92 cells to generate NK92-44wt and NK92-44mut. Expression of mCherry was stably detected in nearly 100% of sorted NK92 cells, and the level of cell membrane-expressed NKp44 was similar between NK92-44wt and NK92-44mut (Fig. 1A–C), as assessed by staining with anti-NKp44 mAb (Fig. 1D). NKp44’s HS binding site mutation did not affect the biochemical characteristics of the NKp44 protein [9] or the recognition of the receptor by specific anti-NKp44 mAb (clone 3.43.13; see Materials and Methods section; Fig. 1D).
Figure 1. Characterization of HS binding site mutation and its effect on NKp44-mediated IFN-γ production by NK cells.
A,C. FACS analysis of NKp44 (NKp44-mCherry fusion protein; as gated on corresponding mCherry) expression by NK92 cell lines: (A) NK92-44wt, (B) NK92-44mut, (C) NK92 parental cell line. D. FACS analysis of NKp44 expression by NK92-44wt and NK92-44mut cell lines as assayed by combined staining with anti-NKp44 mAb (clone 3.43.13) and secondary allophycocyanin-conjugated anti-mouse IgG1 Ab staining; NK92-44wt (dark solid line), NK92-44mut (grey solid line), secondary allophycocyanin-conjugated anti-mouse IgG1 Ab single staining (control) of NK92-44wt (dark dotted line; tinted) andNK92-44mut (grey dotted line; tinted). E-H. Representative images of FACS analysis: IFN-γ production by NK92-44wt and NK92-44mut cell lines as performed by combined intracellular staining with biotin-conjugated anti-IFN-γ mAb and SA- allophycocyanin: NK92-44wt (E, F) or NK92-44mut (G, H) cell lines were stimulated overnight with plate-bound anti-NKp44 mAb (clone 3.43.13; F, H) or Mock control (anti-2B4 mAb; E, G). I,J. Summary of FACS analysis: I (E-F; NK92-44wt) and J (G–H; NK92-44mut). Data represent mean ± SD of n=3 biological replicas for each treatment. (A–J) Data are representative of 3 independent experiments. *p ≤ 0.01 (also NK92wt (I) to NKp44mut (J)); t-test.
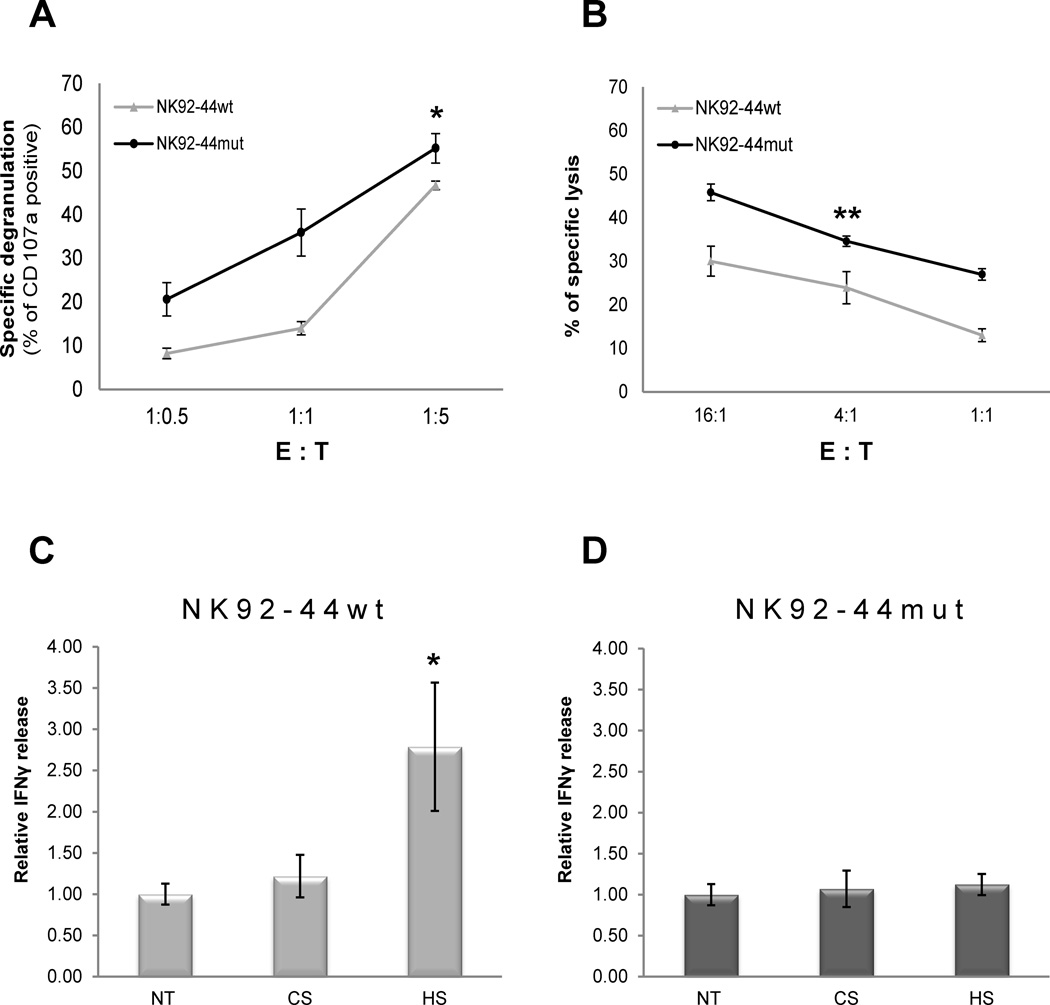
In order to assess the functional outcome of the HS binding site mutation, we initially tested direct NKp44 engagement. We employed plastic-bound anti-NKp44 mAb to activate both NK92-44wt and NK92-44mut cell lines. Engagement of NK92-44mut cell line manifested substantially higher activity compared to NK92-44wt (Fig. 1E–H), as evident in a nearly 3 fold greater induction of IFN-γ-producing NKp44-mCherry positive cells (Fig. 1I, J; summary of 3 replicates). We next investigated whether the effect of NKp44 HS binding site mutation can be explained by either –cis or -trans NKp44 engagement mode. We first examined NK92-44mut effector cell line response in -trans against K562 target cell line. After co-incubation of NKp44 expressing NK effector cell lines with K562 target cells, we observed higher specific degranulation (Fig. 2A) as well as lysis of target cells (Fig. 2B) by NK92-44mut in contrast to the inferior NK92-44wt response. Next, we examined NKp44-mediated functional response in –cis by addition of exogenous HS, followed by specific engagement with anti-NKp44 mAb. Indeed, supplementation of soluble HS, but not chondroitin sulfate A (CS), resulted in potentiation of NKp44 mediated IFN-γ release by NK92-44wt (Fig. 2C), but not NK92-44mut (Fig. 2D). To summarize, mutation of NKp44 in the HS binding site, resulted in significant augmentation of NK92-44mut cells’ activation potential.
Figure 2. Impact of HS binding site mutation on NKp44-mediated functional response of NK cells.
A–B. Impact of HS binding site mutation on NKp44 mediated cytotoxicity. NK92 effector cell lines were co-incubated with K562 target cell line: summary of specific degranulation of NK92-44wt and NK92-44mut cell lines as assayed by anti-CD107a mAb (A); NKp44 mediated cytotoxicity: summary of specific lysis of K562 cell line by NK92-44wt and NK92-44mut cell lines as assayed by multiparametric FACS analysis (B). C-D. Impact of soluble HS on NKp44 mediated IFN-γ secretion: NK92-44wt or NK92-44mut cells were activated overnight by plate-bound anti-NKp44 mAb in the presence of 5µg/ml of either soluble HS or CS or assay medium alone (NT). IFN-γ in culture supernatant was assayed by ELISA: IFN-γ concentration in the sup ranged from 0.1ng/ml (Ctr. Ab) to 4ng/ml (anti-NKp44 mAb). A-D. Data represents mean ± SD of two to three independent experiments in n=6 biological replicas for each treatment. *p ≤ 0.01, **p ≤ 0.05; t-test.
NKp44 and SDC4 co-distribution in non-activated NK cells
To further access the contribution of NK-expressed HSPGs to the membrane distribution and function of NK-expressed NCRs, we co-expressed wild type or mutant NKp44-mCherry with SDC4-GFP fusion protein (SDC4 was cloned in-frame with C-terminal GFP) in NK92 cell line to generate NK92-44wt SDC4 cells and NK92-44mut SDC4 cells respectively. SDC4 HSPG was chosen for this study as it is naturally expressed by human NK cells [25, 26]. The expression levels of cell membrane-associated NKp44 and SDC4 fusion proteins were similar between NK92-44wt SDC4 and NK92-44mut SDC4 (Supporting Information Fig. 1A, B). Additionally, it was assessed by staining with specific anti-NKp44 mAb and anti-SDC4 Ab (see Materials and Methods and Supporting Information Fig. 1C, D).
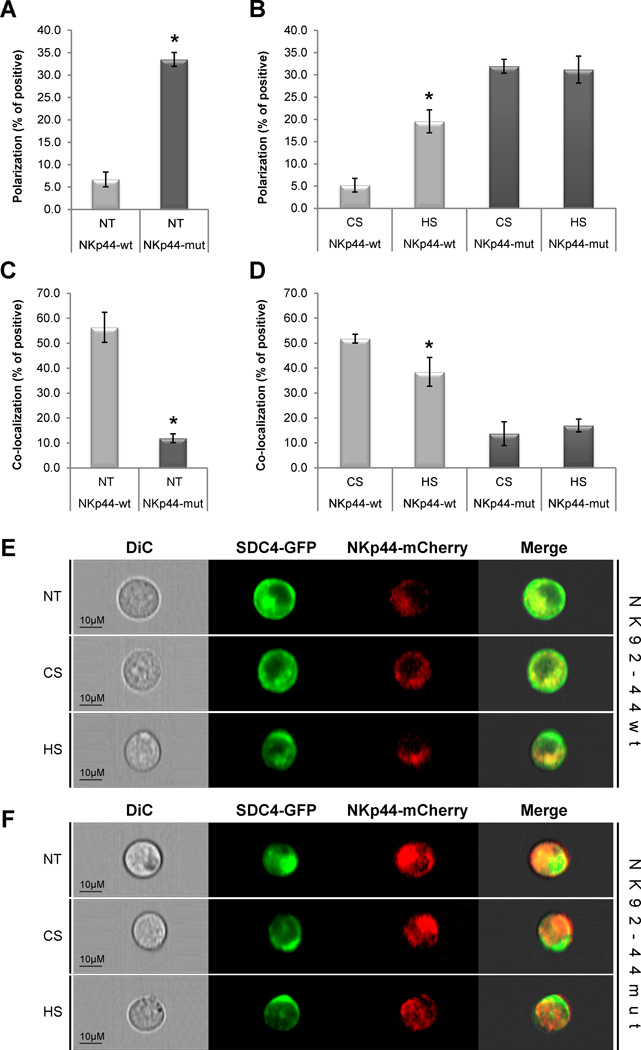
We next examined the co-distribution of NKp44 and SDC4 in non-activated NK92 cell lines (e.g. no specific anti-NKp44 mAb activation was introduced and IL-2 reduced assay medium was used). NK92-44wt SDC4 and NK92-44mut SDC4 cells were treated with either mock medium or medium containing soluble HS or CS and then analyzed by the ImageStream multispectral imaging flow cytometer (Fig. 3). This approach allowed us to assess co-distribution of membrane expressed NKp44 and SDC4 in non-activated NK cells by masking the intracellular portion of relevant fluorescent marker, to eliminate the early ER-expression noise, and measuring the mean polarization and co-localization of membrane-associated mCherry and GFP markers in a large population of NK92 cells in suspension.
Figure 3. NKp44 and SDC4 co-distribution in non-activated NK cells.
(A–F) NK92-44wt and NK92-44mut SDC4-GFP co-expressing cells were complemented with standard assay medium (NT) or with assay medium containing 10µg/ml of either HS or CS. ImageStream analysis of NKp44-mCherry and SDC4-GFP (A, B) polarization and (C, D) co-localization: polarization and co-localization coefficient correspond to the proportion of positive cells. Data represents mean ± SD of n=3 independent experiments and indicated number of biological replicas (n = 3,000 for each treatment). *p < 0.01; t-test. E, F. Representative images of non-treated (NT) and HS- or CS-treated cells are shown. Images are representative of 3 independent experiments.
Indeed, the co-localization of membrane-associated NKp44 conjugated mCherry and intracellular non-conjugated GFP control was found to be below two percent (data not shown). Under standard assay conditions (see methods), we observed no differential polarization of SDC4 and NKp44wt (i.e. no polarization of each marker towards opposite sides of the same cell was observed). Remarkably, the proteins were polarized in almost 6-fold larger portion of NK92-44mut SDC4 cells as compared with their polarization in NK92-44wt SDC4 cells (Fig. 3A; NT). We then analyzed the co-localization of SDC4 and NKp44mut in NK92 cells under the same conditions. The proteins were evenly co-distributed and co-localized in a significant fraction of the cells. In stark contrast, SDC4 co-localized with NKp44mut was observed in almost 5-fold lower fraction of NK92 cells (Fig. 3C; NT). This would suggest that mutating the HS binding site caused NKp44mut and SDC4 separation and therefore enhanced differential membrane polarization of NK cells. We further assessed effect of treatment of NK92-44wt SDC4 cells with either soluble HS or CS. Indeed, supplementation of soluble HS, but not CS, has significantly increased the polarization of SDC4 and NKp44 in NK92-44wt cells, while either glycosaminoglycan treatment caused no change in already highly-polarized NK92-44mut SDC4 cells treated with (Fig. 3B). The already high polarization of both fluorescent markers in NK92-44mut SDC4 cell line under standard assay conditions and enhanced polarization in HS-treated NK92-44wt SDC4 cells were visualized as greater incidence of “capping” of both SDC4-GFP and NKp44-mCherry on the opposite sides of the NK cell (representative images; Fig. 3E, F). Supplementation of soluble HS, but not CS, resulted in reduced co-localization of SDC4 with NKp44wt in accordance; however, the already low levels of NKp44mut co-localization with SDC4 were unchanged by either HS or CS treatment (Fig. 3D).
We next addressed the question whether exogenous HS treatment can affect not only the NKp44 differential distribution, but also the surface density of detectable membrane-expressed fraction of the receptor. We observed no detectable effect of either treatment on the membrane-expressed fraction of NKp44 as the receptor mean surface density in both NK92 and primary NK cells remained unaffected (Supporting Information Fig. 1I, J). To summarize, mutation of NKp44 in the HS binding site or treatment with exogenous HS has significantly induced differential polarization in non-activated NK92 cells, and at the same time has reduced the co-localization of membrane-associated fraction of NKp44 and SDC4.
NKp44 and SDC4 co-localization in activated NK cells
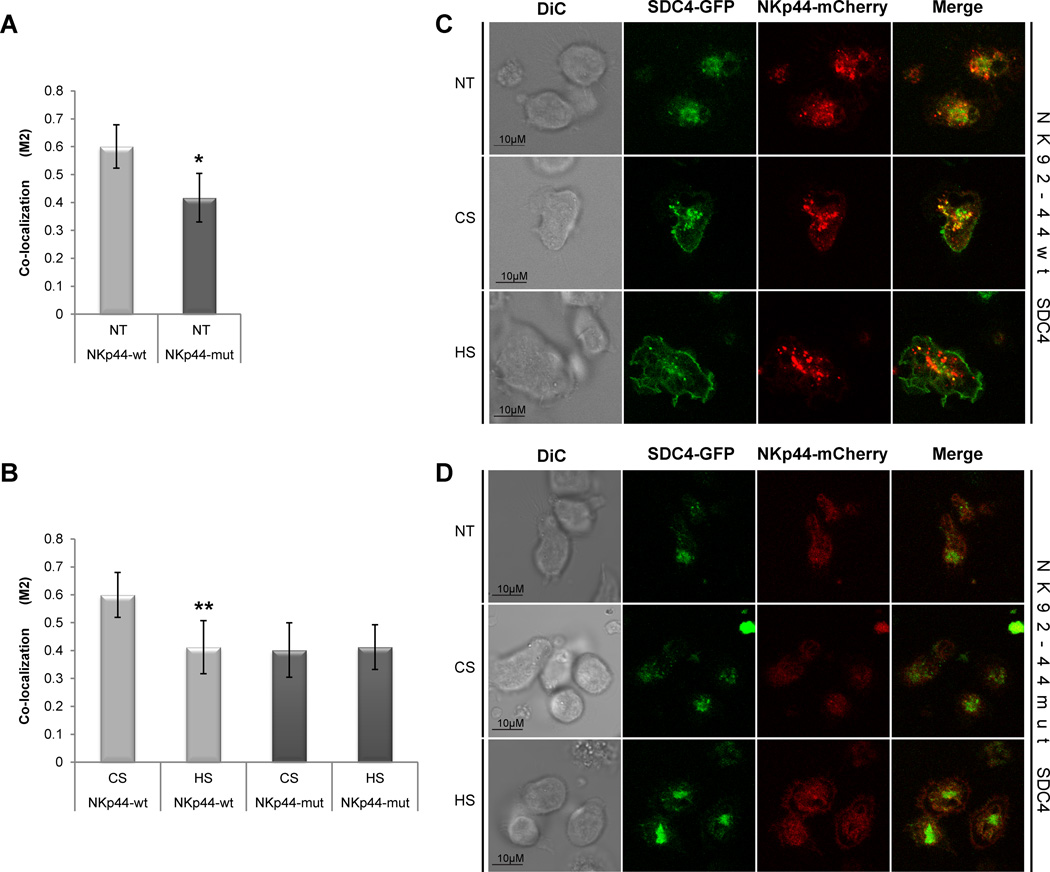
The imaging flow cytometry approach allowed us to assess NKp44 and SDC4 co-distribution in thousands of non-activated NK cells in suspension (Fig. 3). However, to evaluate the precise level of NKp44 and SDC4 interaction, following NKp44 ligation, we first engaged NKp44 with specific surface-bound anti-NKp44 mAb and then measured co-localization at the adhesion focal plane (characterized by high level of refraction in GFP channel, which also ensured the measurements of membrane-expressed, but not golgi/ER- localized NKp44 and SDC4). Moreover, the adhesion focal plane is representing the NK cell surface interaction with an antibody and mimicking the NK immunological synapse (NKIS). Under these conditions, the co-localization of NKp44wt and SDC4 was about 50% more prevalent as compared with co-localization of NKp44mut and SDC4 (Fig. 4A). That is in accordance with our previously described results. We further tested the effect of soluble HS or CS at the artificial synapse-like region. Indeed, treatment of NK92 cells with exogenous HS significantly reduced co-localization of NKp44wt and SDC4, while no change was observed for NKp44mut and SDC4 (Fig. 4B). Treatment of NK cells with exogenous CS;however, did not significantly alter the constitutive high co-localization levels in NK92-44wt SDC4 cells or the low basal co-localization level in NK92-44mut SDC4 cells (Fig. 4B). Representative images showing co-localization in NT samples and following CS or HS treatment are shown in Figure 4C (NK92-44wt SDC4) and 4D (NK92-44mut SDC4). To summarize, mutation of NKp44 in the HS-binding site or exogenous soluble HS treatment of NK92-44wt SDC4 cells significantly reduced co-localization of membrane associated fraction of NKp44 and SDC4 in the artificial NKIS-like region generated at the cell adhesion focal plane.
Figure 4. NKp44 and SDC4 co-localization in activated NK cells.
NK92-44wt and NK92-44mut SDC4-GFP co-expressing cells were complemented with standard assay medium (NT) or with assay medium containing 10µg/ml of either HS or CS. Confocal microscope image analysis of (A) non-treated (NT) and (B) HS- or CS-treated cells: NKp44-mCherry and SDC4-GFP co-localization coefficient (M2; see Materials and Methods) corresponds to the proportion of NKp44-mCherry co-localized with SDC4-GFP. Data represents mean ± SD of n=5 independent experiments and indicated number of biological replicas (N = 100 for each treatment*p ≤ 0.01; **p ≤ 0.05; t-test. C,D. Representative images of non-treated (NT) and HS- or CS-treated cells are shown. Images are representative of 5 independent experiments.
NKp44 and SDC4 proximal interaction in activated NK cells
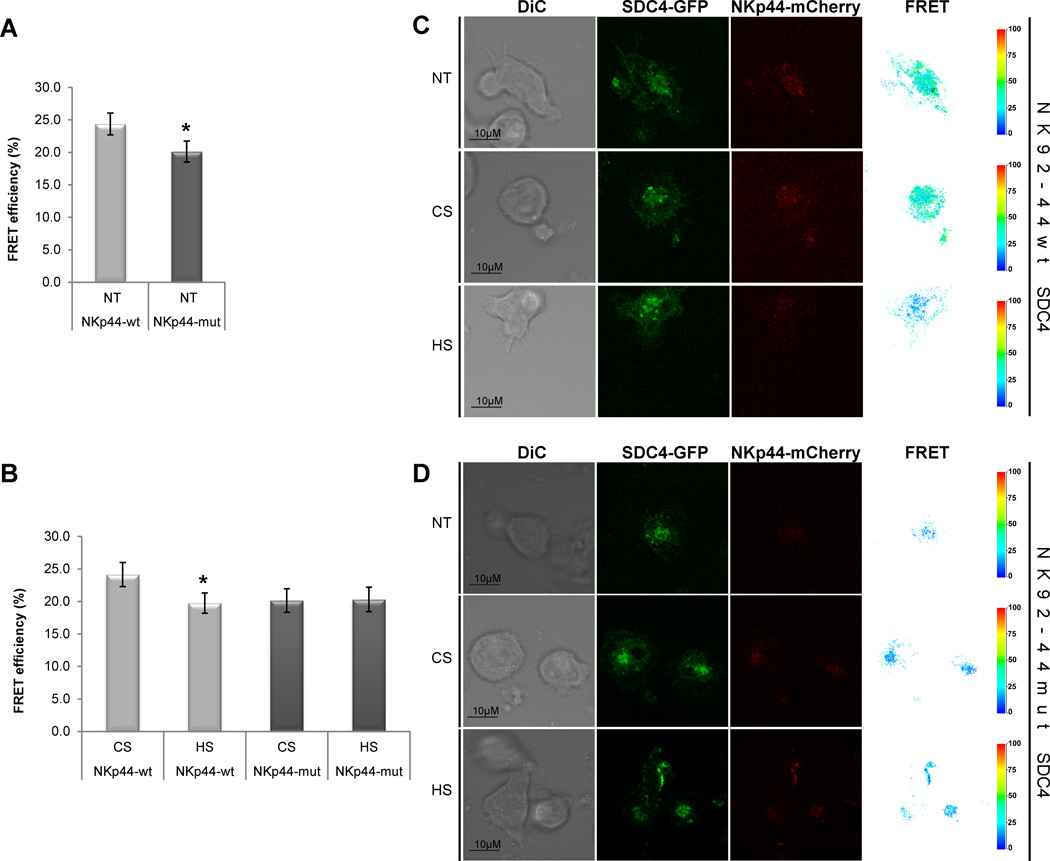
To evaluate levels of proximal interaction between NKp44-mCherry and SDC4-GFP, following NKp44 activation, we employed surface-bound anti-NKp44 mAb as described above and measured fluorescence resonance energy transfer (FRET) at the artificial synapse-like region. NK92-44wt SDC4 cells exhibited considerably higher levels of constitutive FRET efficiency between NKp44-mCherry and SDC4-GFP, as compared to NK92-44mut SDC4 cells (Fig. 5A). Treatment of NK92-44wt SDC4 cells with soluble HS, but not CS, significantly reduced FRET efficiency (Fig. 5B). Although FRET efficiency was unaffected by soluble HS in NK92-44mut SDC4 cells (Fig. 5B). Representative images of cells (incl. FRET channel and diagrams) are shown in Figure 5C (NK92-44wt SDC4) and 5D (NK92-44mut SDC4). To summarize, NKp44 mutation in HS-binding site or soluble HS treatment of NK92-44wt SDC4 cells has significantly reduced FRET efficiency between mCherry and GFP fluorescent markers, thus representing the abrogation of proximal interactions between NKp44wt and SDC4 in the artificial NKIS-like region.
Figure 5. NKp44 and SDC4 proximal interaction in activated NK cells.
NK92-44wt and NK92-44mut SDC4-GFP co-expressing cells were complemented with standard assay medium (NT) or with assay medium containing 10µg/ml of either HS or CS. Confocal microscope images analysis of non-treated (NT) (A) and HS- or CS-treated (B) cells: FRET efficiency is shown for SDC4-GFP (donor) and NKp44-mCherry (acceptor; FRET channel). Data represents mean ± SD of n=3 independent experiments and indicated number of biological replicas (N = 100 for each treatment). P-values were calculated using T-test: *p-value ≤ 0.01. C,D. Representative images of non-treated (NT) and HS- or CS-treated cells are shown (FRET channel diagram and FRET efficiency scale are shown). Images are representative of 3 independent experiments.
NKp44 and SDC4 co-distribution and modulation of activated primary NK cells function
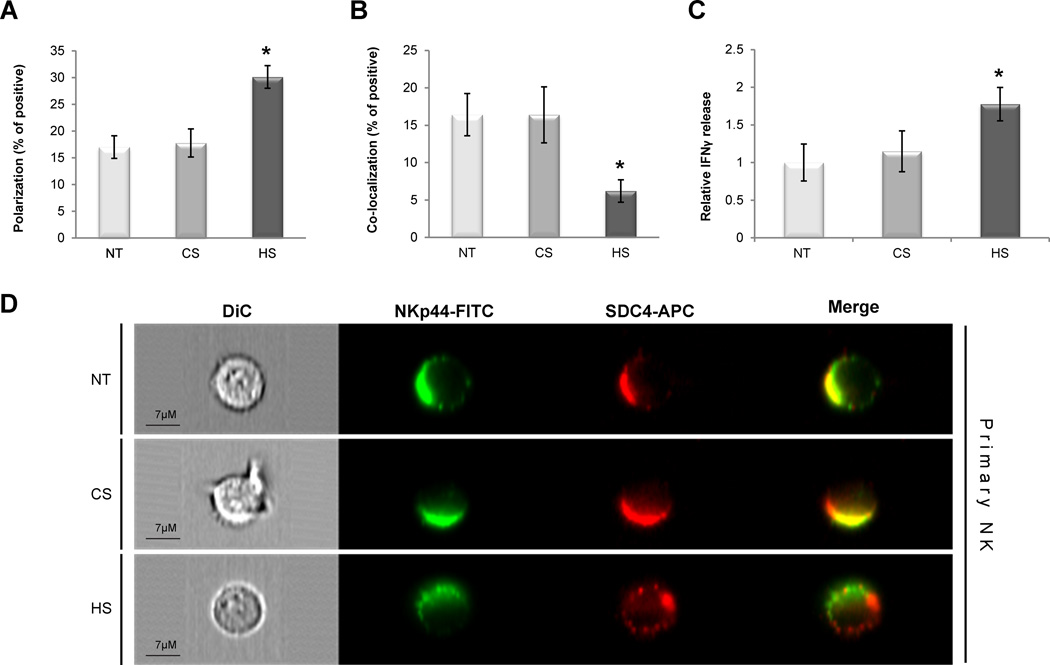
We and others have found high expression of SDC4 in both NK cell lymphomas [25, 26] and primary NK cells (Supporting Information Fig. 1H). Although we have found high expression of the NKp44 and SDC4 in primary NK cells, it was found to be lower as compared with transduced NK92 cells (Supporting Information Fig. 1C,D; G,H). To assess the contribution of SDC4 to the NKp44 mediated response in primary NK cells, we first cultured freshly purified primary NK cells (see Materials and Methods; Supporting Information Fig. 1E,F) with IL-2 to induce NKp44 expression (Supporting Information Fig. 1G). We then examined the effect of exogenous soluble HS or CS treatment on differential co-distribution and polarization of NKp44 and SDC4 by the ImageStream multispectral imaging flow cytometer. Activated (i.e. IL-2 stimulated) primary NK cells were pre-incubated with either assay medium (NT), HS or CS, and then stained with specific anti-NKp44 (FITC channel) and anti-SDC4 (APC channel) antibodies (see Flow Cytometry) in the presence of assay medium only, HS or CS in accordance. The mean polarization and co-localization of membrane-associated FITC and APC markers on the activated primary NK cells’ surface was measured as described elsewhere (Materials and Methods). Under standard assay conditions we observed low levels of NKp44 and SDC4 polarization (Fig. 6A; NT). Next, we analyzed the co-localization of NKp44 and SDC4: proteins were co-localized in the same area of a significant fraction of activated primary NK cells. (Fig. 6B; NT). Further assessment revealed the effect of exogenous soluble HS or CS treatment. Indeed, supplementation of soluble HS, but not CS, resulted in almost two fold increase in the NKp44 and SDC4 polarization (Fig. 6A) and has also reduced co-localization of NKp44 and SDC4 by almost three fold in accordance (Fig. 6B). The polarization and co-localization of both NKp44 and SDC4 in activated primary NK cells is shown in representative images (Fig. 6D).
Figure 6. NKp44 and SDC4 co-distribution and function in activated primary NK cells.
IL-2 activated primary NK cells were complemented with standard assay medium (NT) or with assay medium containing 10µg/ml of either HS or CS (A,B,D). Primary NK cells were stained with specific anti-NKp44 mAb (FITC channel) and goat anti-SDC4 (APC channel) antibodies (see Materials and methods). ImageStream analysis of NKp44 and SDC4 (A) polarization and (B co-localization): polarization and co-localization coefficient correspond to the proportion of positive cells. (A and B) Data represents mean ± SD of two independent experiments and indicated number of biological replicas (N = 15,000 for each treatment). C. Impact of soluble HS on NKp44 mediated IFN-γ secretion: IL-2 activated primary NK cells were stimulated overnight by plate-bound anti-NKp44 mAb in the presence of 5µg/ml of either soluble HS or CS or assay medium alone (NT). IFN-γ in culture supernatant was assayed by ELISA: IFN-γ concentration in the sup ranged from 0ng/ml (not detected; Ctr. Ab) to 0.8ng/ml (anti-NKp44 mAb). Data represent mean ± SD of two independent experiments in n=6 biological replicas for each treatment. *p ≤ 0.01; t-test. D. Representative images of non-treated (NT) and HS- or CS-treated cells are shown. Images are representative of two independent experiments.
In order to assess the functional outcome of NKp44 and SDC4 interaction in activated primary NK cells, we employed soluble HS, followed by NKp44 specific engagement with anti-NKp44 mAb as it was previously described for NKp44-expressing NK92 cell lines. The supplementation of exogenous soluble HS, but not CS, resulted in significant increase in IFN-γ secretion (Fig. 6C). In summary, treatment with exogenous HS has significantly induced differential polarization of NKp44 and SDC4 in activated primary NK cells and at the same time has reduced the co-localization of both proteins. We have also observed superior functional response of activated primary NK cells, following treatment with HS and specific NKp44 engagement.
Discussion
A growing body of evidence indicates that NK cell responses that are mediated by various activating receptors are often influenced by receptor-glycosaminoglycan (GAG) interactions. KIR2DL4, NKG2D [25, 27–29], and all three NCRs [5, 30] have been reported to recognize both their primary ligands and HSGAGs. As aforementioned, each of the NCRs has been shown to recognize distinct HS structures with fine specificity [9, 10]. Moreover, we and others have found high expression of HSGAGs also in NK cells ([25, 26]; Supporting Information Fig. 1H). We have previously reported that exogenous HS can potentiate IFN-γ secretion in NK cells stimulated with specific anti-NCRs mAbs in the absence of target cells. From these data, we speculated that the exogenous HS is interfering with endogenous interactions between NCR and HSGAG on the NK cell surface. Hence, the core hypothesis of this study is that HSPGs on the NK cell surface could play roles as allosteric regulators of the NCRs [25]. An analogous mode of “-cis” interaction between NK cell receptor and its ligand reportedly occurs between the Ly49 receptors and their MHC class I (MHC-I) ligands on the surface of mouse NK cells. It was recently shown that Ly49 is “masked” by the NK cells’ own MHC-I, which thereby modulates receptor function in –cis [31]. Another example of receptor-ligand –cis interaction is Siglec 7 (CD328) and α2,8-linked disialic acid structures, both widely expressed on NK cell surfaces [32–34]. Therefore, we conclude that receptor “masking” in –cis is a common mechanism on NK cells that manages receptor accessibility and consequently modulates receptor function [25, 30, 31, 35, 36].
We researched this hypothesis for the NKp44 receptor by mutating the HS-binding site and subsequently monitoring cell surface co-distribution of NKp44 and SCD4, following challenge with exogenous HS. We first demonstrated that SDC4 and wild type NKp44 are evenly distributed in NK cells and their co-localization is significantly higher as compared with SDC4 and mutated NKp44. Soluble HS manipulation resulted in decreased co-localization of SDC4 and wild type NKp44 in non-activated NK cells, but had no effect on the lower co-localization with mutated NKp44. Similar observations were made for NK cells stimulated with anti-NKp44 mAb, since SDC4 and wild type NKp44 co-localized and this was disrupted by soluble HS. In contrast, SDC4 and mutated NKp44 were only marginally co-localized in this context, whereas exogenous HS had no effect on this co-localization. We then showed that non-activated NK cells expressing mutated NKp44 and SDC4 are extremely polarized as compared with cells expressing wild type NKp44 and SDC4, while exogenous HS also induced polarization in NK cells expressing wild type NKp44 and SDC4. However, the same treatment had no detectable effect on NK cells expressing mutated NKp44 and SDC4.
We further verified the physiological relevance of our findings employing IL-2 activated primary NK cells. , NKp44 and SDC4 have exhibited high degree of co-localization, while soluble HS manipulation resulted in a decrease of co-localization and have induced polarization of both proteins. Furthermore, neither treatment has affected the membrane-expressed NKp44 mean surface density in both NK92 and primary NK cells (Supporting Information Fig. 1I, J). Taken together, we used multiple experimental methods to demonstrate a constitutive interaction between NKp44 and HSGAG on SDC4 on the NK cell surface.
Our data also makes evident that NKp44-HSGAG (SDC4) interactions can modulate the function of NKp44, and therefore may directly impact the activation potential of NK cells. Indeed, the stimulation of NK cells with specific anti-NKp44 mAb resulted in an almost 3 fold greater increase in IFN-γ production in NK cells expressing NKp44mut as compared with the same stimulation of NK cells expressing NKp44wt. Moreover, co-incubation of NKp44mut expressing effector NK cells with K562 target cells resulted in significantly improved functional response (e.g. specific degranulation and lysis of target cells) of NK92-44mut effector cells. Next, we addressed the question whether superior functional response of NK92-44mut cell line can be attributed to NKp44 interaction with HS on NK cells membrane, acting in –cis rather than receptor-ligand interaction on target cells membrane, acting in -trans. We employed exogenous soluble HS, followed by NKp44 specific engagement with anti-NKp44 mAb. Indeed, supplementation of soluble HS, but not CS, resulted in NKp44-mediated potentiation of IFN-γ secretion by NK92-44wt, but not NK92-44mut, this is in accordance to the previously published data [9, 10].
Moreover, we were able to further confirm these findings employing primary NK cells: supplementation of exogenous HS resulted in significant augmentation of IFN-γ secretion, following NKp44 specific engagement. This is also in accordance with previously published data [5, 9, 25, 30]. These results suggests that a -cis interaction between NKp44 and HSGAG on SDC4 constitutively dampens the activating function of the receptor and therefore the disruption of this interaction could enhance the activation potential by freeing the receptor to more efficiently engage with ligands in -trans. A similar mechanism has been proposed for -cis interactions between inhibitory Ly49 and MHC-I suppressing the capacity of the receptor to interact with MHC-I in -trans, thereby diminishing the inhibitory potential [31, 35–39].
It was previously reported that SDC4 ligation in B lymphocytes can result in the formation of long, filopodia-like projections, thereby mimicking an “activated”-like phenotype similar to a phenotype induced by direct BCR stimulation [40]. de Fougerolles et al. [41] reported that the membrane-proximal domain of CD19, CD19-domain 3 (D3) directly recognizes heparan sulfate. Moreover, CD19 is known to mediate BCR microcluster formation and to regulate cytoskeleton reorganization in B-cells [42, 43]. Therefore we can speculate that SDC4 HSPG may also be directly involved in BCR microcluster formation and thus might also modulate the threshold of B-cell activation. Our results extend this HSPG-mediated regulatory mechanism to human NK cells, by suggesting a model in which -cis interactions between HSPGs and numerous receptors on the NK cell surface establish a common regulatory mechanism [25, 30].
To summarize, we propose that the NKp44-HS -cis interactions can regulate receptor function through the NK cell-expressed HSPGs to delicately balance the NK cell activation threshold. Based upon our data, we also postulate that the use of heparan sulfate or heparin as a therapeutic agent in patients may in fact be significantly altering the activation threshold of the NK cells that express NKp44, KIR2DL4, NKp46 and NKp30 [25, 30]. Likewise, NKp44 expressing pathogen- or tumor-activated NK cells might exhibit even higher degree of functional modulation in response to aforementioned treatment. Multiple studies have demonstrated that use of HS and structurally similar Low Molecular Weight Heparin can inhibit tumor progression and metastasis [44]. Nevertheless, it remains to be determined to what degree can HS/heparin-based therapies affect the NK cell physiology to improve anti-tumor responses.
Materials and Methods
Cell culture
NK-92 (ATCC CRL2407) and KHYG-1 (JCRB0156; from HSRRB, Japan Health Sciences Foundation, Osaka, Japan) human NK and K562 (CCL-243; from ATCC) cell lines were cultured as recommended by ATCC or according to previously established protocols [45]. NK functional assay medium was supplemented with about 25% of IL-2 only (20U/mL for NK92 and 50U/mL for primary NK). NK92 cells were transduced with mCherry fused wild type NKp44 or mutant NKp44 alone or co-transduced with GFP fused SDC4 to produce NK92-44wt, NK92-44mut, NK92-44wt SDC4 and NK92-44mut SDC4 cell lines respectively. NK92 cell lines were created using mutagenesis and retroviral transduction protocols that were previously described [9, 25, 46].
Primary NK cell purification
A human negative NK cell isolation kit (Miltenyi Biotec) was used to purify NK cells from peripheral blood of healthy volunteer donors, who were recruited by informed consent as approved by the BGU Institutional Review Board. NK cell purity was >95% (CD3−CD56+; Supporting Information Fig. 1 – E,F). Purified NK cells were cultured in CellGro stem cell serum-free growth medium (CellGenix) supplemented with 10% heat-inactivated human AB plasma from healthy donors, 50U penicillin, 50mg streptomycin, 0.1mM non-essential amino acids, 1mM sodium pyruvate, 2mM L-Glutamine, 10 mM HEPES, pH 7.4, 0.1mM 2-mercaptoethanol (Gibco) and 300 IU/ml human IL-2 (Biological Industries).
Conventional and Image Flow Cytometry
The following antibodies were used: unconjugated or biotinylated Goat anti-human SDC4 (clone 2918; R&D); unconjugated 3.43.13 anti-NKp44 mAb (hybridoma generously provided by Dr. Marco Colonna, Washington University, St. Louis, MO); allophycocyanin-- or FITC-conjugated anti-mouse-IgG-Fc; allophycocyanin-conjugated anti-Goat-IgG-Fc; (all minimal cross reactivity; Jackson). Anti-human CD3-PE and anti-human CD56-BrilliantViolet mAbs and corresponding isotype control antibodies (Biolegend) were used to assess primary NK purity. For intracellular cytokine staining, Brefeldin A (cat. 420601, Biolegend) was added to the assay medium medium to the final concentration of 5µg/ml after 6h of initial incubation for another 6h. Intracellular cytokine staining protocol for IFN-γ was performed using Fixation buffer (cat. 420801, Biolegend) and Permeabilization Wash buffer (cat.421002, Biolegend) according to manufacturer specifications. IFN-γ production was assayed with biotin conjugated anti-human IFN-γ mAb (clone 4S.B3, Biolegend) and AlexaFluor-647 conjugated streptavidin (S-32357, Life Technologies). Conventional Flow Cytometry was performed on FACSCanto II (BD Biosciences) and Gallios (Beckman Coulter) flow cytometers and all the data was analyzed with FlowJo software (TreeStar). Imaging Flow Cytometry was performed on ImageStream Mark II high resolution microscopy in flow system (Amnis) using 40× and 60× magnification for NK92 and primary NK cells respectively. All the data was analyzed using the ImageStream Data Analysis and Exploration Software (IDEAS) (Amnis). NKp44 and SDC4 co-distribution analysis in NK cells was performed using co-localization and centroid features (IDEAS; Amnis).
Confocal imaging
8-chamber coverglass µ-slides (iBidi) were pre-coated with 50µg/ml of poly L lysine in Borate pH8.4 buffer and then coated with 10µg/ml of both unconjugated anti-CD45 mAb (clone HI30, Biolegend) and anti-NKp44 mAb (3.43.13) in DPBS at pH 7.4. Cells were cultured in 300µl of assay medium (see Cell culture) alone or medium supplemented with 10µg/ml of either HS (Heparin - low molecular weight; Sigma) or CS (Chondroitin Sulfate A; Sigma) and incubated at 37°C in 5% CO2 for 2h. All images were acquired at these standard cell culture conditions on a FluoView FV1000 confocal system using a 40x UPLSAPO objective (Olympus). The co-localization analysis was performed using the ImageJ software package supplemented with the JACoP plugin. Manders’ overlap coefficients M1 and M2 were used to evaluate the degree of co-localization between two fluorescent labels [47–49]. The threshold value was calculated from the background intensity of the noisiest images [50]. This value of threshold was applied in all subsequent M1 and M2 calculations, setting all below threshold pixels to zero. M1 was defined as the proportion of pixels with non-zero intensities from the green image (SDC4-GFP fusion protein), for which the intensity in the red channel is also above zero. Conversely, M2 was defined as the proportion of pixels with non-zero intensities from the red image (NKp44-mCherry fusion protein), for which the intensity in the green channel is also above zero. Thereby the M2 coefficient corresponds to the proportion of NKp44-mCherry co-localized with the appropriate GFP-labeled marker.
FRET was measured by the donor-sensitized acceptor fluorescence technique [51], as described previously [52–54]. Briefly, three images were acquired for each set of measurements: mCherry excitation/mCherry emission image; GFP excitation/GFP emission image; and GFP excitation/mCherry emission image (FRET channel). A set of reference images was acquired from GFP or mCherry fluorescent proteins single- or double-expressing cells for each set of acquisition parameters, and a calibration curve was derived to allow elimination of the non-FRET components from the FRET channel. The FRET efficiency was calculated on a pixel-by-pixel basis using the following equation: FRETeff = FRETcorr/(FRETcorr + GFP)×100%, where FRETcorr is the pixel intensity in the corrected FRET image, and GFP is the intensity of the corresponding pixel in the GFP channel image.
Cell stimulation and IFN-γ assays
For IFN-γ production assay, cells were stimulated in 96-well U-bottom plates (NUNC) pre-coated with the following antibodies at 2.5µg/ml concentration for 18h at 4°C: anti-NKp44 mAb (3.43.13) or anti-2B4 mAb (clone c1.7, Biolegend). For HS-dependent modulation of IFN-γ secretion NK cells were pre-incubated in assay medium (see Cell culture) supplemented with 5µg/ml of either HS or CS for 2h (37°C, 5% CO2) before plating (HS or CS were present in the assay medium. Antibodies were diluted in Na2HPO4 buffer, pH8 for plate coating. For all experiments 3×105 NK cells were added to each well in 150µl of assay medium (see Cell culture) and incubated for either 6h (intracellular IFN-γ production assay) or for 18h (IFN-γ secretion assay) (37°C, 5% CO2). Cytokine concentration in 100µl of supernatant was assayed by standard ELISA assay according to manufacturer specifications (ELISA MAX, Biolegend).
CD107a degranulation and direct cytotoxicity assays
For both NK cell degranulation and direct cytotoxicity assays NK92-44wt or NK92-44mut, cell lines were co-incubated for total of 4h (37°C, 5% CO2) with K562 target cell line at indicated effector-target (E:T) ratios. For NK cell degranulation assay, medium was supplemented with BrilliantViolet-conjugated anti-human CD107a mAb (Biolegend) (1:300 final dilution) after 1h of co-incubation. After incubation cells were washed and incubated for additional 30min on ice with BrilliantViolet-conjugated anti-human CD107a mAb (Biolegend) (1:200 final dilution). For each E:T ratio, 50,000 cells were acquired and CD107a expression was detected as described elsewhere (see Flow Cytometry). For direct cytotoxicity assay K562 target cells were first labeled with Vybrant DiD cell labeling solution (Life Technologies) according to the manufacturer recommendations and immediately used in the cytotoxicity assay. At the end of 4h co-incubation cells were washed with PBS and labeled with ZombieViolet viability dye according to the manufacturer recommendations (Life Technologies). Flow Cytometry was performed as described elsewhere (see Flow Cytometry), while 50,000 cells were acquired for each E:T ratio. Assay analysis was made in accordance to previously published methods [55, 56] and based on following gating strategy: mCherry-negative (excludes NK cells) and DiD positive (K562 cells) were subjected to Live/Dead discrimination (ZombieViolet negative or positive staining in accordance).
Supplementary Material
Acknowledgement
This work was supported by U.S./Israel Binational Science Foundation grant (AP and KSC), Israel Science Foundation grant 304/12 (A.P.) and NIH grant R01 CA083859 (KSC).
Footnotes
Author Contributions
M.B. and A.P. conceived the project and designed all the experiments. M.B., O.R., L.C. and R.Y. performed experiments and analyzed data. A.S. helped with primary NK purification. A.B., O.M. and K.S.C. provided structural consultation. M.B., K.S.C. and A.P. wrote the paper.
Conflict of interest
The authors declare no financial or commercial conflict of interest.
References
- 1.Yokoyama WM. The search for the missing 'missing-self' receptor on natural killer cells. Scand J Immunol. 2002;55:233–237. doi: 10.1046/j.1365-3083.2002.01054.x. [DOI] [PubMed] [Google Scholar]
- 2.Ljunggren HG, Karre K. In search of the 'missing self': MHC molecules and NK cell recognition. Immunol Today. 1990;11:237–244. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 3.Kruse PH, Matta J, Ugolini S, Vivier E. Natural cytotoxicity receptors and their ligands. Immunol Cell Biol. 2014;92:221–229. doi: 10.1038/icb.2013.98. [DOI] [PubMed] [Google Scholar]
- 4.Koch J, Steinle A, Watzl C, Mandelboim O. Activating natural cytotoxicity receptors of natural killer cells in cancer and infection. Trends Immunol. 2013;34:182–191. doi: 10.1016/j.it.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Brusilovsky M, Rosental B, Shemesh A, Appel MY, Porgador A. Human NK cell recognition of target cells in the prism of natural cytotoxicity receptors and their ligands. J Immunotoxicol. 2012;9:267–274. doi: 10.3109/1547691X.2012.675366. [DOI] [PubMed] [Google Scholar]
- 6.De Maria A, Ugolotti E, Rutjens E, Mazza S, Radic L, Faravelli A, Koopman G, Di Marco E, Costa P, Ensoli B, Cafaro A, Mingari MC, Moretta L, Heeney J, Biassoni R. NKp44 expression, phylogenesis and function in non-human primate NK cells. Int Immunol. 2009;21:245–255. doi: 10.1093/intimm/dxn144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vitale M, Bottino C, Sivori S, Sanseverino L, Castriconi R, Marcenaro E, Augugliaro R, Moretta L, Moretta A. NKp44, a novel triggering surface molecule specifically expressed by activated natural killer cells, is involved in non-major histocompatibility complex-restricted tumor cell lysis. J Exp Med. 1998;187:2065–2072. doi: 10.1084/jem.187.12.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joyce MG, Sun PD. The structural basis of ligand recognition by natural killer cell receptors. J Biomed Biotechnol. 2011;2011:203628. doi: 10.1155/2011/203628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hershkovitz O, Jivov S, Bloushtain N, Zilka A, Landau G, Bar-Ilan A, Lichtenstein RG, Campbell KS, van Kuppevelt TH, Porgador A. Characterization of the recognition of tumor cells by the natural cytotoxicity receptor, NKp44. Biochemistry. 2007;46:7426–7436. doi: 10.1021/bi7000455. [DOI] [PubMed] [Google Scholar]
- 10.Hecht ML, Rosental B, Horlacher T, Hershkovitz O, De Paz JL, Noti C, Schauer S, Porgador A, Seeberger PH. Natural cytotoxicity receptors NKp30, NKp44 and NKp46 bind to different heparan sulfate/heparin sequences. J Proteome Res. 2009;8:712–720. doi: 10.1021/pr800747c. [DOI] [PubMed] [Google Scholar]
- 11.Ito K, Higai K, Shinoda C, Sakurai M, Yanai K, Azuma Y, Matsumoto K. Unlike natural killer (NK) p30, natural cytotoxicity receptor NKp44 binds to multimeric alpha2,3-NeuNAc-containing N-glycans. Biol Pharm Bull. 2012;35:594–600. doi: 10.1248/bpb.35.594. [DOI] [PubMed] [Google Scholar]
- 12.Campbell KS, Yusa S, Kikuchi-Maki A, Catina TL. NKp44 triggers NK cell activation through DAP12 association that is not influenced by a putative cytoplasmic inhibitory sequence. J Immunol. 2004;172:899–906. doi: 10.4049/jimmunol.172.2.899. [DOI] [PubMed] [Google Scholar]
- 13.Rosental B, Brusilovsky M, Hadad U, Oz D, Appel MY, Afergan F, Yossef R, Rosenberg LA, Aharoni A, Cerwenka A, Campbell KS, Braiman A, Porgador A. Proliferating cell nuclear antigen is a novel inhibitory ligand for the natural cytotoxicity receptor NKp44. J Immunol. 2011;187:5693–5702. doi: 10.4049/jimmunol.1102267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cantoni C, Bottino C, Vitale M, Pessino A, Augugliaro R, Malaspina A, Parolini S, Moretta L, Moretta A, Biassoni R. NKp44, a triggering receptor involved in tumor cell lysis by activated human natural killer cells, is a novel member of the immunoglobulin superfamily. J Exp Med. 1999;189:787–796. doi: 10.1084/jem.189.5.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomasello E, Olcese L, Vely F, Geourgeon C, Blery M, Moqrich A, Gautheret D, Djabali M, Mattei MG, Vivier E. Gene structure, expression pattern, and biological activity of mouse killer cell activating receptor-associated protein (KARAP)/DAP-12. J Biol Chem. 1998;273:34115–34119. doi: 10.1074/jbc.273.51.34115. [DOI] [PubMed] [Google Scholar]
- 16.Wilson MJ, Lindquist JA, Trowsdale J. DAP12 and KAP10 (DAP10)-novel transmembrane adapter proteins of the CD3zeta family. Immunol Res. 2000;22:21–42. doi: 10.1385/IR:22:1:21. [DOI] [PubMed] [Google Scholar]
- 17.Arnon TI, Lev M, Katz G, Chernobrov Y, Porgador A, Mandelboim O. Recognition of viral hemagglutinins by NKp44 but not by NKp30. Eur J Immunol. 2001;31:2680–2689. doi: 10.1002/1521-4141(200109)31:9<2680::aid-immu2680>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 18.Ho JW, Hershkovitz O, Peiris M, Zilka A, Bar-Ilan A, Nal B, Chu K, Kudelko M, Kam YW, Achdout H, Mandelboim M, Altmeyer R, Mandelboim O, Bruzzone R, Porgador A. H5-type influenza virus hemagglutinin is functionally recognized by the natural killer-activating receptor NKp44. J Virol. 2008;82:2028–2032. doi: 10.1128/JVI.02065-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hershkovitz O, Rosental B, Rosenberg LA, Navarro-Sanchez ME, Jivov S, Zilka A, Gershoni-Yahalom O, Brient-Litzler E, Bedouelle H, Ho JW, Campbell KS, Rager-Zisman B, Despres P, Porgador A. NKp44 receptor mediates interaction of the envelope glycoproteins from the West Nile and dengue viruses with NK cells. J Immunol. 2009;183:2610–2621. doi: 10.4049/jimmunol.0802806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baychelier F, Sennepin A, Ermonval M, Dorgham K, Debre P, Vieillard V. Identification of a cellular ligand for the natural cytotoxicity receptor NKp44. Blood. 2013;122:2935–2942. doi: 10.1182/blood-2013-03-489054. [DOI] [PubMed] [Google Scholar]
- 21.Bloushtain N, Qimron U, Bar-Ilan A, Hershkovitz O, Gazit R, Fima E, Korc M, Vlodavsky I, Bovin NV, Porgador A. Membrane-associated heparan sulfate proteoglycans are involved in the recognition of cellular targets by NKp30 and NKp46. J Immunol. 2004;173:2392–2401. doi: 10.4049/jimmunol.173.4.2392. [DOI] [PubMed] [Google Scholar]
- 22.Zilka A, Landau G, Hershkovitz O, Bloushtain N, Bar-Ilan A, Benchetrit F, Fima E, van Kuppevelt TH, Gallagher JT, Elgavish S, Porgador A. Characterization of the heparin/heparan sulfate binding site of the natural cytotoxicity receptor NKp46. Biochemistry. 2005;44:14477–14485. doi: 10.1021/bi051241s. [DOI] [PubMed] [Google Scholar]
- 23.Hershkovitz O, Jarahian M, Zilka A, Bar-Ilan A, Landau G, Jivov S, Tekoah Y, Glicklis R, Gallagher JT, Hoffmann SC, Zer H, Mandelboim O, Watzl C, Momburg F, Porgador A. Altered glycosylation of recombinant NKp30 hampers binding to heparan sulfate: a lesson for the use of recombinant immunoreceptors as an immunological tool. Glycobiology. 2008;18:28–41. doi: 10.1093/glycob/cwm125. [DOI] [PubMed] [Google Scholar]
- 24.Carey DJ. Syndecans: multifunctional cell-surface co-receptors. Biochem J. 1997;327(Pt 1):1–16. doi: 10.1042/bj3270001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brusilovsky M, Cordoba M, Rosental B, Hershkovitz O, Andrake MD, Pecherskaya A, Einarson MB, Zhou Y, Braiman A, Campbell KS, Porgador A. Genome-wide siRNA screen reveals a new cellular partner of NK cell receptor KIR2DL4: heparan sulfate directly modulates KIR2DL4-mediated responses. J Immunol. 2013;191:5256–5267. doi: 10.4049/jimmunol.1302079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fadnes B, Husebekk A, Svineng G, Rekdal O, Yanagishita M, Kolset SO, Uhlin-Hansen L. The proteoglycan repertoire of lymphoid cells. Glycoconj J. 2012;29:513–523. doi: 10.1007/s10719-012-9427-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cerwenka A, Lanier LL. Ligands for natural killer cell receptors: redundancy or specificity. Immunol Rev. 2001;181:158–169. doi: 10.1034/j.1600-065x.2001.1810113.x. [DOI] [PubMed] [Google Scholar]
- 28.Higai K, Matsumoto K. [Glycan ligand specificity of killer lectin receptors] Yakugaku Zasshi. 2012;132:705–712. doi: 10.1248/yakushi.132.705. [DOI] [PubMed] [Google Scholar]
- 29.Higai K, Suzuki C, Imaizumi Y, Xin X, Azuma Y, Matsumoto K. Binding affinities of NKG2D and CD94 to sialyl Lewis X-expressing N-glycans and heparin. Biol Pharm Bull. 2011;34:8–12. doi: 10.1248/bpb.34.8. [DOI] [PubMed] [Google Scholar]
- 30.Brusilovsky M, Radinsky O, Yossef R, Campbell KS, Porgador A. Carbohydrate-mediated modulation of NK cell receptor function: structural and functional influences of Heparan sulfate moieties expressed on NK cell surface. Frontiers in Oncology. 2014;4 doi: 10.3389/fonc.2014.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scarpellino L, Oeschger F, Guillaume P, Coudert JD, Levy F, Leclercq G, Held W. Interactions of Ly49 family receptors with MHC class I ligands in trans and cis. J Immunol. 2007;178:1277–1284. doi: 10.4049/jimmunol.178.3.1277. [DOI] [PubMed] [Google Scholar]
- 32.Avril T, Freeman SD, Attrill H, Clarke RG, Crocker PR. Siglec-5 (CD170) can mediate inhibitory signaling in the absence of immunoreceptor tyrosine-based inhibitory motif phosphorylation. J Biol Chem. 2005;280:19843–19851. doi: 10.1074/jbc.M502041200. [DOI] [PubMed] [Google Scholar]
- 33.Avril T, North SJ, Haslam SM, Willison HJ, Crocker PR. Probing the cis interactions of the inhibitory receptor Siglec-7 with alpha2,8-disialylated ligands on natural killer cells and other leukocytes using glycan-specific antibodies and by analysis of alpha2,8-sialyltransferase gene expression. J Leukoc Biol. 2006;80:787–796. doi: 10.1189/jlb.1005559. [DOI] [PubMed] [Google Scholar]
- 34.Nicoll G, Avril T, Lock K, Furukawa K, Bovin N, Crocker PR. Ganglioside GD3 expression on target cells can modulate NK cell cytotoxicity via siglec-7-dependent and -independent mechanisms. Eur J Immunol. 2003;33:1642–1648. doi: 10.1002/eji.200323693. [DOI] [PubMed] [Google Scholar]
- 35.Chalifour A, Scarpellino L, Back J, Brodin P, Devevre E, Gros F, Levy F, Leclercq G, Hoglund P, Beermann F, Held W. A Role for cis Interaction between the Inhibitory Ly49A receptor and MHC class I for natural killer cell education. Immunity. 2009;30:337–347. doi: 10.1016/j.immuni.2008.12.019. [DOI] [PubMed] [Google Scholar]
- 36.Doucey MA, Scarpellino L, Zimmer J, Guillaume P, Luescher IF, Bron C, Held W. Cis association of Ly49A with MHC class I restricts natural killer cell inhibition. Nat Immunol. 2004;5:328–336. doi: 10.1038/ni1043. [DOI] [PubMed] [Google Scholar]
- 37.Held W, Mariuzza RA. Cis-trans interactions of cell surface receptors: biological roles and structural basis. Cell Mol Life Sci. 2011;68:3469–3478. doi: 10.1007/s00018-011-0798-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Held W, Mariuzza RA. Cis interactions of immunoreceptors with MHC and non-MHC ligands. Nat Rev Immunol. 2008;8:269–278. doi: 10.1038/nri2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bessoles S, Angelov GS, Back J, Leclercq G, Vivier E, Held W. Education of murine NK cells requires both cis and trans recognition of MHC class I molecules. J Immunol. 2013;191:5044–5051. doi: 10.4049/jimmunol.1301971. [DOI] [PubMed] [Google Scholar]
- 40.Yamashita Y, Oritani K, Miyoshi EK, Wall R, Bernfield M, Kincade PW. Syndecan-4 is expressed by B lineage lymphocytes and can transmit a signal for formation of dendritic processes. J Immunol. 1999;162:5940–5948. [PubMed] [Google Scholar]
- 41.de Fougerolles AR, Batista F, Johnsson E, Fearon DT. IgM and stromal cell-associated heparan sulfate/heparin as complement-independent ligands for CD19. Eur J Immunol. 2001;31:2189–2199. doi: 10.1002/1521-4141(200107)31:7<2189::aid-immu2189>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 42.Batista FD, Treanor B, Harwood NE. Visualizing a role for the actin cytoskeleton in the regulation of B-cell activation. Immunol Rev. 2010;237:191–204. doi: 10.1111/j.1600-065X.2010.00943.x. [DOI] [PubMed] [Google Scholar]
- 43.Depoil D, Fleire S, Treanor BL, Weber M, Harwood NE, Marchbank KL, Tybulewicz VL, Batista FD. CD19 is essential for B cell activation by promoting B cell receptor-antigen microcluster formation in response to membrane-bound ligand. Nat Immunol. 2008;9:63–72. doi: 10.1038/ni1547. [DOI] [PubMed] [Google Scholar]
- 44.Sasisekharan R, Shriver Z, Venkataraman G, Narayanasami U. Roles of heparan-sulphate glycosaminoglycans in cancer. Nat Rev Cancer. 2002;2:521–528. doi: 10.1038/nrc842. [DOI] [PubMed] [Google Scholar]
- 45.Purdy AK, Campbell KS. Natural killer cells and cancer: regulation by the killer cell Ig-like receptors (KIR) Cancer Biol Ther. 2009;8:2211–2220. doi: 10.4161/cbt.8.23.10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yusa S, Catina TL, Campbell KS. SHP-1- and phosphotyrosine-independent inhibitory signaling by a killer cell Ig-like receptor cytoplasmic domain in human NK cells. J Immunol. 2002;168:5047–5057. doi: 10.4049/jimmunol.168.10.5047. [DOI] [PubMed] [Google Scholar]
- 47.Manders EM, Hoebe R, Strackee J, Vossepoel AM, Aten JA. Largest contour segmentation: a tool for the localization of spots in confocal images. Cytometry. 1996;23:15–21. doi: 10.1002/(SICI)1097-0320(19960101)23:1<15::AID-CYTO3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 48.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 49.Manders EM, Stap J, Brakenhoff GJ, van Driel R, Aten JA. Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J Cell Sci. 1992;103(Pt 3):857–862. doi: 10.1242/jcs.103.3.857. [DOI] [PubMed] [Google Scholar]
- 50.Hoebe RA, van Noorden CJ, Manders EM. Noise effects and filtering in controlled light exposure microscopy. J Microsc. 2010;240:197–206. doi: 10.1111/j.1365-2818.2010.03400.x. [DOI] [PubMed] [Google Scholar]
- 51.Zal T, Gascoigne NR. Using live FRET imaging to reveal early protein-protein interactions during T cell activation. Curr Opin Immunol. 2004;16:674–683. [PubMed] [Google Scholar]
- 52.Braiman A, Barda-Saad M, Sommers CL, Samelson LE. Recruitment and activation of PLCgamma1 in T cells: a new insight into old domains. EMBO J. 2006;25:774–784. doi: 10.1038/sj.emboj.7600978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barda-Saad M, Braiman A, Titerence R, Bunnell SC, Barr VA, Samelson LE. Dynamic molecular interactions linking the T cell antigen receptor to the actin cytoskeleton. Nat Immunol. 2005;6:80–89. doi: 10.1038/ni1143. [DOI] [PubMed] [Google Scholar]
- 54.Jaron-Mendelson M, Yossef R, Appel MY, Zilka A, Hadad U, Afergan F, Rosental B, Engel S, Nedvetzki S, Braiman A, Porgador A. Dimerization of NKp46 receptor is essential for NKp46-mediated lysis: characterization of the dimerization site by epitope mapping. J Immunol. 2012;188:6165–6174. doi: 10.4049/jimmunol.1102496. [DOI] [PubMed] [Google Scholar]
- 55.Lecoeur H, Fevrier M, Garcia S, Riviere Y, Gougeon ML. A novel flow cytometric assay for quantitation and multiparametric characterization of cell-mediated cytotoxicity. J Immunol Methods. 2001;253:177–187. doi: 10.1016/s0022-1759(01)00359-3. [DOI] [PubMed] [Google Scholar]
- 56.Wilkinson RW, Lee-MacAry AE, Davies D, Snary D, Ross EL. Antibody-dependent cell-mediated cytotoxicity: a flow cytometry-based assay using fluorophores. J Immunol Methods. 2001;258:183–191. doi: 10.1016/s0022-1759(01)00474-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.