Abstract
The β-barrel outer membrane proteins constitute one of the two known structural classes of membrane proteins. Whereas there are several different web-based predictors for α-helical membrane proteins, currently there is no freely available prediction method for β-barrel membrane proteins, at least with an acceptable level of accuracy. We present here a web server (PRED-TMBB, http://bioinformatics.biol.uoa.gr/PRED-TMBB) which is capable of predicting the transmembrane strands and the topology of β-barrel outer membrane proteins of Gram-negative bacteria. The method is based on a Hidden Markov Model, trained according to the Conditional Maximum Likelihood criterion. The model was retrained and the training set now includes 16 non-homologous outer membrane proteins with structures known at atomic resolution. The user may submit one sequence at a time and has the option of choosing between three different decoding methods. The server reports the predicted topology of a given protein, a score indicating the probability of the protein being an outer membrane β-barrel protein, posterior probabilities for the transmembrane strand prediction and a graphical representation of the assumed position of the transmembrane strands with respect to the lipid bilayer.
INTRODUCTION
Integral membrane proteins are divided into two distinct structural classes, the α-helical membrane proteins and the β-barrel membrane proteins. The α-helical membrane proteins are found mostly in the cell membranes of both prokaryotic and eukaryotic organisms, performing a variety of biologically important functions. Their membrane spanning regions form α-helices, which consist mainly of hydrophobic residues (1). A variety of computational techniques have been proposed for the prediction of the transmembrane segments of α-helical membrane proteins, with high levels of accuracy and precision. Furthermore, there are several freely accessible web servers for the prediction of α-helical membrane spanning segments. On the other hand, the members of the β-barrel membrane protein class are located in the outer membrane of Gram-negative bacteria, and presumably in the outer membrane of chloroplasts and mitochondria. These proteins have membrane spanning segments formed by antiparallel β-strands, creating a channel in the form of a barrel that spans the outer membrane (2). It is of great importance to possess powerful, freely available tools to predict the transmembrane topology since only a few outer membrane proteins have known three-dimensional structures. During the last few years, some methods have been proposed for the prediction of beta-barrel outer membrane proteins based on statistical analyses (3,4), neural networks (5,6) and Hidden Markov Models (HMMs) (7,8), but so far none of them has been freely available to the scientific community, with the exception of some older methods based on neural networks trained on smaller datasets (5,6), which demonstrated moderate performance. In this work, we present a web server based on a Hidden Markov Model capable of predicting the transmembrane topology of β-barrel outer membrane proteins. The model is retrained in order to include newly solved three-dimensional structures. The application offers the choice between three different decoding algorithms, and additionally outputs a graphical representation of the assumed topology with respect to the membrane.
MATERIALS AND METHODS
PRED-TMBB is based on a Hidden Markov Model (9), a probabilistic model consisting of several states connected by means of the transition probabilities. The architecture of the model is designed to fit as much as possible to the limitations imposed by the known structures. For training, we used Conditional Maximum Likelihood (CML) training for labelled data, as proposed by Krogh (10). This kind of training, often referred to as discriminating training, seeks to maximize the probability of the correct prediction rather than the probability of the sequences, given the model. The parameters of the model (transition and emission probabilities) are updated simultaneously, using the gradients of the likelihood function as described in (11), and the training process terminates when the likelihood does not increase beyond a pre-specified threshold. To reduce the number of the free parameters of the model, and thus improve the generalization capability, states expected to have the same emission probabilities were tied together. Furthermore, to avoid overfitting, the iterations started from emission probabilities corresponding to the initial amino acid frequencies observed in the known protein structures, and small pseudocounts were added at each step. For decoding we implemented three well-known algorithms, the standard Viterbi algorithm (9), the N-best algorithm (12) and posterior decoding using a dynamic programming algorithm. The complete details of the model, the training and the decoding procedure are described elsewhere (13).
The model was retrained in order to include some recently solved three-dimensional structures of β-barrel outer membrane proteins deposited in the Protein Data Bank (PDB) (14). The total number of sequences in the training set is 16, all belonging to the fold ‘beta-barrel transmembrane proteins’ of the SCOP database (Table 1) (15). The sequences have been submitted to a redundancy check, removing chains with a sequence identity above some threshold. We consider two sequences as being homologues if they possess identical residues >30% in a pairwise alignment in a sequence longer than 80 residues. For the pairwise local alignment we used BlastP (16) with default parameters, and the homologous sequences were removed by implementing Algorithm 2 from Hobohm et al. (17). For training and testing the model, we considered only the part of the beta-strand that is inserted in the lipid bilayer, and not the whole beta-strand, which in some cases extends far away from the membrane. For the transmembrane strand predictions, we report the well-known SOV (measure of the segment's overlap), which is considered to be the most reliable measure for evaluating the performance of secondary structure prediction methods (18). We also report the total number of correctly predicted topologies, i.e. when both the strands' localization and the loops' orientation have been predicted correctly. As measures of the per-residue accuracy, we report here both the total fraction of the correctly predicted residues (Qβ) in a two-state model (transmembrane versus non-transmembrane) and the well-known Matthews Correlation Coefficient (Cβ) (19). For reasons of fair comparison with other existing methods, all measures of performance were evaluated against the manually derived annotations for the transmembrane segments used by our team for training (13), and also against the annotations of the transmembrane strands as deposited in the PDB, even though, in some cases, these clearly extend beyond the lipid bilayer. Finally, the model produces a score used to discriminate β-barrel membrane proteins from globular ones (13). This score is just the negative log-likelihood of the sequence given the model, normalized by dividing by the sequence length. Proteins producing a score lower than a predefined threshold (see below) are considered to be beta-barrel membrane proteins.
Table 1. The non-redundant dataset of 16 outer membrane proteins used for training the model.
| Protein name | Number of β-strands | PDB code | Organism |
|---|---|---|---|
| OmpA | 8 | 1QJP | E.coli |
| OmpX | 8 | 1QJ8 | E.coli |
| OmpT | 10 | 1I78 | E.coli |
| OpcA | 10 | 1K24 | Neisseria meningitidis |
| OmpLA | 12 | 1QD5 | E.coli |
| Omp32 | 16 | 1E54 | Comamonas acidovorans |
| OmpF | 16 | 2OMF | E.coli |
| Porin | 16 | 2POR | Rhodobacter capsulatus |
| Porin | 16 | 1PRN | Rhodobacter blasticus |
| Sucrose porin | 18 | 1A0S | Salmonella typhimurium |
| Maltoporin | 18 | 2MPR | S.typhimurium |
| FepA | 22 | 1FEP | E.coli |
| NspA | 8 | 1P4T | N.meningitidis |
| BtuB | 22 | 1NQE | E.coli |
| FhuA | 22 | 2FCP | E.coli |
| FecA | 22 | 1KMO | E.coli |
RESULTS
The performance of the model is summarized in Table 2, where we list the results obtained by comparing the three different decoding algorithms. We note that the posterior decoding method using the dynamic programming algorithm to locate the transmembrane strands performs marginally better than the Viterbi or the N-best algorithm, as already noted in (20), and should perhaps be preferred. In the self-consistency test (when the model is trained and tested on the whole dataset at the same time) the percentage of correctly predicted residues is 92.2%, the correlation coefficient = 0.84 and SOV = 0.94. When we tested the model using the jackknife procedure (i.e. removing a protein from the training set, training the model with the remaining proteins and performing the test on the protein removed), the percentage of correctly predicted residues is 87.5%, the correlation coefficient = 0.74 and SOV = 0.85. The number of correctly predicted topologies is 12 (15 when counting 3 strands that were predicted misplaced) in the self-consistency test and 8 (11 when counting 3 strands that were predicted misplaced) in the jackknife test. All results reported here are computed according to the posterior decoding method. The results in the jackknife test clearly outperform the original version of the algorithm (13), and also all other methods reported in the literature (6–8), even though we use only single-sequence information. In Table 3, we also report the prediction performance of two other publicly available predictors on the same dataset. B2TMPRED is the neural network developed in (6), using evolutionary information derived from multiple alignments, and TM-BETA is a newly developed neural network method (21) using single-sequence information. PRED-TMBB, even in the jackknife test reported in Table 2, performs significantly better, although the majority of the proteins in the dataset were also present in the sets used for training these methods. The superiority of our method is due to the model design and the training scheme, and also in part to the complete training set, which is the largest non-redundant set reported in the literature so far. We speculate that other, more refined methods, such as those reported in (7) or (8), would perform comparable to PRED-TMBB, but they are not publicly available. Furthermore, PRED-TMBB is the only available method which is capable of not only predicting the membrane spanning strands but also discriminating beta-barrel membrane proteins. At a fixed score threshold of 2.965, the model correctly discriminated 88% of a non-redundant set of 133 well-annotated outer membrane proteins and 89% of a set of 1100 globular proteins from PDB-Select [for details on these datasets, refer to (13)]. Using the same threshold, it correctly predicted 95% of the 149 beta-barrel membrane proteins deposited in TCDB (22) and 73% of a non-redundant set of 82 alpha-helical membrane proteins with structures known at atomic resolution. Clearly, the model discriminates, with the highest level of accuracy and precision reported so far, beta-barrel membrane proteins from globular ones. However, when it comes to alpha-helical membrane proteins, more reliable predictors already exist (23), and their use should be preferred to filtering completely unknown sequences or screening large datasets (13).
Table 2. Overall measures of accuracy obtained in the self-consistency and jackknife tests for the three different decoding algorithms.
| Decoding method | Qβ | Cβ | SOV | TOP | TOPs |
|---|---|---|---|---|---|
| Self-consistency | |||||
| Viterbi | 92.6% (79.9%) | 0.84 (0.62) | 0.93 (0.86) | 12 (12) | 13 (13) |
| N-Best | 92.8% (80.0%) | 0.85 (0.62) | 0.94 (0.86) | 12 (12) | 14 (14) |
| Posterior | 92.2% (80.1%) | 0.84 (0.62) | 0.94 (0.86) | 12 (12) | 15 (15) |
| Jacknife | |||||
| Viterbi | 86.0% (75.6%) | 0.70 (0.53) | 0.82 (0.76) | 9 (10) | 12 (12) |
| N-Best | 86.0% (75.6%) | 0.70 (0.53) | 0.82 (0.76) | 9 (10) | 12 (12) |
| Posterior | 87.5%(77.0%) | 0.74 (0.56) | 0.85 (0.80) | 8 (8) | 11 (11) |
Qβ: percentage of correctly predicted residues (19). Cβ: Matthews Correlation Coefficient (19). SOV: Segment Overlap measure (18). TOP: proteins with correctly predicted topologies (strand localization and orientation of the loops). TOPs: proteins with correctly predicted topologies, with the inclusion of shifted strand predictions. Values in parentheses correspond to the measures of accuracy obtained when using, as observed, the annotations for the transmembrane strands taken from PDB [see also (13)].
Table 3. Overall measures of the accuracy of PRED-TMBB and comparison with other available predictors.
| Method | Qβ | Cβ | SOV | TOP | TOPs |
|---|---|---|---|---|---|
| PRED-TMBB | 92.2% (80.1%) | 0.84 (0.62) | 0.94 (0.86) | 12 (12) | 15 (15) |
| B2TMPREDa | 78.3% (80.1%) | 0.57 (0.60) | 0.69 (0.73) | 4c (4c) | 5c (5c) |
| TM-BETAb | 71.9% (72.6%) | 0.44 (0.45) | 0.62 (0.63) | 1c (1c) | 1c (1c) |
For definitions of accuracy measures see Table 2.
aB2TMPRED is available at http://gpcr.biocomp.unibo.it/cgi/predictors/outer/pred_outercgi.cgi.
bTM-BETA is available at http://psfs.cbrc.jp/tmbeta-net/.
cThese predictors do not report the full topology but only the location of the transmembrane strands.
THE SERVER
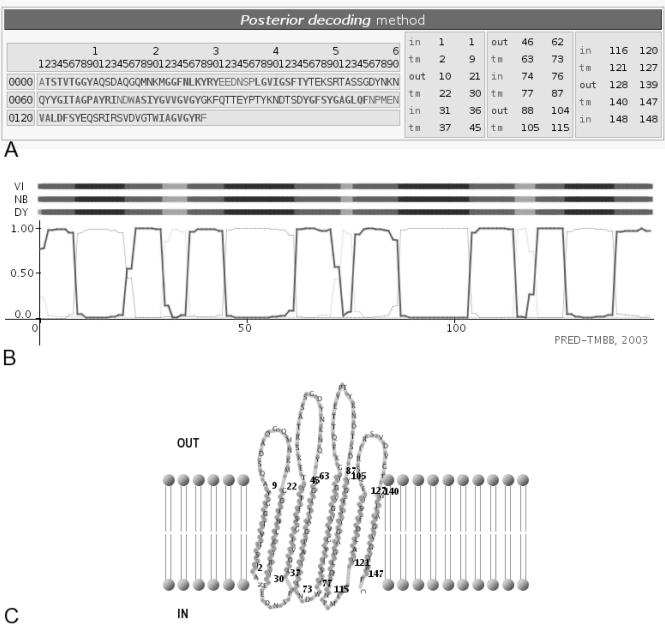
On the initial page, the user may submit a sequence in FASTA format and has the option of choosing between the three different decoding methods currently available. Decoding can be performed using the N-best algorithm, the standard Viterbi algorithm or ‘a posteriori’ with the aid of a dynamic programming algorithm. The three alternative algorithms can be run simultaneously, but this may slow down the server's reporting time. We should mention here that although the accuracy of PRED-TMBB is not significantly affected by the existence of a signal peptide, the presence of a signal peptide is a strong indication of the protein's localization in the outer membrane. Thus, when it comes to precursor sequences such as those of genome projects, this aspect should also be considered. The final output consists of the prediction for the transmembrane strands (Figure 1). Optionally, the user may obtain a graphical plot showing the posterior probabilities in a three-state mode (extracellular, periplasmic and transmembrane), which may be useful in the case of ambiguously defined topologies. The application also returns the score used for discrimination purposes, thus helping the user to identify possible β-barrel outer membrane proteins. Another useful feature of the application is the option to produce (after the decoding process) a graphical representation showing the relative position of the predicted transmembrane strands with respect to the lipid bilayer. Such a depiction might be useful for presentation and publication purposes.
Figure 1.
Output of the prediction obtained from PRED-TMBB for the OmpX of E.coli. (A) The prediction of the transmembrane strands according to posterior decoding. (B) Plot of the posterior probabilities for the transmembrane strands, along the sequence. (C) Graphical representation of the predicted topology with respect to the lipid bilayer.
CONCLUSIONS
We present here a web sever based on a Hidden Markov Model for the prediction of the transmembrane β-strands of the outer membrane proteins of Gram-negative bacteria. To our knowledge, this is the first time that such a web server accessible to the public has been made. Furthermore, the method performs better than any previously published method and is the only method not only able to predict the strands' localization and the location of the loops (periplasmic/extracellular), but also capable of discriminating beta-barrel membrane proteins from globular ones. The server outputs the prediction of the transmembrane β-strands, posterior probabilities for the prediction, the discrimination score and a graphical depiction of the protein's orientation with respect to the lipid bilayer, thus making this server a unique and complete approach for the prediction of the transmembrane topology of outer membrane proteins. The Hidden Markov Model parameters will be updated on a regular basis whenever new crystallographically solved structures become available, and we plan to enrich the application with additional new services in the future.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank the anonymous referees for their valuable comments and constructive criticism. P.G.B. was supported by a grant from the IRAKLEITOS fellowships programme of the Greek Ministry of National Education, supporting basic research in the National and Kapodistrian University of Athens.
REFERENCES
- 1.Von Heijne G. (1999) Recent advances in the understanding of membrane protein assembly and function. Quart. Rev. Biophys., 32, 285–307. [DOI] [PubMed] [Google Scholar]
- 2.Schulz G.E. (2002) The structure of bacterial outer membrane proteins. Biochim. Biophys. Acta, 1565, 308–317. [DOI] [PubMed] [Google Scholar]
- 3.Zhai Y. and Saier,M.H.,Jr (2002) The beta-barrel finder (BBF) program, allowing identification of outer membrane beta-barrel proteins encoded within prokaryotic genomes. Protein Sci., 11, 2196–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wimley W.C. (2002) Toward genomic identification of beta-barrel membrane proteins: composition and architecture of known structures. Protein Sci., 11, 301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diederichs K., Freigang,J., Umhau,S., Zeth,K. and Breed,J. (1998) Prediction by a neural network of outer membrane beta-strand protein topology. Protein Sci., 7, 2413–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacoboni I., Martelli,P.L., Fariselli,P., De Pinto,V. and Casadio,R. (2001) Prediction of the transmembrane regions of beta-barrel membrane proteins with a neural network-based predictor. Protein Sci., 10, 779–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Q., Zhu,Y.S., Wang,B.H. and Li,Y.X. (2003) A HMM-based method to predict the transmembrane regions of beta-barrel membrane proteins. Comput. Biol. Chem., 27, 69–76. [DOI] [PubMed] [Google Scholar]
- 8.Martelli P.L., Fariselli,P., Krogh,A. and Casadio,R. (2002) A sequence-profile-based HMM for predicting and discriminating beta barrel membrane proteins. Bioinformatics, 18(Suppl 1), S46–S53. [DOI] [PubMed] [Google Scholar]
- 9.Rabiner L. (1989) A tutorial on hidden Markov models and selected applications in speech recognition. Proc. IEEE, 77, 257–286. [Google Scholar]
- 10.Krogh A. (1994) Hidden Markov Models for labelled sequences. Proceedings of the 12th IAPR International Conference on Pattern Recognition, Israel, pp. 140–144.
- 11.Baum L. (1972) An inequality and associated maximization technique in statistical estimation for probabilistic functions of Markov processes. Inequalities, 3, 1–8. [Google Scholar]
- 12.Krogh A. (1997) Two methods for improving performance of an HMM and their application for gene finding. Proc. Int. Conf. Intell. Syst. Mol. Biol., 5, 179–186. [PubMed] [Google Scholar]
- 13.Bagos P.G., Liakopoulos,T.D., Spyropoulos,I.C. and Hamodrakas,S.J. (2004) A Hidden Markov Model method, capable of predicting and discriminating beta-barrel outer membrane proteins. BMC Bioinformatics, 5, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berman H.M., Battistuz,T., Bhat,T.N., Bluhm,W.F., Bourne,P.E., Burkhardt,K., Feng,Z., Gilliland,G.L., Iype,L., Jain,S. et al. (2002) The Protein Data Bank. Acta Crystallogr. D Biol. Crystallogr., 58, 899–907. [DOI] [PubMed] [Google Scholar]
- 15.Andreeva A., Howorth,D., Brenner,S.E., Hubbard,T.J., Chothia,C. and Murzin,A.G. (2004) SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res., 32, D226–D229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hobohm U., Scharf,M., Schneider,R. and Sander,C. (1992) Selection of representative protein data sets. Protein Sci., 1, 409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zemla A., Venclovas,C., Fidelis,K. and Rost,B. (1999) A modified definition of Sov, a segment-based measure for protein secondary structure prediction assessment. Proteins, 34, 220–223. [DOI] [PubMed] [Google Scholar]
- 19.Baldi P., Brunak,S., Chauvin,Y., Andersen,C.A. and Nielsen,H. (2000) Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics, 16, 412–424. [DOI] [PubMed] [Google Scholar]
- 20.Fariselli P., Finelli,M., Marchignoli,D., Martelli,P.L., Rossi,I. and Casadio,R. (2003) MaxSubSeq: an algorithm for segment-length optimization. The case study of the transmembrane spanning segments. Bioinformatics, 19, 500–505. [DOI] [PubMed] [Google Scholar]
- 21.Gromiha M.M., Ahmad,S. and Suwa,M. (2004) Neural network-based prediction of transmembrane beta-strand segments in outer membrane proteins. J. Comput. Chem., 25, 762–767. [DOI] [PubMed] [Google Scholar]
- 22.Busch W. and Saier,M.H.,Jr (2002) The transporter classification (TC) system, 2002. Crit. Rev. Biochem. Mol. Biol., 37, 287–337. [DOI] [PubMed] [Google Scholar]
- 23.Pasquier C., Promponas,V.J. and Hamodrakas,S.J. (2001) PRED-CLASS: cascading neural networks for generalized protein classification and genome-wide applications. Proteins, 44, 361–369. [DOI] [PubMed] [Google Scholar]