Abstract
The entomopathogenic fungi of the genus Metarhizium have several subtilisin-like proteases that are involved in pathogenesis and these have been used to investigate genes that are differentially expressed in response to different growth conditions. The identification and characterization of these proteases can provide insight into how the fungus is capable of infecting a wide variety of insects and adapt to different substrates. In addition, the pr1A gene has been used for the genetic improvement of strains used in pest control. In this study we used quantitative RT-PCR to assess the relative expression levels of the pr1A gene in M. anisopliae and M. acridum during growth in different culture conditions and during infection of the sugar cane borer, Diatraea saccharalis Fabricius. We also carried out a pathogenicity test to assess the virulence of both species against D. saccharalis and correlated the results with the pattern of pr1A gene expression. This analysis revealed that, in both species, the pr1A gene was differentially expressed under the growth conditions studied and during the pathogenic process. M. anisopliae showed higher expression of pr1A in all conditions examined, when compared to M. acridum. Furthermore, M. anisopliae showed a greater potential to control D. saccharalis. Taken together, our results suggest that these species have developed different strategies to adapt to different growing conditions.
Keywords: entomopathogen, Diatraea saccharalis, quantitative RT-PCR, expression pattern
Introduction
In Brazil, sugarcane monoculture forms the basis of the sugar export and biofuel industries (Alves et al., 2008). The sugarcane borer Diatraea saccharalis Fabricius is considered one of the major sugarcane pests in the Americas. Due to its cryptic lifestyle, conventional control measures by deploying chemical insecticides targeting the larvae are ineffective.
Entomopathogenic fungi are used in biological pest control as a promising alternative to chemical pesticides due to a number of advantages, including lower environmental impact, lower costs, higher specificity and lesser risk of development of resistance (Samuels et al., 1989; Frazzon et al., 2000). Species that belong to the genus Metarhizium are collectively able to infect a broad range of insects (Samuels et al., 1989) and are adapted to life in the root rhizosphere (Roberts and St. Leger, 2004). However, little is known about the molecular and physiological mechanisms involved in their adaptation to different growth conditions, and this lack of understanding has prevented the development of new strategies to improve their effectiveness in biological pest control (Zhang et al., 2011).
The taxonomy of Metarhizium was reassessed based on multigenic phylogenetic analysis covering the tef-1 gene (translation elongation factor 1-α) rpb1 (large subunit of RNA polymerase II) rpb2 (second largest subunit of RNA polymerase II) and β-tub (β-tubulin). This revaluation led to the recognition of different Metarhizium species: M. anisopliae (Mestchnikoff) Sorokin, M. guizhouense Q.T. Chen & H.L. Guo, M. pingshaense Q.T. Chen & H.L. Guo, M. acridum (Driver and Milner) Bischoff, Rehner & Humber, M. lepidiotae (Driver and Milner) Bischoff, Rehner & Humber and M. majus (Johnston) Bischoff, Rehner & Humber (Bischoff et al., 2009).
A large and diverse range of enzymes and toxins have been identified in several studies that are thought to be critical for the ability of the fungus to infect diverse groups of insects and ticks, and to grow in different types of substrate (Freimoser et al., 2003, 2005; Wang et al., 2005). In particular, subtilisin protease PR1A is the predominant protein produced during degradation of insect cuticle (Bagga et al., 2004). Thus, the gene encoding this protein (pr1A) is investigated for its use in the development of advanced engineered biopesticides (St. Leger et al., 1989).
Identifying genes that are up- or down-regulated in response to a given host insect or growth condition should increase our understanding of the genetic mechanisms involved in host specificity and adaptation (Pathan et al., 2007; He et al., 2012; Luo et al., 2013; Jin et al., 2014). M. anisopliae has been reported to have a large host range of over 200 insect species (Samuels et al., 1989). In contrast, M. acridum has a very limited host-range and is only known to attack orthopteran insects (Bischoff et al., 2009). A more thorough understanding of this behavioral flexibility may be obtained by comparatively studying the two species, and such studies should contribute to the development of more effective pest control. Thus, the objectives of this study were to analyze the expression of the pr1A gene in M. anisopliae and M. acridum during growth in different culture conditions and during the pathogenic process of D. saccharalis infection. We then went on to correlate the expression of pr1A with the pathogenicity of M. anisopliae and M. acridum, against D. saccharalis.
Materials and Methods
Fungal culture and identification
Metarhizium anisopliae URM 4921, originally isolated from Mahanarva pos-ticata Stal from Brazil, and M. acridum URM 4412, originally isolated from Austracnis guttulosa Walker from Australia, were obtained from the mycological collection of the Department of Mycology, Federal University of Pernambuco (URM-UFPE). Cultures were grown on potato dextrose agar at 28 °C for 12 days to obtain conidia. For confirmation of the species identity the partial sequence of tef-1 gene (translation elongation factor 1-α) was amplified and sequenced. The primers used for amplification and sequencing were designed for the EF-1a intron region: EF1T (5′-ATGGGTAAGGARGACAAGAC) and EF2T (5′-GGAAGTACCAGTGATCATGTT) (Bischoff et al., 2009). Sequence data were manually adjusted by Staden software (Staden et al., 1998) and compared to the GenBank database using BLASTn (Altschul et al., 1990).
Growth conditions for expression analysis of the pr1A gene
Analysis of the pr1A gene expression pattern was performed after growth of the two fungi species in YPD medium (0.2% yeast extract, 1% peptone, 2% dextrose), in liquid minimal medium (MM) (Pontecorvo et al., 1953) without glucose and supplemented with 1% casein (MM + casein), and in liquid MM without glucose and supplemented with 1% (w/v) insect cuticle (MM + cuticle). To obtain the cuticle, we used third-instar larvae of D. saccharalis. The insects were crushed to remove internal material, dried to a constant weight in an oven at 80 °C, and then the exoskeleton was macerated. The resulting powder was sieved and stored frozen at −25 °C. To obtain a cuticle suspension (1%), we resuspended the cuticle powder in an aqueous solution of potassium tetraborate (1%) and subjected the mixture to flowing steam for 20 min (Andersen, 1980). The cuticle extract was then added to sterile liquid MM.
Conidia were harvested in 0.01% Tween 80 aqueous solution, and the conidia suspen-sion was filtered through glass wool to remove mycelia. Conidia (2 × 108 conidia/mL) were inoculated in 20 mL of culture medium as described above and incubated as shake cultures at 150 rpm, 28 °C. The mycelia were collected at 24 h and 72 h after inoculation, immediately frozen in liquid nitrogen, and maintained for 24 h at −80 °C for subsequent extraction of RNA. The experimental design followed a 2×3 randomized factorial scheme (two fungal strains + three culture medium) for a total of six treatments with two replicates each.
Preparation of insects for expression analysis of the pr1A gene
Third-instar larvae of D. saccharalis were infected by immersion for 1 min in a conidia suspension of 2 × 108 conidia/mL. Each infected larva was then placed in a Petri dish containing a small piece of sugarcane stalk, and the plates were incubated at 28 °C. The process of fungal pathogenesis is commonly divided into the following five phases: uninfected insect, 20 h after infection, dead infected insect, emergent mycelia from the insect cadaver, and insect cadaver mummified with conidia. The insects collected in the phases described above were immediately frozen in liquid nitrogen and maintained for 24 h at −80 °C for subsequent RNA extraction. Two biological replicates were performed for each phase analyzed. Eight insects were used per biological replicate.
Total RNA isolation and cDNA synthesis
The frozen samples of mycelia grown in different culture media and the insect samples described above were ground in liquid nitrogen. For each sample, 100–150 mg of powdered sample was placed in a cooled 2-mL tube. RNA was extracted with Trizol reagent (Invitrogen) according to the manufacturer’s instructions. RNA was suspended in 50 μL DEPC-treated water. The purity of the total RNA was determined based on the absorbance at 260/280 nM, and RNA integrity was verified by electrophoresis in a 1% agarose gel. Residual DNA was removed by treating RNA with RNase-free DNase I according to the manufacturer’s instructions (Invitrogen). RNA was stored at −80 °C until further use. An aliquot of 2 μg DNase-treated RNA was transcribed into cDNA using the SuperScriptTM First-Strand Synthesis System for RT-PCR (Invitrogen) and oligo-dT primers (Invitrogen).
Quantitative real-time reverse transcription polymerase chain reaction (RT-qPCR)
To obtain qPCR products, the Platinum® SYBR® Green qPCR SuperMix-UDG kit (Invitrogen) was used. Each 25-μL qPCR reaction contained 12.5 μL of qPCR SuperMix-UDG Kit (Invitrogen), 0.5 μL of MgCl2 (50 mM), 0.1 μL each of the forward and reverse primers (each at 100 μM) (Bioneer), 10.8 μL nuclease-free water and 1 μL cDNA (20 ng/μL cDNA in each sample). Negative controls (no DNA template) for each primer set were included in each run to ensure that there was no contamination in any of the qPCR reagents. Uninfected insect cDNA was also used to control for amplification of insect DNA. Two qPCR assays were performed per biological replicate. The asays were done using an iCycler system (Bio-rad). qPCR with the following protocol: a 1.5-min activation/denaturation step at 95 °C, followed by 40 cycles of 15 s at 95 °C, 30 s at 60 °C, and 30 s at 72 °C. Subsequently, the specificity of the primers was verified by melting curve analyses done at 72 °C to 96 °C. All PCR assays were performed in duplicate, and the mean of these values was calculated for the final analysis.
The try gene, which encodes an enzyme involved in the tryptophan synthesis, was used as the reference gene for these studies due to its constitutive expression across tissue types (Fang and Bidochka, 2006). Indeed, we found that qPCR of try transcripts generated highly similar quantification cycle (Cq) values across all of the cDNA samples analyzed. We then went on to assess the expression of the pr1A gene (encoding the subtilisin protease PR1A) in M. anisopliae and M. acridum during growth in different culture conditions and during D. saccharalis pathogenesis. Try transcripts were amplified using the forward primer 5′-TGCAATGCATGTTTGATGTC-3′ and the reverse primer 5′-CAAAGAGTGGTATCGAGTTAC-3′. Pr1A transcripts were amplified using the forward primer 5′-GATTGGTGGCAGCACTAAC-3′ and the reverse primer 5′-TCCTGGATCTTCTTGCAAAG-3′ (Fang and Bidochka, 2006).
Generation of quantitative data by real-time qPCR is based on the number of cycles required for the optimal amplification fluorescence to reach a specific threshold of detection (Cq value) (Bustin et al., 2009). The relative expression ratios were calculated using a mathematical model that includes an efficiency correction for real-time qPCR efficiency of the individual transcripts (Pfaffl, 2001). Real-time qPCR amplification efficiencies (E) were determined for each set of primers using the slope of a linear regression model (Pfaffl, 2001). The cDNA samples were diluted to 50, 25, 5, 1, and 0.25 ng/μL and used as template for RT-qPCR reactions. Standard curves were generated by plotting the log of the cDNA values against Cq values obtained over the range of dilutions. The slope of the curves was used to determine the reaction efficiency (E), as E=10[−1/slope]. When assessing gene expression during fungal growth in different culture media, the average Cq value of conidia collected at 24 h was considered the control treatment, and the average Cq value of conidia collected at 72 h was considered the experimental treatment. Similarly, when assessing gene expression during pathogenesis, the average Cq value of an insect 20 h after infection was considered the control treatment, and the average Cq value of dead infected insects, insects with emergent mycelia from insect cadavers and insect cadavers mummified with conidia were all considered experimental treatments.
After an R value was calculated for each biological replicate, the relative gene expression levels of pr1Ain M. anisopliae and M. acridum were expressed as Log2 R. In this way, gene expression levels can be expressed as a numerical value that directly correlates with the induc-tion or repression of that gene’s expression. Thus, if zero is considered to be “no expression”, the genes analyzed are “induced” if they are associated with positive values or “repressed” if they are associated with negative values. Subsequently, the data were subjected to analysis of variance (ANOVA) and means compared by the Tukey test at 5% probability using the Assistat 7.4 software (Silva and Azevedo, 2002). For the analysis of pr1A gene expression across all stages of pathogenesis in D. saccharalis, the data were normalized by transformation (x = 0.4053 + c) before analysis of variance (ANOVA), using ASSISTAT 7.4 Beta software (Silva and Azevedo, 2002).
Pathogenicity test
A randomized design was used in a split plot and subdivided by time (from 1 to 10 days survival). The design included two species of Metarhizium plus one control in the plots. The evaluation days were used as entries in the sub-plots. Each evaluation day was replicated five times. Third-instar larvae of D. saccharalis were immersed in 20 mL of a 1×108 conidias/mL suspension of each variety for 1 min and then transferred to Petri dishes containing a small piece of sugar cane. The samples were then maintained at 28 °C. In the control treatment, larvae were immersed in sterile water containing Tween 80 (0.01% v/v). For each treatment, five Petri dishes were used, each containing 10 larvae for a total of 50 larvae per treatment group. The experiment was performed in triplicate. Observations were done daily for 10 days. The larval cadavers were placed in Petri dishes containing moist filter paper to confirm the causal agent. The data were subjected to analysis of variance (ANOVA), and the means were compared by Tukey test at the 5% probability level using Assistat 7.4 Beta software (Silva and Azevedo, 2002). The Pearson correlation coefficient was used to analyze the degree of correlation between pathogenicity and pr1A gene expression in culture medium supplemented with cuticle and during pathogenesis (at the phase in which mycelium are emerging from the insect corpse) induced by the two species.
Results
Denaturation curve and efficiency of amplification
Analysis of the melting curves confirmed that all corresponded to a unique amplification product. According to the slope obtained from the standard curve of dilutions evaluated for each gene, the PCR efficiency (E) varied from 98% to 106.2%, and regression coefficient values (R2) varied from 0.994 to 0.998 (Table 1).
Table 1. E and R2 values of linear regression for dilutions of the reference and target gene of Metarhizium anisopliae and Metarhizium acridum.
| Gene | E value (%) | R2 value | ||
|---|---|---|---|---|
|
|
|
|||
| Metarhizium anisopliae | Metarhizium acridum | Metarhizium anisopliae | Metarhizium acridum | |
| pr1 | 103% | 104% | 0.998 | 0.996 |
| try | 106.2% | 98% | 0.995 | 0.994 |
E = efficiency value; R2 = regression coefficient value.
Expression of the pr1A gene in different culture conditions
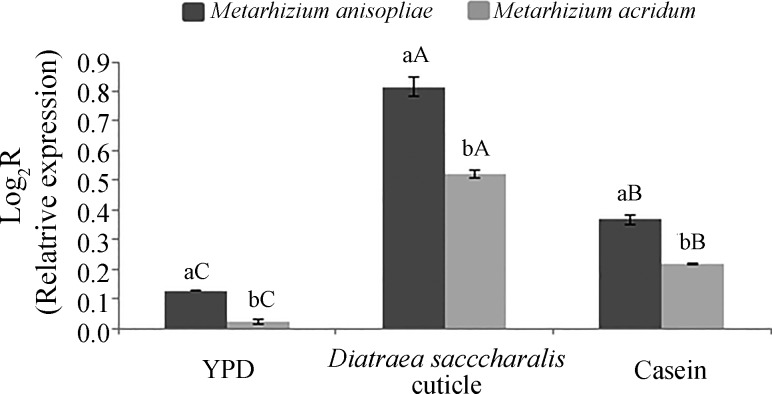
In both M. anisopliae and M. acridum, the pr1A gene was induced after 72 h of culture in the three types of media tested (saprophytic phase) when compared with 24 h of culture. There was a significant interaction between the Metarhizium species and the different culture media factors on the relative expression of the pr1A gene. Specifically, there was significantly less pr1A expression in YPD medium and higher expression of pr1A in MM supplemented with D. saccharalis cuticle (F = 1206.595, p < 0.001). A significantly higher expression of pr1A was also observed in M. anisopliae (F = 332.6593, p < 0.001) (Figure 1).
Figure 1. Relative expression levels of pr1A gene in Metarhizium anisopliae and Metarhizium acridum after 72 h of growth in different culture media. Means followed by distinct letters differ by the Tukey test at the 5% probability level; lower-case letters represent differences between isolates, and upper-case letters represent differences between culture media.
Expression of the pr1A gene during pathogenicity
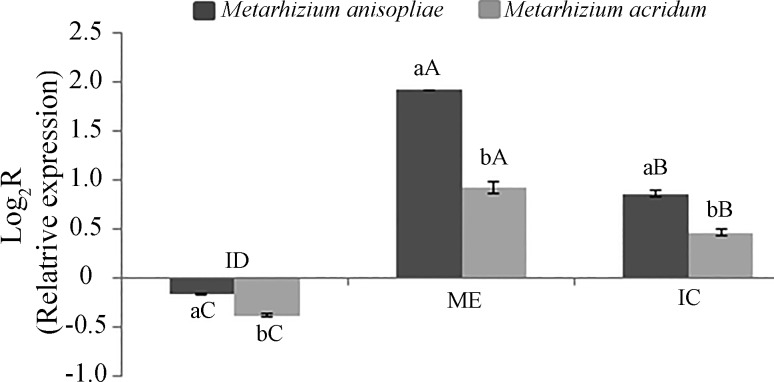
For both M. anisopliae and M. acridum, pr1A expression was repressed when the fungi were grown in dead insects compared to expression in infectious conditions (20 h post-infection of insects). The pr1A gene was induced in insects covered with mycelia or conidia compared to insects 20 h post-infection. We found that fungal species and phases of fungal pathogenesis all had significant effects on the relative expression of pr1A. By comparing different pathogenesis phases we observed higher pr1A expression in insects covered with mycelia than in insects covered with conidia (F = 2224.833, p < 0.001) (Figure 2). We also observed differences in pr1A expression among fungal species. Specifically, we found that M. anisopliae caused less pr1A repression in the dead insect than did M. acridum and greater pr1A induction in mycelia- and conidia-covered insects (F = 668.4208, p < 0.001) (Figure 2).
Figure 2. Relative expression levels of pr1A during pathogenesis of Metarhizium anisopliae and Metarhizium acridum. Means followed by distinct letters differ by the Tukey test at 5% probability; lower-case letters represent differences between isolates at each phase, and upper-case letters represent the differences between phases for each isolate. ID = dead infected insect; ME = emergent mycelia from insect cadavers; IC = insect cadavers completely covered with conidia.
Pathogenicity
The pathogenicity test revealed that M. anisopliae caused an 88% mortality rate in D. saccharalis larvae, whereas M. acridum caused a mortality of 38%. We also observed that M. anisopliae caused an initial mortality (12%) on the third day after infection, whereas M. acridum caused mortality (7%) only on the fourth day (Table 2). The Pearson correlation analysis showed a strong correlation between mortality and pr1A gene expression in medium containing cuticle (r = 0.9835, p < 0.0004) and during pathogenesis (r = 0.9987, p < 0.0001).
Table 2. Accumulated mortality of Diatraea saccharalis infected by Metarhizium anisopliae and Metarhizium acridum during 10 days of evaluation.
| Fungus | Days (% accumulated mortality) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| Metarhizium anisopliae | 0aH | 0aH | 9aG | 21aF | 34aE | 51aD | 63aC | 73aB | 83aA | 88aA |
| Metarhizium acridum | 0aF | 0aF | 0bF | 7bE | 15bD | 24bC | 30bB | 34bAB | 37bA | 38bA |
| Control | 0aA | 0aA | 0bA | 0cA | 0cA | 0cA | 0cA | 0cA | 0cA | 0cA |
Means followed by the same letter (upper case letters in columns and lower case letters in lines) did not differ significantly among each other by the Tukey test at 5% probability.
Discussion
Presumably, Metarhizium has different subsets of genes that are responsive to different growth conditions. Thus, identifying these genes should help to elucidate mechanisms of adaptation (Zhang et al., 2011; Liu et al., 2013; Jin et al., 2014). In this study, we found that expression of the pr1A gene was affected by different growth conditions, indicating that pr1A expression may vary in the presence of different carbon sources. Previous work has shown that subtilisin PR1A protein is produced when grown on minimal medium, and this production is enhanced by the addition of cuticle to the media; however, PR1A synthesis is repressed in rich medium (Freimoser et al., 2003, 2005). Our finding of a significant increase in pr1A expression in medium supplemented with D. saccharalis cuticle compared to either medium supplemented with casein or rich medium (YPD) corroborates these reports. The demonstration of differential expression of pr1A in different culture media suggests that the two fungal species can regulate pr1A gene expression during saprophytic growth. It is likely that differences in pr1A expression are related to factors that allow M. anisopliae and M. acridum to flexibly respond to the conditions of the surrounding environment.
During the pathogenicity process, adhesion to the host and penetration through the cuticle are decisive stages in establishing the infection, and these involve major changes in gene expression patterns that result in dramatic changes in the amount of available nutrients from the host (Roberts and St. Leger, 2004; He et al., 2012). In nutrient-deprived conditions, subtilisin PR1 is produced in the appressorium where it hydrolyzes cuticular proteins, thus facilitating hyphal penetration through the insect cuticle (Goettel et al., 1989; St. Leger et al., 1989).
Once the hyphae have breached the cuticle, the fungus grows in the hemolymph, which is rich in nutrients, and here PR1 is down-regulated by the presence of accessible carbon and nitrogen sources (Small and Bidochka, 2005). This process explains why pr1A gene is repressed in dead insects infected by either of the two species. The death of the insect occurs after the fungus has penetrated the cuticle, formed the blastospores in the hemolymph and invaded the various internal organs that are rich in nutrients (Samuels et al., 1989). Metarhizium then utilizes and depletes the available nutrients of the insect. Subsequently, PR1 is induced again and performs functions that facilitate the penetration of hyphae through the cuticle, allowing the fungus to emerge and produce conidia in large masses on the surface of the insect cadaver, resulting in mummification (Scholte et al., 2007) Thus, induction of pr1A expression in insects covered with either mycelia or conidia suggests that PR1A may not only be directly involved in the formation of conidia but may also be required for the completion of the pathogenic cycle.
Regarding virulence factors, there is evidence that mutant strains of M. anisopliae that have increased expression of the enzyme PR1A are more pathogenic to the host Manduca sexta (St. Leger et al., 1989). Furthermore, spontaneous pr1A− deficient mutants of M. anisopliae demonstrated a reduction in their ability to infect Tenebrio molitor compared to wild-type M. anisopliae (Wang et al., 2005). Gillespie et al. (1998) reported that differences in the in vivo production of proteases among Metarhizium isolates can influence the virulence of the individual isolates. In the current study we found a correlation between the level of pr1A expression in MM supplemented with cuticle and during pathogenesis and observed pathogenesis. When comparing the two fungal species, we observed that M. anisopliae, besides having higher expression of pr1A in culture medium and in all phases of pathogenicity, showed better pathogenic action against D. saccharalis, and herein we considered both the initial and final mortality caused by the infection. Taken together, our data suggest that the level of pr1A gene expression may predict the ability of the fungus to cause disease.
Surface structure and the chemical composition of the host cuticle are believed to affect the adhesion of fungal spores and, consequently, pathogenicity. Metarhizium recognizes specific host signals that induce the secretion of different host-specific proteins. As a consequence, this selectivity for host signals is most likely to define the specificity for a particular isolate/host (Lazzarini et al., 2006; Pedrini et al., 2007; Santi et al., 2010). Thus, although pr1A is expressed by two different species, we speculate that the higher level of expression detected in M. anisopliae occurred in response to the host, indicating that induction of pr1A gene expression is one of the factors that determines the ability of this pathogen to infect D. saccharalis. The difference in pathogenicity factors between the species of Metarhizium may exist due to differential regulation of the same set of genes, rather than variations in the number and types of gene for each host species.
Although many studies involving genes related to pathogenicity have been reported in the literature (Fang and Bidochka, 2006; Scholte et al., 2007; Bischoff et al., 2009), much can still be learned about the functions of these genes. Gene functionality can be inferred from the differential expression of these genes in response to different growing conditions and hosts. In support of this premise, Fang et al. (2010) showed that Metarhizium can induce genes differently for each type of environment or host. We observed such differences in our assays with M. anisopliae and M. acridum, species that have not previously been evaluated under these conditions. Our findings that pr1A is differentially expressed in M. anisopliae and M. acridum in different culture media, as well as during pathogenesis, indicates that these species have different strategies to adapt to different growth conditions. This conclusion is consistent with the saprophytic and/or parasitic lifestyle of these pathogens and may be a sign of their versatility, which favors adaptability to environmental conditions.
Our results provide a better understanding of the roles that PR1A plays during the saprophytic and parasitic phases in the two species of Metarhizium studied. This study serves to emphasize the importance of pr1A gene expression in the biology of these fungi and also suggests that this gene is a potential virulence factor for the development of advanced engineered biopesticides.
Acknowledgments
This project received financial support by the Conselho Nacional de Desenvolvimento Científico e Tecnológico - CNPq (PhD fellowship to M.P. Carneiro-Leão).
Footnotes
Associate Editor: Célia Maria Soares
References
- Alves SB, Lopes RB, Vieira SA, Tamai MA. Fungos entomopatogênicos usados no controle de pragas na América Latina. In: Alves SB, Lopes RB, editors. Controle Microbiano de Pragas na América Latina: Avanços e Desafios. FEALQ; Piracicaba: 2008. 69–110 [Google Scholar]
- Altschul SF, Gish W, Miller E, Myers EW, Lipton DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andersen SO. Cuticular sclerotization. In: Miller TA, editor. Cuticle Techniques in Arthropods. Springer-Verlag; New York: 1980. 185–215 [Google Scholar]
- Bagga S, Hu G, Screen SE, St. Leger RJ. Reconstructing the diversification of subtilisins in the pathogenic fungus Metarhizium anisopliae . Gene. 2004;324:159–169. doi: 10.1016/j.gene.2003.09.031. [DOI] [PubMed] [Google Scholar]
- Bischoff JF, Rehner SA, Humber RA. A multilocus phylogeny of the Metarhizium anisopliae lineage. Mycologia. 2009;101:512–530. doi: 10.3852/07-202. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;4:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Fang W, Bidochka MJ. Expression of genes involved in germination, conidiogenesis and pathogenesis in Metarhizium anisopliae by quantitative real-time RT-PCR. Mycol Res. 2006;110:1165–1171. doi: 10.1016/j.mycres.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Fang W, Fernandes EKK, Robert DW, Bidochka MJ, St. Leger RJ. A laccase exclusively expressed by Metarhizium anisopliae during isotropic growth is involved in pigmentation, tolerance to abiotic stresses and virulence. Fungal Genet Biol. 2010;47:602–607. doi: 10.1016/j.fgb.2010.03.011. [DOI] [PubMed] [Google Scholar]
- Frazzon APG, Da Silva V, Jr I, Masuda A, Schrank A, Vainstein MH. In vitro assessment of Metarhizium anisopliae isolates to control the cattle tick Boophilus microplus . Vet Parasitol. 2000;94:117–125. doi: 10.1016/s0304-4017(00)00368-x. [DOI] [PubMed] [Google Scholar]
- Freimoser FM, Hu G, St. Leger RJ. Variation in gene expression patterns as the insect pathogen Metarhizium anisopliae adapts to different host cuticles or nutrient deprivation in vitro. Microbiology. 2005;151:361–371. doi: 10.1099/mic.0.27560-0. [DOI] [PubMed] [Google Scholar]
- Freimoser FM, Screen S, Bagga S, Hu G, St Leger RJ. Expressed sequence tag (EST) analysis of two subspecies of Metarhizium anisopliae reveals a plethora of secreted proteins with potential activity in insect hosts. Microbiology. 2003;149:239–247. doi: 10.1099/mic.0.25761-0. [DOI] [PubMed] [Google Scholar]
- Gillespie JP, Bateman R, Charnley AK. Role of cuticle-degrading proteases in the virulence of Metarhizium spp. for the desert locust, Schistocerca . J Invertebr Pathol. 1998;71:128–137. doi: 10.1006/jipa.1997.4733. [DOI] [PubMed] [Google Scholar]
- Goettel MK, St Leger RJ, Rizzo NW, Staples RC, Roberts DW. Ultrastructural localization of a cuticle degrading protease produced by the entomopathogenic fungus Metarhizium anisopliae during penetration of host (Manduca sexta) cuticle. J Gen Microbiol. 1989;135:2233–2239. [Google Scholar]
- He M, Hu J, Xia Y. Large scale expressed sequence tag (EST) analysis of Metarhizium acridum infecting Locusta migratoria reveals multiple strategies for fungal adaptation to the host cuticle. Curr Genet. 2012;58:265–279. doi: 10.1007/s00294-012-0382-6. [DOI] [PubMed] [Google Scholar]
- Jin K, Han L, Xia Y. MaMk1, a FUS3/KSS1-type mitogen-activated protein kinase gene, is required for appressorium formation, and insect cuticle penetration of the entomopathogenic fungus Metarhizium acridum . J Invertebr Pathol. 2014;115:68–75. doi: 10.1016/j.jip.2013.10.014. [DOI] [PubMed] [Google Scholar]
- Lazzarini GMJ, Rocha LFN, Luz C. Impact of moisture on in vitro germination of Metarhizium anisopliae and Beauveria bassiana and their activity on Triatoma infestans . Mycol Res. 2006;110:485–492. doi: 10.1016/j.mycres.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Liu Q, Ying SH, Li JG, Tian CG, Feng MG. Insight into the transcriptional regulation of Msn2 required for conidiation, multi-stress responses and virulence of two entomopathogenic fungi. Fungal Genet Biol. 2013;54:42–51. doi: 10.1016/j.fgb.2013.02.008. [DOI] [PubMed] [Google Scholar]
- Luo S, He M, Cao Y, Xia Y. The tetraspanin gene MaPls1 contributes to virulence by affecting germination, appressorial function and enzymes for cuticle degradation in the entomopathogenic fungus, Metarhizium acridum . Environ Microbiol. 2013;15:2966–2979. doi: 10.1111/1462-2920.12166. [DOI] [PubMed] [Google Scholar]
- Pathan AAK, Devi KU, Vogel H, Reineke A. Analysis of differential gene expression in the generalist entomopathogenic fungus Beauveria bassiana (Bals.) Vuillemin grown on different insect cuticular extracts and synthetic medium through cDNA-AFLPs. Fungal Genet Biol. 2007;44:1231–1241. doi: 10.1016/j.fgb.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Pedrini N, Crespo R, Juárez MP. Biochemistry of insect epicuticle degradation by entomopathogenic fungi. Comp Biochem Physiol C Pharmacol Toxicol Endocrinol. 2007;146:124–137. doi: 10.1016/j.cbpc.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontecorvo G, Roper JA, Hemons LM, MacDonald KD, Bufton AWJ. The genetics of Aspergillus nidulans . Adv Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- Roberts DW, St Leger RJ. Metarhizium spp., cosmopolitan insect-pathogenic fungi: Mycological aspects. Adv Appl Microbiol. 2004;54:1–70. doi: 10.1016/S0065-2164(04)54001-7. [DOI] [PubMed] [Google Scholar]
- Samuels KDZ, Pinnock DE, Allsopp PG. The potential of Metarhizium anisopliae (Metschnikoff) Sorokin (Deutermycotina, Hyphomycetes) as a biological control-agent of Inopus rubriceps (Macquart) (Diptera, Stratiomyidae) J Aust Entomol Soc. 1989;28:69–74. [Google Scholar]
- Santi L, Silva WOB, Pinto AFM, Schrank A, Vainstein MH. Metarhizium anisopliae host-pathogen interaction: Differential immunoproteomics reveals proteins involved in the infection process of arthropods. Fungal Biol. 2010;114:312–319. doi: 10.1016/j.funbio.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Scholte EJ, Takken W, Knols BGJ. Infection of adult Aedes aegypti and Ae. albopictus mosquitoes with the entomopathogenic fungus Metarhizium anisopliae . Acta Trop. 2007;102:151–158. doi: 10.1016/j.actatropica.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Silva FAS, Azevedo CAV. Assistant computational program version for the windows operating system. Braz J Agro-Industr Prod. 2002;4:71–78. [Google Scholar]
- Small CN, Bidochka MJ. Up-regulation of Pr1, a subtilisin-like protease, during conidiation in the insect pathogen Metarhizium anisopliae . Mycol Res. 2005;3:301–313. doi: 10.1017/s0953756204001856. [DOI] [PubMed] [Google Scholar]
- Staden R, Beal KF, Bonfield JK. The Staden Package. Computer Methods in Molecular Biology. Bioinform Meth Prot. 1998;132:115–130. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- St Leger RJ, Butt TM, Staples RC, Roberts DW. Synthesis of proteins including a cuticle-degrading protease during differentiation of the entomopathogenic fungus Metarhizium anisopliae . Exp Mycol. 1989;13:253–262. [Google Scholar]
- Wang C, Gang Hu, St. Leger RJ. Differential gene expression by Metarhizium anisopliae growing in root exudate and host (Manduca sexta) cuticle or hemolymph reveals mechanisms of physiological adaptation. Fungal Genet Biol. 2005;42:704–718. doi: 10.1016/j.fgb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Zhang C, Xia Y, Li Z. Identification of genes differentially expressed by Metarhizium anisopliae growing on Locusta migratoria wings using suppression subtractive hybridization. Curr Microbiol. 2011;62:1649–1655. doi: 10.1007/s00284-011-9909-1. [DOI] [PubMed] [Google Scholar]