Abstract
The WUSCHEL (WUS)-related homeobox (WOX) gene family coordinates transcription during the early phases of embryogenesis. In this study, a putative WOX2 homolog was isolated and characterized from Aegilops tauschii, the donor of D genome of Triticum aestivum. The sequence consisted of 2045 bp, and contained an open reading frame (ORF), encoded 322 amino acids. The predicted protein sequence contained a highly conserved homeodomain and the WUS-box domain, which is present in some members of the WOX protein family. The full-length ORF was subcloned into prokaryotic expression vector pET-30a, and an approximately 34-kDa protein was expressed in Escherichia coli BL21 (DE3) cells with IPTG induction. The molecular mass of the expressed protein was identical to that predicted by the cDNA sequence. Phylogenetic analysis suggested that Ae. tauschii WOX2 is closely related to the rice and maize orthologs. Quantitative PCR analysis showed that WOX2 from Ae. tauschii was primarily expressed in the seeds; transcription increased during seed development and declined after the embryos matured, suggesting that WOX2 is associated with embryo development in Ae. tauschii.
Keywords: WOX2, Aegilops tauschii, phylogenetic analysis, real-time quantitative PCR
Introduction
WOX genes comprise a large plant-specific gene family that belongs to the homeodomain (HD) class of transcription factors. WUS HDs are critical for early-phase embryogenesis and lateral organ development (Haecker et al., 2004). HD proteins typically contain a highly conserved motif of 60 amino acids encoded by a characteristic DNA fragment called the homeobox and consisting of a helix-loop-helix-turn-helix structure (Kamiya et al., 2003). Phylogenetic analyses have divided the WOX gene family into three major clades: the ancient clade with representatives from all lineages of land plants as well as green algae, and the intermediate and modern clades with representatives found only in ferns and seed plants (Nardmann et al., 2009; van der Graaff et al., 2009). In addition to the HD, the primary structures of WOX proteins in the modern clade also possess conserved structures called the WUS box and/or EAR-like motif (Kieffer et al., 2006; Lin et al., 2013).
Previous studies have identified 15 WOX gene family members in Arabidopsis thaliana: the WUS gene is specifically expressed in the shoot apical meristem (SAM) where it regulates stem cell fate (Mayer et al., 1998; Haecker et al., 2004; Lian et al., 2014; Zhang et al., 2010) and WOX5 performs similar functions in the root apical meristem (RAM) (Sarkar et al., 2007). PRESSED FLOWER/AtWOX3 is required for the development of lateral stipules in the leaves and the lateral sepals and stamens of flowers (Matsumoto and Okada, 2001), while AtWOX4 acts as a key regulator of cambium activity in the main stem of Arabidopsis (Suer et al., 2011). PRETTY FEW SEEDS2/AtWOX6 transcripts accumulate in developing primordia and differentiating organs and regulate ovule development (Park et al., 2005). STIMPY/AtWOX9 maintains cell division and prevents premature differentiation in the center zone of the SAM (Wu et al., 2005). Moreover, OsWOX11 mediates activation of crown root emergence and growth in rice (Zhao et al., 2009). AtWOX2 and AtWOX8 are co-expressed in egg cells and zygotes and are involved in embryo formation and differentiation in Arabidopsis (Haecker et al., 2004). In maize, ZmWOX2A, a homolog of AtWOX2, is expressed in the zygote, where it marks the apical cell fate and prepatterns the SAM position (Nardmann et al., 2007). PaWOX2, which is the Picea abies homolog of AtWOX2, is highly expressed during early growth of the zygote and somatic embryo (Palovaara et al., 2010).
Although there have been several studies of the WOX gene family in monocots, such as rice and maize (Kamiya et al., 2003; Nardmann et al., 2007), little is known about WOX genes in Triticeae plants. In this study, we characterized the WOX2 gene structure and its expression from Aegilops tauschii, the D-genome progenitor species of hexaploid wheat. The objective of this study was to investigate the structural features of WOX2, determine its expression pattern at different tissues in Ae. tauschii, and examine the evolutionary history of the WOX genes.
Material and Methods
Plant material
Ae. tauschii AS2407 (2n = 2× = 14, DD) was cultivated in the field for sample collection. The seeds were provided by the Triticeae Research Institute of Sichuan Agriculture University, ChengDu, Sichuan, China. Genomic DNA was extracted from fresh leaves by using a CTAB procedure (Murray and Thompson, 1980). For gene expression analysis, root tissue was harvested from 1-week old seedlings. Leaf and stem tissues were from 3-weeks old seedlings. Stamina and ovaries were harvested before anthesis. Developing seeds were harvested from between 2 and 24 days after flowering at 2-day intervals. Immature embryos were cultured on MS callus induction medium (Özgen et al., 1998) supplemented with 2 mg/L 2,4-dichlorophenoxyacetic acid. The cultures were maintained at 26 °C in the dark. Calli were collected after twelve days’ culture. All samples were immediately frozen in liquid nitrogen and stored at −80 °C.
Database analysis and cloning of WOX2 from Ae. tauschii
We searched Arabidopsis, rice, and maize WOX2 DNA sequences from the National Centre for Biotechnology Information. After alignment using the ClustalW program (version 1.83) (Thompson et al., 1994), the conserved sequences were used to design a pair of degenerate primers (HDF/R) (Table 1) for amplifying part of the Ae. tauschii WOX2 gene. The amplified PCR product was purified and cloned into a pMD19-T vector (TaKaRa Biotechnology Co. Ltd. Dalian, China) for sequencing. BLAST searching of the GenBank database with the partial Ae. tauschii WOX2 sequence identified one WOX ortholog. Based on this WOX ortholog, a pair of primers (WOX2_F/R) (Table 1) was designed to amplify the full-length gene from Ae. tauschii using cDNA and gDNA templates. PCR was performed with the LA PCR Kit Ver. 2.1 (TaKaRa Biotechnology Co. Ltd. Dalian, China). Sequence alignment and protein prediction were performed in DNAMAN software (Ver. 5.2.10; Lynnon Biosoft).
Table 1. Primer sequences.
| Primer name | Primer sequence (5′→3′) |
|---|---|
| HDF | TGGACVCCGACGVCGGAGCAGRT |
| HDR | GCCTTGTGGTTCTGGAACCAGTAGAA |
| WOX2_F | ACAGAGCAGATCCGTCCTCCTTAGCC |
| WOX2_R | CTCCTTGAGTCGGTGTTCAGCTTAGTGA |
| PET30a_F | TATCATATGAGAGCAGATCCGTCCTCCTTAGCC |
| PET30a_R | GTCGAATTCCTTGAGTCGGTGTTCAGCTTAGTGA |
| qWOX2F1 | CCCCATCGAGGGCAAGAACG |
| qWOX2R1 | CGGCGGAACTGCTTGGAGAA |
| actinF1 | GTTCCAATCTATAAGGGATACACGC |
| actinR1 | GAACCTCCACTGAGAACAACATTACC |
Phylogenetic analysis
To examine the evolutionary history of the WOX genes and the evolutionary relationship of the Ae. tauschii WOX2 with other plants, we screened sequences homologous to AtWUS and AtWOX2 using TBLASTN in the NCBI database, with default parameters and the WUS/WOX2 homeodomain sequences as queries. We retrieved 76 WOX genes including 15 in Arabidopsis, 10 in rice, 15 in maize, 17 in Populus tomentosa, six in Theobroma cacao, and six in Ginkgo biloba (GenBank accession numbers are listed in the Supplemental Table S1). One WOX2 gene was identified in each of the following species: Glycine max, Vitis vinifera, Petunia x hybrida, Picea abies, Cicer arietinum, Fragaria vesca, and Solanum tuberosum (Table S1). These sequences were used to construct a phylogenetic tree. Multiple sequence alignment was performed in ClustalW (version 1.83) (Thompson et al., 1994). MEGA 5.0 was used to construct a Maximum Likelihood (ML) tree by using the Jones-Taylor-Thornton (JTT) model and with 1000 bootstrap replicates (Jones et al., 1992; Tamura et al., 2011).
Bacterial expression of cloned Ae. tauschii WOX2 ORFs
To express the Ae. tauschii WOX2 open reading frame (ORF) in bacteria, the PET30a_F/R primer (Table 1) sets were designed to amplify an ORF from the cDNA sequence. We modified the WOX2 ORF using PCR mutagenesis, which is almost the same as general PCR except the primer added two restriction sites for NdeI and EcoRI. PCR conditions were identical to those described above except the template was plasmid DNA purified from the respective clones. The ORFs were cloned into the pET-30a expression vector (Novagen) and then expressed in BL21 Escherichia coli (DE3) cells. To induce bacterial expression, the cells were treated with 0.5 mM isopropyl β-d-thiogalactopyranoside (IPTG) for 3–5 h. The proteins were extracted with extraction buffer (1% SDS, 5% 2-mercaptoethanol, and 0.25 M Tris-HC1, pH 6.8) (Santambrogio et al., 1993), heated at 96 °C for 10 min, separated by 12% SDS-PAGE, and stained with Coomassie Blue R-250.
Expression analysis by real-time quantitative PCR (RT-qPCR)
Total RNA was extracted with Trizol reagent (Tiangen Biotech Co. Ltd, Beijing, China) according to the manufacturer’s protocol. First-strand cDNA was synthesized with PrimeScript RT and gDNA Eraser (TaKaRa Biotechnology Co. Ltd. Dalian, China). RT-qPCR was performed with diluted cDNA and the SYBR Premix Ex Taq II Kit (TaKaRa Biotechnology Co. Ltd. Dalian, China). Reactions were performed in triplicate in a BioRad CFX96 thermocycler under the following cycling conditions: 5 min at 95 °C, followed by 40 cycles of 30 s at 95 °C, 30 s at 60 °C, 40 s at 72 °C, and a final extension of 10 min at 72 °C. Negative (distilled water) and no-template (total RNA) controls were included in each run. The data were analyzed with the BioRad CFX Manager software. Relative expression was calculated based on the standard curve method described by Pfaffl (2001). Gene-specific primers for WOX2 (qWOX2F1/R1) were used to detect WOX2 RNA levels normalized to actin (actinF1/R1; GenBank accession number, AY145451) (Li et al., 2010). Primer sequences are shown in Table 1.
Results
Isolation and sequence analysis of WOX2 from Ae. tauschii
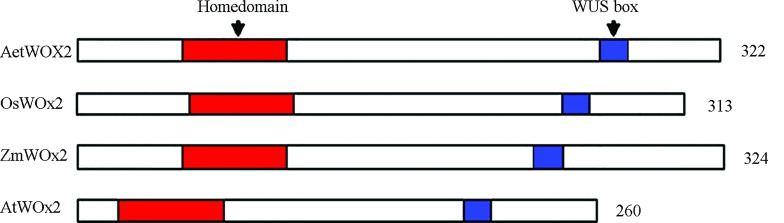
Based on the multiple alignments of homologous genes from Arabidopsis, rice, and maize, we amplified the HD region of Ae. tauschii WOX2 with degenerate primers. After blasting against the GenBank database, we found a cDNA clone from wheat (AK335177) in which the HD is the same as the partial WOX2 sequence we cloned in Ae. tauschii and highly homologous to WOX2 in other species. This clone contained the complete ORF. To determine the coding sequence (CDS) of the putative Ae. tauschii WOX2 gene, we isolated the gDNA and cDNA sequences from Ae. tauschii. Sequence analysis revealed that the WOX2 gene is 2,045 bp in length, with a 969 bp ORF, a 29 bp 5′ upstream sequence, and a 104 bp 3′ downstream sequence (GenBank accession numbers KJ459391). Two exons and one intron were identified for WOX2 after alignment of gDNA with the respective ORF. The putative WOX2 gene in Ae. tauschii was predicted to encode a polypeptide of 322 amino acids. Our BlastP results against public databases indicated that the primary HD region of Ae. tauschii WOX2 (amino acids 40–105) is conserved in all WOX family members. The WOX2 HD in Ae. tauschii shared 89% and 85% similarity with OsWOX2 in rice and ZmWOX2A in maize, respectively. Its similarity to Arabidopsis AtWOX2 was, however, only 69%. Domain prediction analysis revealed that the putative WOX2 protein of Ae. tauschii also contains the WUS-box domain, which is present in the modern clade of the WOX protein family (Figure 1). We also compared the full protein sequences of Ae. tauschii WOX2 with O. sativa, Z. mays, G. max, Arabidopsis, V. vinifera, P. hybrida, G. biloba, S. tuberosum, C. arietinum, F. vesca, and P. abies (Figure 2). Multiple alignments showed that the WOX2 HD was highly conserved across all 12 species whereas the flanking sequences varied.
Figure 1. Structure of WOX2 proteins in different species. Abbreviations are Ae. tauschii, Aet; O. sativa, Os; Z. mays, Zm; A. thaliana, At; The homeodomain (red) is the most prominent and defining feature of the family. The WUS-box motif (blue) was defined in a strict sense, as T-L-[DEQP]-L-F-P-[GITVL]-[GSKNTCV], consensus TLELFPLH.
Figure 2. Multiple sequence alignment of WOX2. Abbreviations are G. biloba, Gb; G. max, Gm; V. vinifera, Vv; Petunia x hybrida, Ph; S. tuberosum, St; C. arietinum, Ca; F. vesca, Fv; and P. abies, Pa.
Expression of Ae. tauschii WOX2 in E. coli
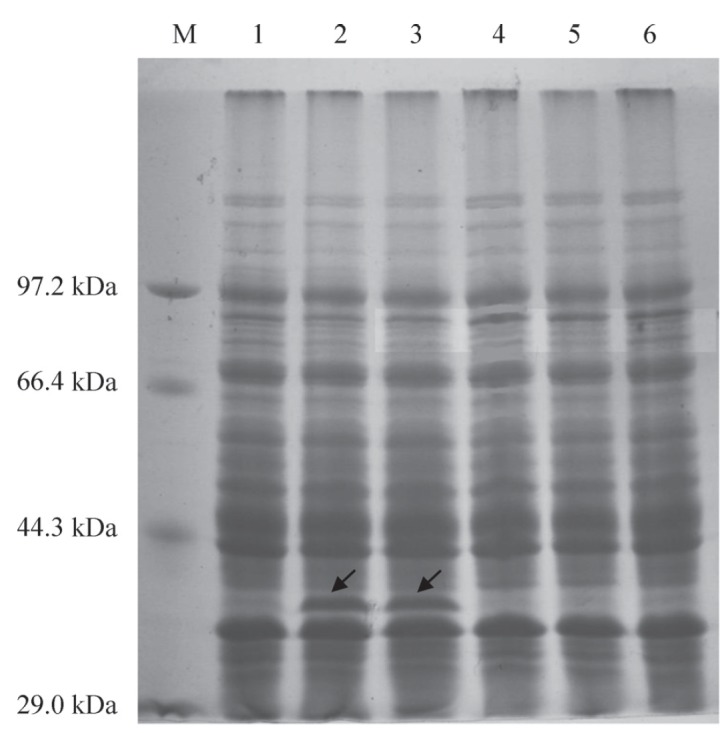
We used the clone of the Ae. tauschii WOX2 ORF to analyze protein expression so as to verify whether the cloned gene expresses the corresponding protein product. Furthermore this also will allow the recombinant protein to be used as antigen to develop antibodies, which could be used to detect the expression of the transgenic progeny at the protein level in future studies. We ligated the WOX2 ORF from Ae. tauschii into pET-30a by using modified restriction sites. The recombinant plasmid, pET30a-AetWOX2, was then expressed in E. coli BL21 (DE3) and protein expression was induced with IPTG. SDS-PAGE revealed a single protein band of approximately 34 kDa in the induced extract (Figure 3). The molecular mass of the expressed protein was identical to that predicted by the AetWOX2 cDNA sequence, so the single protein band is accurate representation of the AetWOX2 encoding protein.
Figure 3. Expression of a pET30a-WOX2 fusion protein. Lane 1, uninduced pET30a-WOX2; lanes 2–3, pET30a-WOX2 induced by 0.5 mM IPTG for 3 and 5 h, respectively; lane 4, uninduced pET30a; lane 5, induced pET30a for 3 h; lane 6, negative control E. coli BL21 (DE3); M, Protein Molecular Weight Marker (Low) (TaKaRa). The target protein is indicated with arrows.
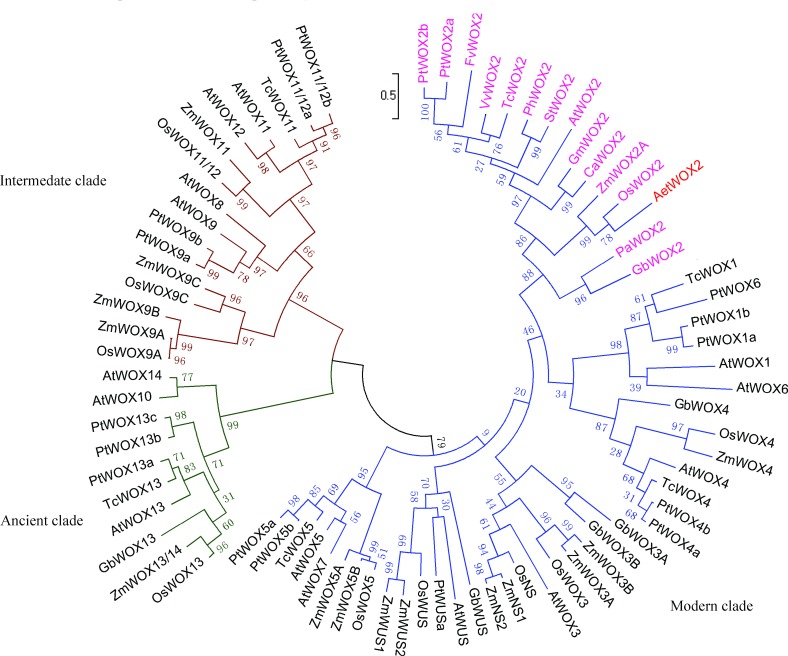
Phylogenetic analysis of Ae. tauschii WOX2
To confirm the classifications and analyze the phylogenetic relationships of the WOX proteins, multiple alignment analyses of the 77 WOX proteins were performed. The resulting ML tree divided the 77 WOX members into three clades: modern, intermediate, and ancient (Figure 4). There were 51 WOX proteins in the modern clade, which can be subdivided into five subgroups including WUS, WOX1/WOX4, WOX2, WOX3/NS, and WOX5; ten WOXs in the ancient clade of WOX10, WOX13, and WOX14 homologues; and sixteen WOXs in the intermediate clade consisting of WOX8, WOX9, WOX11, and WOX12. The WOX2 subgroup in the modern clade contained one member from each of twelve species. AetWOX2 in the WOX2 subgroup clustered with OsWOX2 and ZmWOX2A. WOX2 from Ae. tauschii is closely related to the rice and maize orthologs.
Figure 4. Phylogenetic analysis of plant WUS/WOX proteins based on the Maximum Likelihood method. Abbreviations are P. tomentosa, Pt; and T. cacao, Tc. The three clades are color-coded, WUS/WOX1-7 (modern) in blue, WOX8, 9, 11, 12 (intermediate) in brown and WOX10, 13, 14 (ancient) in green. Protein names of the WOX2 subclade are in purple and AetWOX2 is red.
Expression pattern analysis
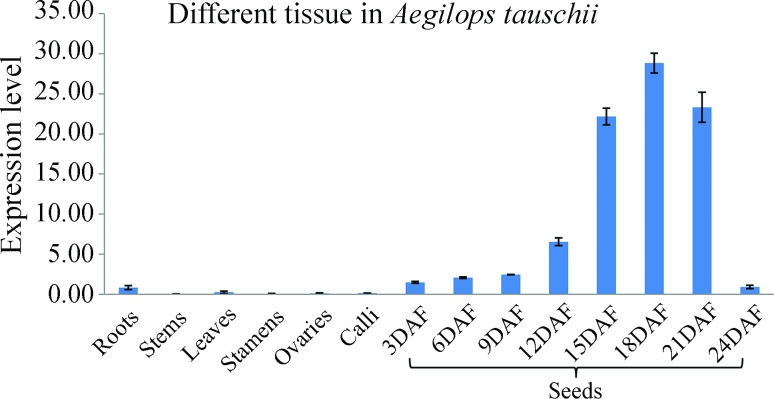
To understand the function of WOX2 from Ae. tauschii, we investigated its expression in different tissues of Ae. tauschii, and in seeds at different stages of development. We set the expression level in roots as equal to 1, and calculated the expression in other tissues as relative to roots. The qRT-PCR results revealed that WOX2 is primarily expressed in seeds, with lower levels transcript levels in the other tissues (Figure 5). Expression of Ae. tauschii WOX2 continuously increased from 12 day after flowering (DAF), peaked at 18 DAF, and then declined until 24 DAF. Around 18 DAF is the stage at which embryos become mature.
Figure 5. Real-time PCR analysis of WOX2 transcript levels in different tissues of Ae. tauschii and seed growth stages. Expression was normalized to an actin gene.
Discussion
The coding region of AetWOX2 shared 99% similarity with the clone AK335177 from wheat, a result agrees with the well-established fact that Ae. taushii is the donor of the D genome to the hexaploid wheat, T. aestivum (Mc and Sears, 1946; Gill et al., 1996). As Ae. taushii is one of most important gene resources for improving common wheat, exploiting the WOX gene in Ae. tauschii may contribute to understanding the differentiation and evolution of the WOX gene in Triticeae species.
The WOX2 gene from Ae. tauschii encodes a WUS/WOX-like HD protein that is highly conserved. The WOX2 HD consists of about 65 amino acids with one extra amino acid in the loop between helices 1 and 2, and four extra amino acids in the turn between helices 2 and 3 (Figure 2). The WUS-box, another domain in Ae. tauschii WOX2, is located downstream of the HD and is found in most WOX proteins (Haecker et al., 2004). We did not find an EAR-like motif in any the examined WOX2 sequences, but it is present in WUS/WOX5. The sequence similarity of AetWOX2 with OsWOX2 and ZmWOX2A is 89% and 85%, respectively. The orthology of AetWOX2 with OsWOX2 and ZmWOX2A in the modern clade (Figure 4) is well supported by the phylogenetic tree. Taken together, these results suggest that WOX2 is a bona fide member of the WOX family in Ae. tauschii.
The close phylogenetic relationship among WOXs in Arabidopsis, rice, maize, P. tomentosa, T. cacao, and G. biloba orthologs (Figure 4) suggested that WOXs are evolutionarily conserved. It is interesting that no rice and maize orthologs of Arabidopsis WOX1, 6, 7, 8, 10, and 14 were found in the phylogenetic tree. Arabidopsis has at least 15 WOX members, there are nine independent clones encoding WUS-type proteins from rice (Kamiya et al., 2003), and research in maize indicated that orthologs of WOX1, 6, 7, 8, 10, and 14 are not present in grass genomes (Nardmann et al., 2007). Therefore, the WOX family and functions of its member are distinct in different species.
WOX2 is associated with embryo patterning in Arabidopsis, maize, and spruce (Haecker et al., 2004; Nardmann et al., 2007; Palovaara et al., 2010). In Ae. tauschii, the process of embryonic growth is consistent with seed development, so it is possible that the expression of WOX2 in seeds corresponds to that in the embryo. Transcript levels of Ae. tauschii WOX2 increased during the early process of seed development and declined after the embryo matured (Figure 5). Based on the function of members of the WOX2 gene family already described in different plant species, and the expression pattern during seed development, the data suggest that WOX2 in Ae. tauschii may indeed be associated with embryo development.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31230053) and the China Transgenic Research Program (2011ZX08002-005).
Supplementary Material.
Accession numbers of WOX proteins.
The following online material is available for this article:
Table S1 - Accession numbers of WOX proteins.
This material is available as part of the online article from http://www.scielo.br/gmb.
Footnotes
Associate Editor: Adriana S. Hemerly
References
- Gill KS, Gill BS, Endo TR, Boyko EV. Identification and high-density mapping of gene-rich regions in chromosome group 5 of wheat. Genetics. 1996;143:1001–1012. doi: 10.1093/genetics/143.2.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana . Development. 2004;131:657–668. doi: 10.1242/dev.00963. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kamiya N, Nagasaki H, Morikami A, Sato Y, Matsuoka M. Isolation and characterization of a rice WUSCHEL-type homeobox gene that is specifically expressed in the central cells of a quiescent center in the root apical meristem. Plant J. 2003;35:429–441. doi: 10.1046/j.1365-313x.2003.01816.x. [DOI] [PubMed] [Google Scholar]
- Kieffer M, Stern Y, Cook H, Clerici E, Maulbetsch C, Laux T, Davies B. Analysis of the transcription factor WUSCHEL and its functional homologue in Antirrhinum reveals a potential mechanism for their roles in meristem maintenance. Plant Cell. 2006;18:560–573. doi: 10.1105/tpc.105.039107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QF, Sun SS, Yuan DY, Yu HX, Gu MH, Liu QQ. Validation of candidate reference genes for the accurate normalization of real-time quantitative RT-PCR data in rice during seed development. Plant Mol Biol Rep. 2010;28:49–57. [Google Scholar]
- Lian G, Ding Z, Wang Q, Zhang D, Xu J. Origins and evolution of WUSCHEL-related homeobox protein family in plant kingdom. Sci World J. 2014;2014:e534140. doi: 10.1155/2014/534140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Niu L, McHale NA, Ohme-Takagi M, Mysore KS, Tadege M. Evolutionarily conserved repressive activity of WOX proteins mediates leaf blade outgrowth and floral organ development in plants. Proc Natl Acad Sci USA. 2013;110:366–371. doi: 10.1073/pnas.1215376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto N, Okada K. A homeobox gene, PRESSED FLOWER, regulates lateral axis-dependent development of Arabidopsis flowers. Genes Dev. 2001;15:3355–3364. doi: 10.1101/gad.931001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- McFadden ES, Sears ER. The origin of Triticum spelta and its free-threshing hexaploid relatives. J Hered. 1946;37:107–116. doi: 10.1093/oxfordjournals.jhered.a105590. [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardmann J, Zimmermann R, Durantini D, Kranz E, Werr W. WOX gene phylogeny in Poaceae: A comparative approach addressing leaf and embryo development. Mol Biol Evol. 2007;24:2474–2484. doi: 10.1093/molbev/msm182. [DOI] [PubMed] [Google Scholar]
- Nardmann J, Reisewitz P, Werr W. Discrete shoot and root stem cell-promoting WUS/WOX5 functions are an evolutionary innovation of angiosperms. Mol Biol Evol. 2009;26:1745–1755. doi: 10.1093/molbev/msp084. [DOI] [PubMed] [Google Scholar]
- Özgen M, Türet M, Altinok S, Sancak C. Efficient callus induction and plant regeneration from mature embryo culture of winter wheat (Triticum aestivum L.) genotypes. Plant Cell Rep. 1998;18:331–335. doi: 10.1007/s002990050581. [DOI] [PubMed] [Google Scholar]
- Palovaara J, Hallberg H, Stasolla C, Hakman I. Comparative expression pattern analysis of WUSCHEL-related homeobox 2 (WOX2) and WOX8/9 in developing seeds and somatic embryos of the gymnosperm Picea abies . New Phytol. 2010;188:122–135. doi: 10.1111/j.1469-8137.2010.03336.x. [DOI] [PubMed] [Google Scholar]
- Park SO, Zheng Z, Oppenheimer DG, Hauser BA. The PRETTY FEW SEEDS2 gene encodes an Arabidopsis homeodomain protein that regulates ovule development. Development. 2005;132:841–849. doi: 10.1242/dev.01654. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:45e. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santambrogio P, Levi S, Cozzi A, Rovida E, Albertini A, Arosio P. Production and characterization of recombinant heteropolymers of human ferritin H and L chains. J Biol Chem. 1993;268:12744–12748. [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T. Conserved factors regulate signaling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007;446:811–814. doi: 10.1038/nature05703. [DOI] [PubMed] [Google Scholar]
- Suer S, Agusti J, Sanchez P, Schwarz M, Greb T. WOX4 imparts auxin responsiveness to cambium cells in Arabidopsis . Plant Cell. 2011;23:3247–3259. doi: 10.1105/tpc.111.087874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaff E, Laux T, Rensing SA. The WUS homeobox-containing (WOX) protein family. Genome Biol. 2009;10:e248. doi: 10.1186/gb-2009-10-12-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Dabi T, Weigel D. Requirement of homeobox gene STIMPY/WOX9 for Arabidopsis meristem growth and maintenance. Curr Biol. 2005;15:436–440. doi: 10.1016/j.cub.2004.12.079. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zong J, Liu J, Yin J, Zhang D. Genome-wide analysis of WOX gene family in rice, sorghum, maize, Arabidopsis and poplar. J Integr Plant Biol. 2010;52:1016–1026. doi: 10.1111/j.1744-7909.2010.00982.x. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Hu Y, Dai M, Huang L, Zhou DX. The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell. 2009;21:736–748. doi: 10.1105/tpc.108.061655. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Accession numbers of WOX proteins.