Abstract
The Genotyping tool at the National Center for Biotechnology Information is a web-based program that identifies the genotype (or subtype) of recombinant or non-recombinant viral nucleotide sequences. It works by using BLAST to compare a query sequence to a set of reference sequences for known genotypes. Predefined reference genotypes exist for three major viral pathogens: human immunodeficiency virus 1 (HIV-1), hepatitis C virus (HCV) and hepatitis B virus (HBV). User-defined reference sequences can be used at the same time. The query sequence is broken into segments for comparison to the reference so that the mosaic organization of recombinant sequences could be revealed. The results are displayed graphically using color-coded genotypes. Therefore, the genotype(s) of any portion of the query can quickly be determined. The Genotyping tool can be found at: http://www.ncbi.nih.gov/projects/genotyping/formpage.cgi.
INTRODUCTION
Determining the genotype of viruses is important not only in epidemiology studies, but also for efficient vaccine development and treatment of major viral diseases such as HIV–AIDS and viral hepatitides. For example, correlating HIV-1 genotypes from clinical samples with existing clinical data is important for detecting and quantifying viral drug resistance in AIDS patients (1,2). Genotyping of hepatitis B virus (HBV) has been used primarily in molecular epidemiology; however, several recent studies have demonstrated that the severity of the hepatitis, in terms of both acute liver damage and chronic disease status, can vary between genotypes (3). For another major cause of chronic liver disease, the hepatitis C virus (HCV), the mode and effectiveness of therapeutic treatment also depend on the viral genotype (4).
Phylogenetic analysis has been used to distinguish viral genotypes and/or subtypes from each other, and for subtyping newly isolated strains by comparing them to existing alignments and trees. However, phylogenetic analysis often cannot distinguish between inter-subtype recombinants and new subtypes, as both can branch out between clusters in a similar way. To reveal the mosaic organization of recombinant viruses, new methods that process the genotype in segments along a sequence were designed (5–7). These methods rely upon multiple alignments of a query sequence and the reference sequences of known viral subtypes. However, the high variability of viral genomes often makes it impossible to align viral sequences automatically. In these cases, the alignments have to be done laboriously by hand.
The NCBI Genotyping tool introduces a new algorithm that does not require a multiple alignment as input; rather, it uses scored BLAST pairwise alignments between overlapping segments of a query sequence and a reference sequence for each virus. The results are obtained for multiple segments and displayed in a graphical format that allows clear determination of the genotype of the query sequence, or of all genotypes involved and recombination breakpoints if the query is a recombinant. The NCBI Genotyping resource has predefined sets of reference genomes for HIV-1, HCV and HBV and is available at http://www.ncbi.nih.gov/projects/genotyping/formpage.cgi.
METHODS
Genotyping procedure
The algorithm works by sliding a ‘window’ along the query sequence and processing each window/sequence segment separately. Each segment is compared to a set of reference sequences using BLAST (8), which returns the similarity scores for the local alignments. The reference sequence genotype that matches the query with the highest similarity score is assigned to the query segment. The process is repeated for the next window, and so on, until the whole length of the query sequence has been covered by overlapping BLAST alignments. The results from all windows are combined and displayed graphically. If the same genotype is assigned to most segments, then the query sequence is considered a single genotype. If a query sequence is recombinant, both the genotypes of the predecessors and the region of probable recombination breakpoint(s) are displayed.
There are three key parameters required by the algorithm: (i) window size, (ii) the incremental step by which the window is shifted along the sequence and (iii) the similarity threshold for a match. The default settings can be changed by the user if desired (see below).
Reference sequences
The reference sequence sets for HIV-1, HCV and HBV are complete genomes and were created in collaboration with expert advisors (Table 1). To take into account genetic polymorphism, the predefined reference sets typically contain two to four reference sequences for each genotype and include both ancient and more recent sequence data.
Table 1. Currently available sets of reference sequences for genotyping.
| Virus | Reference set source |
|---|---|
| Human immunodeficiency virus 1 (HIV-1) | HIV Database at Los Alamos National Laboratory (http://hiv-web.lanl.gov) |
| Human immunodeficiency virus 2 (HIV-2) | HIV Database at Los Alamos National Laboratory (http://hiv-web.lanl.gov) |
| Human T-lymphotropic virus 1 (HTLV-1) | HIV Database at Los Alamos National Laboratory (http://hiv-web.lanl.gov) |
| Human T-lymphotropic virus 2 (HTLV-2) | HIV Database at Los Alamos National Laboratory (http://hiv-web.lanl.gov) |
| Hepatitis B virus (HBV) | Reference (14) and V. Chulanov (personal communication) |
| Hepatitis C virus (HCV) | M.R., A.K. and T.T. (this report); V. Chulanov and C. Kuiken (personal communication); the HCV Sequence Database (http://hcv.lan/gov/) |
| Poliovirus (complete genomes) | E. Cherkasova and K. Chumakov (personal communication). Helps highlight recombination events in the complete genomes of polioviruses |
| Human enterovirus C (HEV-C) viruses (P1 region only) | Under construction. E. Cherkasova and K. Chumakov (personal communication). Is being designed for identification of serotype and genotype in HEV-C viruses |
A total of 117 reference sequences represent 14 HIV-1 genotypes and subtypes and 16 circulating recombinant forms (CRFs) (9,10). HCV is classified into six genotypes, corresponding to the major branches of a phylogenetic tree (clades) constructed from alignments of complete HCV genomes. Smaller branches of the tree correspond to subtypes. Genotypes are numbered 1 to 6; subtypes are designated by letters (11,12). Recombinant genotypes have also been suggested for this virus (13). Numerous HBV isolates have been grouped into eight genotypes (A through H) based on alignments of complete HBV genomes (14). Additional reference sets for HIV-2, primate lymphotropic viruses and poliovirus will soon be available.
Either the entire genome or portions of the genome (depending on the virus) can be used to assign a genotype to a query sequence. The predefined reference sets for HIV-1, HCV and HBV consist of complete sequences. This allows a query sequence that contains any phylogenetically informative portion of the viral genome to be analyzed. Indeed, genotyping analyses performed separately on different genes of the HIV-1, HCV and HBV genomes revealed similar clusters corresponding to the genotypes and/or subtypes. However, genetic distances between genotypes are greater in more polymorphic genes (such as the HIV-1 envelope gene or HCV E2 gene) than in others. Some genes of some viruses are not be suitable at all for genotyping. For example, non-structural genes from different serotypes of poliovirus and other members of the human enterovirus species C (HEV-C) have been apparently exchanged by recombination (15); therefore, these genes are not particularly useful for distinguishing between poliovirus serotypes, or between poliovirus and HEV-C. It is recommended that the current literature is checked to make sure that a particular genomic fragment can be used for genotyping.
The predefined reference sequence sets exist as separate local databases, making the algorithm fast. If user-provided reference sequences are also supplied as GenBank accession/gi numbers, then the ‘nr’ (non-redundant) database is used. Any number of reference sequences can be added.
INTERFACE
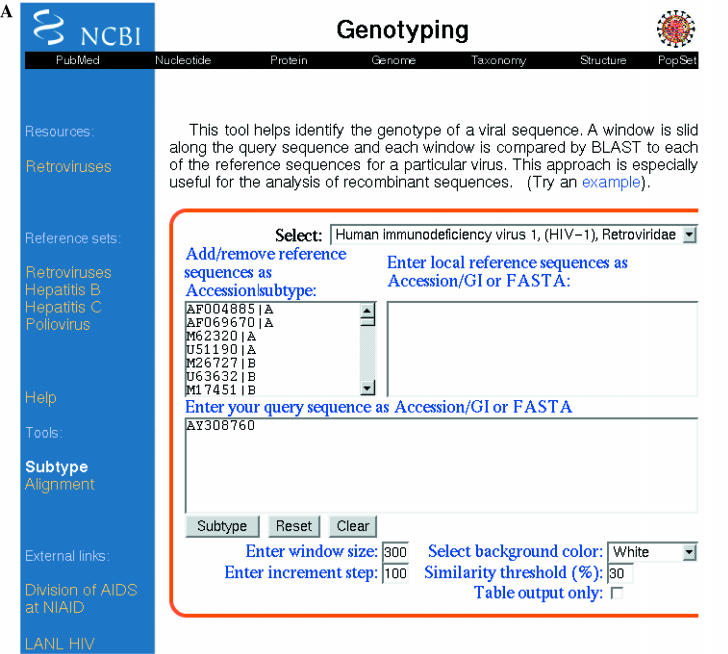
The Genotyping tool can be found at http://www.ncbi.nih.gov/projects/genotyping/formpage.cgi. To use it, enter the query sequence into the search box (either as a GenBank accession/gi number or as a FASTA-formatted sequence), select the reference sequence from the menu and click the ‘Subtype’ button. Additional options can be found under the ‘Advanced’ button (Figure 1A). Here, the predefined reference sets can be edited and additional reference sequences provided. The parameters may also be changed from the default settings (window size: 300 nt; incremental step: 100 nt; and similarity threshold: 30%). A smaller window size (50 nt minimum) and a lower incremental step (10 nt minimum) help to locate more precisely the recombination breakpoints. However, lower parameter values result in less reliable BLAST results and, consequently, less confidence in the identification of each fragment's genotype.
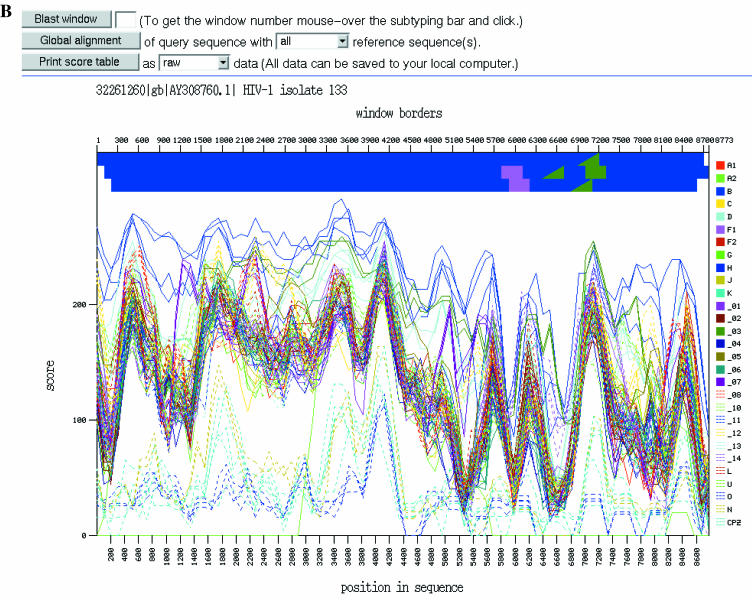
Figure 1.
Subtyping of the complete genome of HIV-1 isolate 1333_d2 from the USA. (A) Advanced interface. GenBank accession number AY308760 is pasted into the query box. Default parameters were used. (B) Output. The dominant blue color of the horizontal top bar unequivocally suggests genotype B for this HIV-1 sequence (AY308760).
OUTPUT
The output is a plot of the BLAST similarity scores for the alignments between the query segment and each reference sequence within the overlapping windows (Figure 1B). Above the plot, a series of color-coded rectangles represent the windows. Each rectangle is colored corresponding to the genotype of the reference sequence that obtained the highest BLAST score against the query (see right of graph for key). This allows the genotype(s) of the query to be inferred from the dominant color(s) on the graphics. Results can be saved on a local computer in the form of BLAST scores and further processed with a spreadsheet program. Query-anchored alignments generated by BLAST can also be saved in a format that highlights mismatches between sequences.
SAMPLE ANALYSIS OF RECOMBINANT GENOMES
A/G recombinant HIV-1
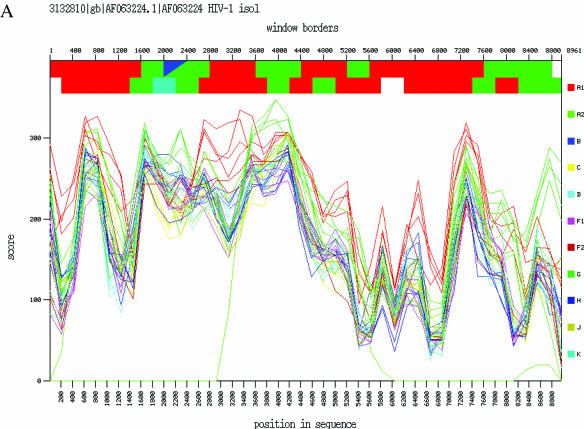
CRF02_AG is a subtype A/G recombinant form of HIV-1 that is circulating widely in West and Central Africa and has also been reported in Taiwan. The NCBI Genotyping tool was used to investigate the genotype of the full-length genome sequence of an A/G recombinant virus from Djibouti (GenBank accession number AF063224) (16). A window size of 400 nt shifted by 200 positions was used. The output shows that this viral sequence has regions of subtype A and subtype G with multiple cross-over points, and that the mosaic structure of the Djibouti strain is identical to that of another A/G recombinant virus from Nigeria, IbNG (accession number L39106) (17) (Figure 2). This is a demonstration of the geographic spread of the A/G recombinant in Africa.
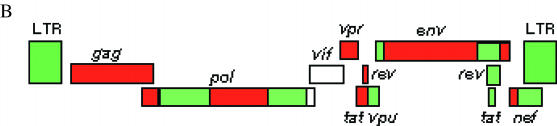
Figure 2.
Mosaic structure of two African full-length A/G recombinant viruses. (A). Graphical output of the local genotyping of the genome sequence from Djibouti (accession number AF063224) as obtained by the NCBI Genotyping resources. (B) Genome structure overview of a virus from Nigeria, IbNG (accession number L39106), published on the website of the Los Alamos HIV Sequence Database (http://www.hiv.lanl.gov/content/hiv-db/CRFs/CRFs.html). Red: subtype A; green: subtype G.
Recombinant 2a/1a HCV sequences
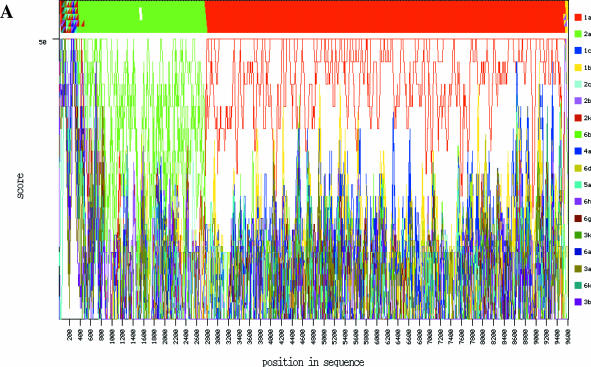
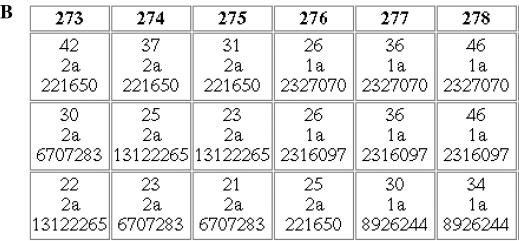
Artificially constructed inter-genotypic recombinants of HCV (GenBank accession numbers AF177037 and AF177038) (17) were analyzed with the Genotyping tool using the minimum allowed values for the parameters ‘window’ and ‘increment’ (50 and 10 nt, respectively) (Figure 3). For AF177037 (clone pH77CV-J6S), the majority of query segments (windows) 1 through 275 (positions 2741–2790) correspond to genotype 2a. All the windows downstream of that (starting from the 276th with positions 2751–2800), with the exception of a few at the very 3′ end, correspond to genotype 1a. In full agreement with the author's description, indicating position 2765 for the artificial restriction site used to create the chimera, the result demonstrates that AF177037 is a 2a/1a recombinant with the recombination breakpoint located between (or in close vicinity to) nucleotides 2751 and 2790. The recombination breakpoint for AF177038 [clone pH77(p7)CV-J6S] was similarly found between nucleotides 2571 and 2610.
Figure 3.
Analysis of 2a/1a recombinant clones of Hepatitis C virus, AF177037 (clone pH77CV-J6S) and AF177038 [clone pH77(p7)CV-J6S] (18). (A). Output for AF177037 with the parameters ‘window size’ and ‘incremental step’ set to the minimal values, 50 and 10 nt, respectively. (B). A fragment of tabular output, sorted by score, in the vicinity of the recombination breakpoint. Each square contains the BLAST score, the identifier of the genotype and subtype of the reference sequence which produced this score, and the gi number of this sequence.
DISCUSSION
The NCBI Genotyping tool allows almost instant identification of the genotype of a virus by comparison of its nucleotide sequence to reference sequences from previously characterized viruses. The method is based on local pairwise alignments generated by BLAST in real time. Making use of overlapping sequence segments as BLAST queries increases the reliability of the results; this is especially important for the analysis of recombinant sequences. If the results suggest recombination events over a short length of sequence, (e.g. within the scope of the sliding window), we strongly recommend that the user conducts additional runs with various parameter values (to reduce the possible effects of non-significant local fluctuation in BLAST scores). If the results seem inconsistent or fluctuate dramatically with different parameter values, check that the correct set of reference sequences is selected, and supplement this set with new sequences, if necessary. To draw careful conclusions, any recombination events and breakpoints suggested by the analysis should be further analyzed/confirmed using models that reconstruct the phylogenies from the aligned sequences (19). The Genotyping tool is not designed for delineating new genotypes or subtypes; even so, useful information can usually be gleaned, even for queries belonging to previously uncharacterized viral strains or types.
Questions are to be directed to info@ncbi.nlm.nih.gov.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Vladimir Chulanov, Brian Foley, Carla Kuiken, Elena Cherkasova and Konstantin Chumakov for fruitful discussions and their contributions to the creation or updates of the reference sequence sets, and to Jo McEntyre for preparing this paper for publication.
REFERENCES
- 1.Sturmer M., Doerr,H.W. and Preiser,W. (2003) Variety of interpretation systems for human immunodeficiency virus type 1 genotyping: confirmatory information or additional confusion? Curr. Drug Targets Infect. Disord., 3, 373–382. [DOI] [PubMed] [Google Scholar]
- 2.Parkin N.T. and Schapiro,J.M. (2004) Antiretroviral drug resistance in non-subtype B HIV-1, HIV-2 and SIV. Antiviral Therapy, 9, 3–12. [PubMed] [Google Scholar]
- 3.Tsubota A., Arase,Y., Ren,F., Tanaka,H., Ikeda,K. and Kumada,H. (2001) Genotype may correlate with liver carcinogenesis and tumor characteristics in cirrhotic patients infected with hepatitis B virus subtype adw. J. Med. Virol., 65, 257–265. [DOI] [PubMed] [Google Scholar]
- 4.National Institutes of Health Consensus Development Conference Statement (2002) Management of hepatitis C 2002 (June 10–12, 2002). Gastroenterology, 123, 2082–2099. [DOI] [PubMed] [Google Scholar]
- 5.Siepel A.C., Halpern,A.L., Macken,C. and Korber,B.T. (1995) A computer program designed to screen rapidly for HIV type 1 intersubtype recombinant sequences. AIDS Res. Hum. Retroviruses, 11, 1413–1416. [DOI] [PubMed] [Google Scholar]
- 6.Salminen M.O., Carr,J.K., Burke,D.S. and McCutchan,F.E. (1995) Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res. Hum. Retroviruses, 11, 1423–1425. [DOI] [PubMed] [Google Scholar]
- 7.Lole K.S., Bollinger,R.C., Paranjape,R.S., Gadkari,D., Kulkarni,S.S., Novak,N.G., Ingersoll,R., Sheppard,H.W. and Ray,S.C. (1999) Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol., 73, 152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 26, 3986–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coffin J.M., Hughes,S.H. and Varmus,H.E. (1997) Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, NY. [PubMed] [Google Scholar]
- 10.Leitner T., Korber,B., Robertson,D.L., Gao,F. and Hahn,B.H. (1997) Updated proposal of reference sequences of HIV-1 genetic subtypes. In Korber,B., Hahn,B.H, Foley,B., Mellors,J.W., Leitner,T., Meyers,G., McCutchan,F.E. and Kuiken,C.L. (eds) Human Retroviruses and AIDS: A Compilation and Analysis of Nucleic Acid and Amino acid Sequences. Los Alamos National Laboratory, Los Alamos, NM, pp. 1–6. [Google Scholar]
- 11.Robertson B., Myers,G., Howard,C., Brettin,T., Bukh,J., Gaschen,B., Gojobori,T., Maertens,G., Mizokami,M., Nainan,O., Netesov,S., Nishioka,K., Shin-i,T., Simmonds,P., Smith,D., Stuyver,L. and Weiner,A. (1998) Classification, nomenclature, and database development for hepatitis C virus (HCV) and related viruses: proposals for standardization. International Committee on Virus Taxonomy. Arch. Virol., 143, 2493–2503. [DOI] [PubMed] [Google Scholar]
- 12.van Regenmortel M.H.V., Fauquet,C.M, Bishop,D.H.L., Carstens,E.B., Estes,M.K., Lemon,S.M., Maniloff,J., Mayo,M.A., McGeoch,D.J., Pringle,C.R. and Wickner,R.B. (2000) Virus Taxonomy. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, New York.
- 13.Kalinina O., Norder,H., Mukomolov,S. and Magnius,L.O. (2002) A natural intergenotypic recombinant of Hepatitis C virus identified in St. Petersburg. J. Virol., 76, 4034–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arauz-Ruiz P., Norder,H., Robertson,B.H. and Magnius,L.O. (2002) Genotype H: a new Amerindian genotype of hepatitis B virus revealed in Central America. J. Gen. Virol., 83, 2059–2073. [DOI] [PubMed] [Google Scholar]
- 15.Brown B., Oberste,M.S., Maher,K. and Pallansch,M.A. (2003) Complete genomic sequencing shows that polioviruses and members of human enterovirus species C are closely related in the noncapsid coding region. J. Virol., 77, 8973–8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carr J.K., Salminen,M.O., Albert,J., Sanders-Buell,E., Gotte,D., Birx,D.L. and McCutchan,F.E. (1998) Full genome sequences of human immunodeficiency virus type 1 subtypes G and A/G intersubtype recombinants. Virology, 247, 22–31. [DOI] [PubMed] [Google Scholar]
- 17.Gao F., Robertson,D.L., Morrison,S.G., Hui,H., Craig,S., Decker,J., Fultz,P.N., Girard,M., Shaw,G.M., Hahn,B.H. and Sharp,P.M. (1996) The heterosexual human immunodeficiency virus type 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J. Virol., 70, 7013–7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yanagi M., Purcell,R.H., Emerson,S.U. and Bukh,J. (1999) Hepatitis C virus: an infectious molecular clone of a second major genotype (2a) and lack of viability of intertypic 1a and 2a chimeras. Virology, 262, 250–263. [DOI] [PubMed] [Google Scholar]
- 19.Grassly N.C. and Holmes,E.C. (1997) A likelihood method for the detection of selection and recombination using nucleotide sequences. Mol. Biol. Evol., 14, 239–247. [DOI] [PubMed] [Google Scholar]