Abstract
The functional role of IGFBP5 in breast cancer is complicated. Experimental and bioinformatics studies have shown that IGFBP5 is targeted by miR-140-5p and miR-193b, although this has not yet been proven in clinical samples. The aim of this study was to evaluate the expression of miR-140-5p and miR-193b in breast cancer and adjacent normal tissue and assess its correlation with IGFBP5 and the clinicopathological characteristics of the tumors. IGFBP5 protein expression was analyzed immunohistochemically and IGFBP5, miR-140 and miR-193b mRNA expression levels were analyzed with real-time RT-PCR. Tumor tissue had higher miR-140-5p expression than adjacent normal tissue (p = 0.015). Samples with no immunohistochemical staining for IGFBP5 showed increased miR-140-5p expression (p = 0.009). miR-140-5p expression was elevated in invasive ductal carcinomas (p = 0.002), whereas basal-like tumors had decreased expression of miR-140-5p compared to other tumors (p = 0.008). Lymph node-positive samples showed an approximately 13-fold increase in miR-140-5p expression compared to lymph node-negative tissue (p = 0.049). These findings suggest that miR-140-5p, but not miR-193b, could be an important determinant of IGFBP5 expression and clinical phenotype in breast cancer patients. Further studies are needed to clarify the expressional regulation of IGFBP5 by miR-140-5p.
Keywords: breast cancer, ER alpha, IGFBP5, micro RNA, miR-140, miR-193b
Introduction
Breast cancer is the most common cancer and the leading cause of cancer-related deaths among women worldwide, according to the World Health Organization (WHO). The treatment of breast cancer remains largely ineffective, primarily because of the complex etiology of this disease, acquired drug resistance and our incomplete understanding of the molecular pathways involved (Eroles et al., 2011).
The insulin-like growth factor (IGF) signaling pathway has an important role in cell growth, differentiation, apoptosis regulation (Valentinis and Baserga, 2001) and tumor development (Khandwala et al., 2000). IGF signaling involves two growth factors (IGF-I, IGF-II), two IGF receptors (IGF-IR, IGF-IIR), seven well-defined IGF-binding proteins (IGFBPs), a group of IGFBP-related proteins that bind IGFs with low affinity and IGFBP proteases. IGFBP5, the most conserved member of the IGFBP family, (Mohan and Baylink, 2002) is involved in carcinogenesis via IGF-dependent and independent pathways (Beattie et al., 2006; Kricker et al., 2010), and influences the rate of apoptosis, cell motility and survival (Akkiprik et al., 2008; Güllü et al., 2012).
The functional role of IGFBP5 in breast cancer is complex, with numerous studies examining its involvement in cell survival and apoptosis in normal and cancer cells. Butt et al. (2005) showed that IGFBP5 activated caspases 8 and 9 and caused apoptosis through Bcl-2 in the intrinsic apoptotic pathway in MDA-MB-231 breast cancer cells. IGFBP5 has been reported to inhibit cell growth and cause G2/M arrest in human breast cancer and PANC-1 pancreatic cancer cells (Butt et al., 2005; Johnson and Haun, 2009). IGFBP5 protein levels in breast cancer patients are related to metastasis, poor prognosis, drug sensitivity and limited response to endocrine treatment, although some studies have suggested anti-metastatic and anti-migratory effects for this protein (Becker et al., 2012). Identification of the molecular regulators of IGFBP5 expression in breast cancer patients is critical to understanding the developmental mechanisms of breast cancer.
Epigenetic factors, such as micro RNAs (miRNA), may regulate IGFBP5 expression level in breast cancer. miRNAs are small, non-protein coding RNA gene products 20–24 nucleotides long that mediate target mRNA degradation or translation (Palmero et al., 2011). miRNAs can serve as oncogenes or tumor suppressors. Bioinformatics analyses have shown that miR-193b targets IGFBP5 but this association has not yet been demonstrated experimentally. miR-140 expression was recently shown to be reduced in response to estrogen stimulation of ERα-positive breast cancer cells (Zhang et al., 2012). Promoter analyses revealed that ERα binds to a specific estrogen response element flanking the miR-140 promoter and suppresses miR-140 transcription. The stem cell self-renewal regulator SOX2 is a novel target of miR-140 and the miR-140/SOX2 pathway that critically regulates the survival of breast tumor cells. This finding provides a new link between ERα signaling and breast cancer stem cell maintenance (Zhang et al., 2012). miRNA microarray analysis of breast cancer tissue samples revealed that miR-140 is down-regulated more than two-fold in primary breast cancer samples than in adjacent normal tissues (Yan et al., 2008). The transfection of chondrocytes with pre-miR-140 has shown that IGFBP5 is a direct target of miR-140 (Tardif et al., 2009). Inconsistent expression analysis results of mesenchymal stromal cells suggest that miR-140 has tissue-specific effects on IGFBP5 expression (Buechli et al., 2013).
ER-negative breast cancer patients have lower IGFBP5 mRNA levels than ER-positive patients (Li et al., 2007), although some studies have reported no association between IGFBP5 levels and ER expression (Mita et al., 2007). ER-positive breast cancer patients with low levels of IGFBP5 mRNA have a better disease-free survival rate (Mita et al., 2007). One of the best-known ERα-regulating miRNAs is miR-193b and ERα-negative tumors have lower miR-193b expression than ERα-positive tumors (Yoshimoto et al., 2011). Increased expression of miR-193b decreases tumor migration, invasion and proliferation. Leivonen et al. (2009) reported that miR-193b directly targets ERα by binding to the 3-UTR region of ERα to inhibit estrogen-induced proliferation; consequently, higher levels of miR-193b expression led to better disease-free survival in breast cancer patients.
IGFBP5 expression, which is known to be regulated by miR-140-5p, has variable expression in breast cancer tissue. miR-140-5p can be regulated by ER expression and ER expression is known to be altered by miR-193b. Our aim in this study was to analyze the expression levels of IGFBP5, miR-140-5p, miR-193b and ERα in breast cancer tissues, and their relation to each other and to the patients clinicopathological characteristics.
Materials and Methods
Specimen collection
Human breast cancer Formalin-fixed, paraffin-embedded (FFPE) sections were provided by Gülhane Military Medical Academy Haydarpasa Training Hospital (Istanbul, Turkey). All patients provided written informed consent prior to participation in this study. All patients were diagnosed with breast cancer between 2005 and 2011 and had undergone mastectomy. Forty-eight tumor samples were examined by pathologists and divided into subgroups based on their expression of ER, PgR, Her2, CK5/6 and EGFR: 11 were classified as Luminal A, 11 as Luminal B, 12 as Her2 and 14 as Basal-like (www.cap.org). The Her2 status of tumor samples was determined by fluorescence in situ hybridization (FISH).
This study was done with the understanding and written consent of each subject, and conformed to The Code of Ethics of the World Medical Association (Declaration of Helsinki), as published in the British Medical Journal (18 July 1964). Marmara University Clinical Research Ethical Committee approved this study on 03/21/2012 (protocol no: 53).
RNA, miRNA isolation and cDNA synthesis
FFPE sections of the tissue samples were deparaffinized prior to miRNA isolation. miRNA was extracted using High Pure miRNA isolation kits (Roche, Germany), according to the manufacturer’s instructions. The concentration and purity of RNA were determined spectrophotometrically based on the absorption at 260 to 320 nm. DEPC-treated water used in all spectrophotometric analyses. 20 ng of miRNA was used for cDNA synthesis in a reaction volume of 20 μL. cDNA was synthesized using a Universal cDNA synthesis kit (Exiqon, USA), according to the manufacturer’s instructions. cDNA samples were stored at −20 °C until used.
The isolation of total RNA from FFPE sections was done using a Roche High Pure RNA paraffin kit (Roche), according to the manufacturer’s instructions. 500 ng of total RNA was used for cDNA synthesis in a reaction volume of 20 μL, in conjunction with a Transcriptor High Fidelity cDNA synthesis kit (Roche).
Immunohistochemistry
Three-micrometer thick FFPE sections of breast tumor tissue and adjacent normal tissue were analyzed for IGFBP5 protein expression by an immunohistochemical method using a Peroxidase Detection System kit (Novacastra Laboratories Ltd., UK), according to the manufacturer’s instructions. After deparaffinization and antibody retrieval, tissue sections were incubated overnight with IGFBP5 primary antibody (antibody C-18, diluted 1:50; Santa Cruz Biotechnology, Santa Cruz, CA, USA), washed with phosphate-buffered saline (PBS) and incubated with secondary antibody for 30 min. IGFBP5 protein was visualized using streptavidin-HRP and diaminobenzidine (DAB).
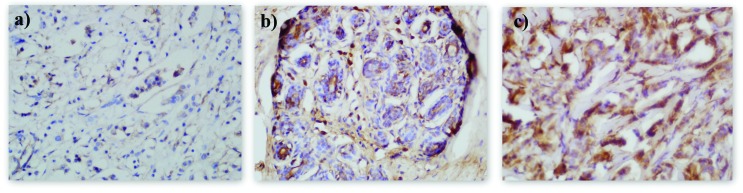
Two independent pathologists performed immunohistochemistry on 48 tumor samples and 32 samples of adjacent normal tissue. Sections that contained no stained cells were classified as negative (−), sections with 1–3 stained cells out of 10 cells examined were classified as low positive (+), those with 4–7 stained cells out of 10 cells were classified as medium positive (++) and those with 8–10 stained cells out of 10 cells were classified as strongly positive (+++). Positive staining was observed in 34 of the tumor and 22 of the normal tissue samples and all positive staining was cytoplasmic. Negative, low and strong stainings are shown in Figure 1.
Figure 1. Immunohistochemical staining for IBFBP5 in breast tissue. (A) Low (+), (B) medium (++) and (C) strong (+++) positive cytoplasmic staining for IGFBP5.
Real-time quantitative PCR (Q-PCR)
Real-time quantitative PCR for miR-193b and miR-140 was done using miRCURY LNA Universal RT microRNA PCR SYBR Green master mix (Exiqon, Vedbaek, Denmark) and an RNU-5G PCR primer set for the reference gene, an miR140-5p primer set for miR-140 and an miR-193b primer set for miR-193b (Exiqon, Vedbaek, Denmark). Thermocycling conditions in a Biometra TPersonal system (Goettingen, Germany) were as follows: polymerase activation at 95 °C for 10 min, followed by 50 amplification cycles of 95 °C for 10 s and 60 °C for 1 min. Melting curves were obtained from 40–95°C with continuous ramps of 0.1 °C. All reactions were done in duplicate for the reference gene, miR-140, miR-193b and no-template samples. All analyses and quantifications were done using LightCycler 480 software (Roche, Germany). Relative expression was quantified by the delta delta Ct method subsequent to the normalization of miR-140 and miR-193b expression in relation to RNU-5G.
Real-time quantitative PCR for IGFBP5 and β-actin (reference housekeeping gene) were done using LightCycler 480 Probes Master (Roche). An aliquot of cDNA (2.5 μL) was used in a reaction volume of 10 μL. All reactions were done in duplicate, and all analyses and quantifications were done using LightCycler 480 software. Relative expression was quantified by the delta delta Ct method, subsequent to the normalization of IGFBP5 expression in relation to β-actin.
Statistical analysis
Statistical analysis was done using SPSS Statistics 17.0 software. miR-140 expression levels in IGFBP5-positive samples were compared with IGFBP5-negative samples, and tumor diameter and vascular invasion were compared between the two groups with the Mann-Whitney U-test. The correlation between miR-140-5p and miR-193b expression was calculated using Spearman’s correlation coefficient, p < 0.05 indicate significance. Table 1 summarizes the statistical significance of the various variables for tumor tissues.
Table 1. miR-140-5p and miR-193b expression in invasive breast carcinomas in relation to clinicopathological characteristics and IGFBP5 expression.
| Variables | No of cases | miR-140-5p expression (median) | p | miR-193b expression (median) | p |
|---|---|---|---|---|---|
| Age (years) | |||||
| ≤ 60 | 24 | 3.41 | 0.433 | 0.80 | 0.184 |
| > 60 | 24 | 1.12 | 1.99 | ||
| ERα status | |||||
| Positive | 22 | 6.92 | 0.200 | 1.51 | 0.542 |
| Negative | 26 | 1.01 | 1.07 | ||
| Histology | |||||
| Invasive ductal | 26 | 41.57 | 0.002 | 1.69 | 0.121 |
| Other | 19 | 0.62 | 0.69 | ||
| Her2 Status | |||||
| Her2 − | 25 | 1.01 | 0.089 | 0.98 | 0.613 |
| Her2 + | 23 | 6.96 | 2.44 | ||
| Subtype | |||||
| Luminal | 22 | 6.92 | 0.200 | 1.51 | 0.542 |
| Other | 26 | 1.09 | 1.07 | ||
| Subtype | |||||
| Basal-like | 14 | 0.53 | 0.008 | 0.87 | 0.252 |
| Other | 34 | 11.36 | 2.13 | ||
| Tumor size (cm) | |||||
| < 2.5 | 21 | 0.72 | 0.126 | 0.59 | 0.011 |
| ≥ 2.5 | 22 | 6.14 | 2.62 | ||
| IHC (+ vs −) | |||||
| IGFBP5− | 14 | 294.6 | 0.006 | 2.63 | 0.252 |
| IGFBP5+ | 34 | 1.01 | 1.03 | ||
| IHC (low vs high) | |||||
| IGFBP5 Low | 33 | 2.00 | 0.648 | 1.46 | 0.155 |
| IGFBP5 High | 15 | 1.01 | 0.80 | ||
| Lymph node status | |||||
| Negative | 18 | 0.80 | 0.049 | 0.84 | 0.127 |
| Positive | 25 | 9.03 | 2.76 | ||
| Vascular invasion | |||||
| Negative | 26 | 1.40 | 0.07 | 1.03 | 0.312 |
| Positive | 16 | 180.9 | 2.63 | ||
| IGFBP5 mRNA | |||||
| Low | 23 | 1.00 | 0.102 | 0.80 | 0.063 |
| High | 23 | 4.81 | 1.83 |
ER - estrogen receptor, IHC - immunohistochemistry. p < 0.05 indicates significance (Mann-Whitney U-test).
Results
Patient demographics and tumor characteristics
Table 2 summarizes the histology, histological grade, tumor diameter, nuclear grade, vascular invasion and lymph node status for the patients examined in this study. Some of the data are missing and these missing items were excluded from the statistical analyses. All statistical analyses of miRNA expression and the clinopathological characteristics are shown in Table 1.
Table 2. Patient demographics and tumor characteristics.
| Variable | Values percentages (%) and number of cases (n) | ||
|---|---|---|---|
| Age (years) | 61.4 ± 15.6 (range: 33–91) | ||
| Missing | n = 0 | ||
| Tumor diameter (cm) | 2.95 ± 1.71 (range: 0.8–10) | ||
| Missing | n = 6 | ||
| n | % | ||
| Histology | |||
| Invasive ductal | 26 | 54.2 | |
| Invasive lobular | 13 | 27.1 | |
| Medullary | 2 | 4.2 | |
| Invasive mixed | 3 | 6.3 | |
| Invasive micropapillary | 1 | 2.1 | |
| Missing | 3 | 6.3 | |
| Histologic grade | |||
| 1 | 2 | 4.2 | |
| 2 | 22 | 45.8 | |
| 3 | 5 | 10.4 | |
| Missing | 19 | 39.6 | |
| Nuclear grade | |||
| 1 | 1 | 2.1 | |
| 2 | 25 | 52.1 | |
| 3 | 3 | 6.2 | |
| Missing | 19 | 39.6 | |
| Vascular invasion | |||
| Positive | 16 | 33.3 | |
| Negative | 26 | 54.2 | |
| Missing | 6 | 12.5 | |
| Lymph node | |||
| Positive | 25 | 52.1 | |
| Negative | 18 | 37.5 | |
| Missing | 5 | 10.4 | |
| Tumor type | |||
| Her2 | 12 | 25.0 | |
| LumA | 11 | 22.9 | |
| LumB | 11 | 22.9 | |
| Basal | 14 | 29.2 | |
| Tumor IGFBP5 IHC | |||
| Positive | 34 | 70.8 | |
| Negative | 14 | 29.2 | |
| Normal tissue IGFBP5 IHC | |||
| Positive | 22 | 68.8 | |
| Negative | 10 | 31.2 | |
miR-140 and miR-193b expression in breast cancer tissue and adjacent non-cancerous tissue
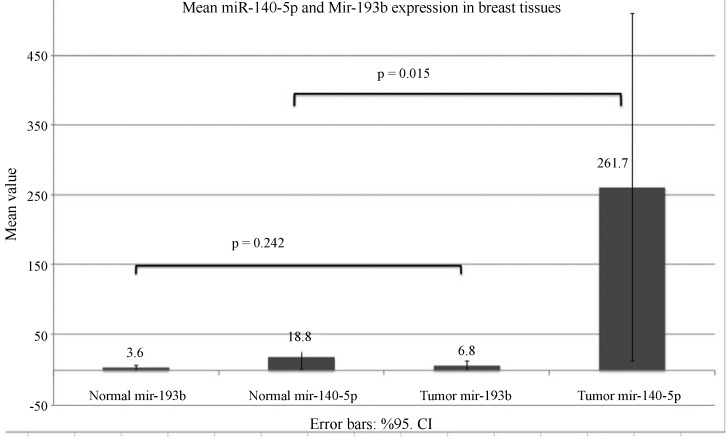
The miRNA expression in tumor (n = 48) and adjacent normal (n = 32) tissues was examined to determine whether miR-140 or miR-193b expression was a tumorigenic property (Figure 2). Tumor tissue samples had higher miR-140-5p expression than adjacent normal tissues (mean = 261.7 and median = 1.8 for tumor samples and mean = 18.79 and median = 0.57 for normal tissues; p = 0.015). These results indicate that enhanced miR-140 expression was characteristic of tumors and could be a potential biomarker for breast cancer. There was no difference in the miR-193b expression of tumors and adjacent normal tissue.
Figure 2. Expression profiling data for miR-140 and miR-193b in breast cancer and normal tissue. miR-140 expression was significantly upregulated in breast cancer tissue (miR-140 expression in breast cancer tissue was ∼14 fold that of normal tissue; p = 0.015) whereas miR-193b expression was not significantly different from that of normal tissue (p = 0.242). The first two columns from left are the mean of normal breast tissue samples, the last two columns are the mean of tumor breast tissue samples and the error bars indicate the 95% confidence intervals (CI).
Spearman’s correlation was used to assess the correlation between miR-140-5p and miR-193b expression in tissue samples and both were found to be positively correlated in tumor samples (n = 48), in normal tissue samples (n = 32) and in all tissue samples (n = 80) (correlation coefficient (CC) = 0.616, p = 0.000 for tumor tissue, CC = 0.507, p = 0.003 for normal tissue, and CC = 0.583, p = 0.0000 for all samples, respectively).
IGFBP5 protein immunohistochemistry and correlation with miR-140 and miR-193b
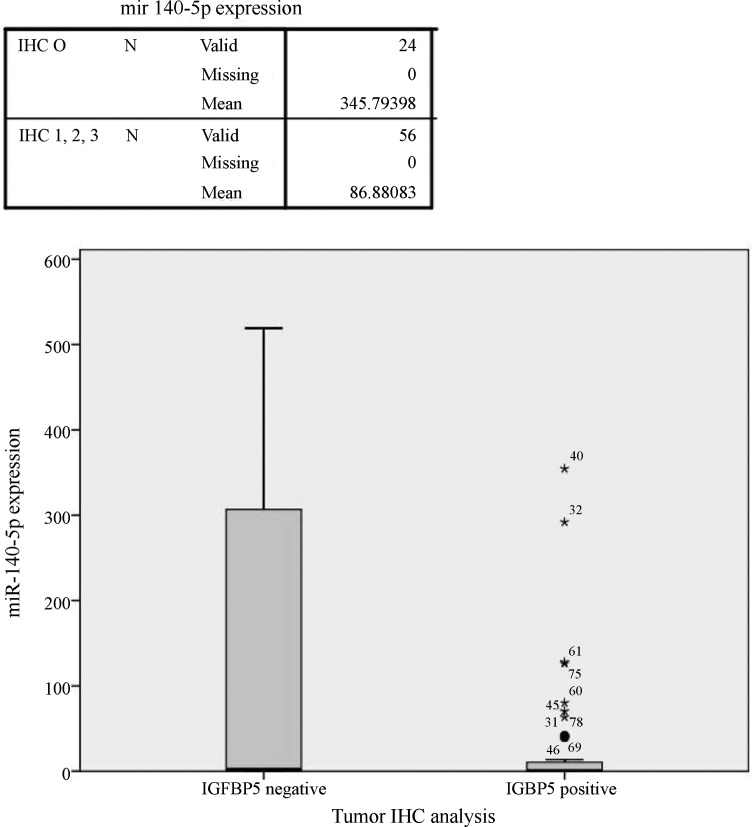
Tumor tissue samples and all tissue samples were classified into two groups as IGFBP5-positive (expressing the protein) and IGFBP5-negative (not expressing the protein). Based on this classification, 29.2% of tumor samples and 31.3% of normal tissue samples were IGFBP5-negative and there was no significant difference between the two groups. Differences in miR-140-5p expression between these groups were analyzed with the Mann-Whitney U-test. Tumor samples (n = 48) and all of the samples that were analyzed immunohistochemically (n = 80; regardless of whether they were normal or tumor tissue) showed greater miR-140-5p expression in IGFBP5-negative than in IGFBP5-positive samples (p = 0.006 and 0.009, respectively) (Figure 3). Immunohistochemically IGFBP5-negative samples showed increased expression of miR-140-5p, but this was not the case for miR-193b.
Figure 3. miR-140-5p expression in tissues with positive and negative immunohistochemical staining for IGFBP5. Of the 80 samples analyzed immunohistochemically, those that were IGFBP5-negative showed significantly greater miR-140-5p expression than IGFBP5-positive samples (p = 0.009; Mann-Whitney U-test); miR-140 expression is 4-fold less than in tissues that do not express IGFBP5 protein. The numbers beside some of the asterisks (*) states the sample number. The error bars indicate the 95% confidence intervals (CI).
mRNA expression of IGFBP5 and correlation with miR-140 and miR-193b
Spearmans correlation coefficient was also used to examine the correlation between the protein (immunohistochemistry) and mRNA (RT-PCR) levels of IGFBP5. Based on this analysis, IGFBP5 protein expression in tumor samples was found to be negatively correlated with mRNA expression of this protein (CC = −0.295; p = 0.047), when protein expression [in which negative and low positive (+) staining were considered as negative results, and medium positive (++) and strong positive (+++) staining were considered as positive results] was compared with IGFBP5 mRNA expression levels, calculated relative to the reference gene [low (< 1) and high (> 1)] (Table 3). Although there was no significance, IGFBP5 mRNA-negative samples showed a tendency to express higher levels of miR-140-5p, which suggested that miR-140-5p could post-transcriptionally regulate IGFBP5 expression.
Table 3. Correlation between IGFBP5 mRNA (RT-PCR) and protein (immunohistochemistry - IHC) expression levels.
| IHC results | ||||
|---|---|---|---|---|
|
| ||||
| Negative | Positive | Total (n) | ||
| mRNA expression | Low | 13 | 11 | 24 |
| High | 18 | 4 | 22 | |
| Total (n) | 31 | 15 | 46 | |
Relationship between the clinicopathological characteristics and miR-140, miR-193b and IGFBP5 protein levels
There was no correlation between miR-193b and miR-140-5p expression and patient age, ERα status, Her2 status, vascular invasion and nuclear grade. However, invasive ductal carcinomas showed greater expression of miR-140-5p than other histological types (p = 0.002, median expression = 41.57 and 0.618 for invasive ductal carcinomas and others, respectively). No such association was seen for miR-193b. On the contrary, basal-like tumors showed decreased expression of miR-140-5p compared to other tumors (median miR-140 expression = 0.525 and 11.36 for basal-like tumors and other tumors, respectively; p = .008). No such relationship was observed for miR-193b. Lymph node-positive samples had an approximately 13-fold greater expression of miR-140-5p than lymph node-negative samples (median expression = 0.799 and 9.031 for lymph node-negative and -positive samples, respectively; p = 0.049).
Comparison of the diameters of vascular invasive and non-invasive tumors based on the Mann-Whitney U-test showed that the former were greater than the latter (n = 42, p = 0.003). The median diameter of vascular non-invasive tumors was 2.05 cm. Tumors > 2.5 cm in diameter had greater expression of miR-193b than tumors < 2.5 cm (median expression = 0.59 and 2.62 for small and large tumors, respectively; p = 0.011).
All of the tumor samples and 27 of the adjacent normal tissue samples were miR-193b-positive and only five of the adjacent normal tissues were negative; all samples of the latter group were immunohistochemically IGFBP5-positive and four were ER-negative. There was no significant correlation between the expression levels of IGFBP5, ER and miR-193b.
Discussion
We analyzed miRNA expression in breast cancer tumors (n = 48) and adjacent normal mammary (n = 32) tissues and found that tumor tissue had higher miR-140-5p expression than adjacent normal tissue. miR-140 has been considered a possible tumor suppressor miRNA because of its regulatory actions in the MAPK/ERK, TGF-β and SOX2 pathways (Zhang et al., 2012; Yang et al., 2013). However, as shown here, breast tumor samples had higher expression of miR-140 than normal breast tissue, although it was found to be down-regulated in some breast tumors. The role of miR-140 in breast tumorigenesis is unclear, but has been associated with drug resistance, tumor growth and metastasis in various cancers. miR-140 and SOX2, a stem cell self-renewal regulator, are crucial components in the maintenance of breast cancer stem cells. The expression of miR-140 in ERα-positive breast cancer cells decreases subsequent estrogen stimulation because ERα suppresses miR-140 transcription by binding to the estrogen response element flanking the miR-140 promoter. miR-140 targets the SOX2 gene, thereby regulating breast tumor cell survival (Zhang et al., 2012). However, we found no association between ERα-positive breast cancer tissue and miR-140 expression levels.
Recent studies have described anti-proliferative and anti-metastatic effects of miR-140 in Hepatocellular carcinoma (HCC), which suggests a possible role in tumor suppression. miR-140 targets the Tissue Growth Factor β receptor 1 (TGFBR1) and Fibroblast Growth Factor 9 (FGF9), thereby inhibiting TGF-β and MAPK/ERK signaling (Yang et al., 2013). Our findings indicate that miR-140 expression has tumorigenic activity and could be a potential biomarker for breast cancer. However, more data about patient demographics and clinical outcome are needed to evaluate whether patients with higher expression of miR-140 have any clinical advantage, such as response to treatment, metastasis-free survival, disease-free survival and overall survival.
Recently, Li et al. (2013) showed that miR-140 expression is down-regulated in ductal carcinoma in situ (DCIS) stem cells compared to normal mammary stem cells, and that SOX9 and ALDH1, direct targets of miR-140, were markedly activated in DSIC stem cells. Restoration of miR-140 expression in ERα-negative/basal-like DCIS cells resulted in a decrease in SOX9 and ALDH1 expression and decreased tumor growth in vivo (Li et al., 2013). As shown here, ERα-negative/basal-like samples had the lowest miR-140 expression (2.5-fold and 4-fold lower than in other tumors and normal tissue samples, respectively). This could explain the aggressiveness of basal-like tumors in which miR-140 is suppressed, stem cell factors are induced and the tumors are more resistant to therapy. Normal tissues had about a 15-fold lower expression of miR-140 compared to tumors, excluding ERα-negative/basal-like tumors. Based on these findings, we conclude that normal breast tissues need optimal expression of miR-140 for cell renewal, and that basal-like tumors, which have lower expression of this protein, have aggressive characteristics compared to other types of breast cancer.
Invasive ductal tumors showed a 70-fold greater miR-140 expression than other tumors (median = 42.57 and 0.618, respectively; p = 0.002). In addition, patients with positive lymph nodes had greater expression of miR-140 than the other groups (median expression = 0.80 and 9.03, respectively; p = 0.049); this finding suggests that miR-140 may be involved in lymph node invasion. To our knowledge, this is the first demonstration of a significant association between miR-140 and lymph node metastasis in breast cancer tissue. Previously, miR-140 had only been reported to be down-regulated in primary breast cancer tissue compared to normal breast tissue, but it was not shown to be associated with lymph node invasion (Yan et al., 2008). Further data are needed to shed light on the relationship between miR-140 and invasiveness.
Tardif et al. (2009) showed that IGFBP5 was directly targeted for mRNA degradation by miR-140, but expression analysis of miR-140 during chondrogenic differentiation did not confirm that IGFBP5 was a direct target of miR-140 (Buechli et al., 2013). These authors indicated that IGFBP5 was not directly targeted by miR-140 for degradation, which would be expected if miR-140 facilitated its decay; the possibility that the translation of IGFBP5 could be inhibited by miR-140 was not tested. As shown here, there was no correlation between IGFBP5 mRNA expression and miR-140. In addition, regardless of the type of tissue (tumor or normal), in samples in which IGFBP5 protein was expressed miR-140 expression was four-fold lower than in samples not expressing IGFBP5 protein (Figure 3). These results indicate that miR-140 post-transcriptionally regulates IGFBP5 by inhibiting its translation rather than by degrading its mRNA. We conclude that miR-140 is more effective at repressing IGFBP5 protein expression in normal tissues than in tumor samples and that the higher expression of miR-140 in IGFBP5-negative tumors reflects this proteins effectiveness.
Although IGFBP5 mRNA is up-regulated in cancer tissue (Li et al., 2007), we found no significant difference among our samples. IGFBP5 mRNA levels have been positively correlated with ER and PR status, and negatively correlated with HER2 overexpression, with low levels of IGFBP5 mRNA in tumors being associated with better prognosis and disease-free survival (Hung et al., 2008). In contrast to this study, we found no significant correlation between IGFBP5 mRNA levels vs. ER and PR status or HER2 overexpression.
There was a significant positive correlation between miR-140-5p and miR-193b expression in all tissue samples. However, there was no significant correlation between ER status and miR-193b, in contrast to the findings of Yoshimoto et al. (2011) who reported that ERα-negative tumors had relatively lower miR-193b expression than ERα-positive tumors. Foekens et al. (2000) reported a relation between miR-193b and uPA that plays a key role in invasion and metastasis in breast cancers. uPA expression is increased in metastatic breast cancer and is negatively correlated with miR-193b. This may mean that miR-193b could be a biomarker for metastatic breast cancer (Noh et al., 2011). On the other hand, miR-193b acts as a tumor suppressor that limits proliferation, migration and invasion in acute myeloid leukemia by regulating c-Kit proto-oncogene (Gao et al., 2011), and also suppresses human hepatocellular carcinoma cells (Xu et al., 2010), melanoma (by regulating Mcl-1) (Chen et al., 2010) and non-small cell lung cancer cells (Hu et al., 2012). In the present study, it was not possible to analyze our data based on metastatic behavior or tumor invasiveness since we did not have the necessary data for the patients.
In conclusion, our results show that breast tumor tissue had higher miR-140-5p expression than adjacent normal tissues and that patients with positive lymph nodes had greater miR-140 expression than other individuals. On the other hand, basal-like breast tumors had lower expression of miR-140-5p than other types of breast tumors. Thus, miR-140 expression is characteristic of breast tumors in general and may be an important molecule for understanding lymph node invasion and tumor differentiation in breast cancer. There was no correlation between IGBFP5 mRNA and miR-140 expression, but miR-140 expression was lower in tumors expressing IGFBP5 protein compared to samples which not expressing this protein. There was a significant positive correlation between miR-140-5p and miR-193b expression in all tissue samples, but there was no association between miR-193b expression and any other clinical features. Further studies, including clinical follow-up of the patients, would be useful in clarifying the role of miR-140, miR-193b and IGFBP5 in breast cancer and in assessing the usefulness of these miRNAs and IGFBP5 as suppressors or inducers of breast carcinogenesis.
Acknowledgments
The authors thank Dr. Nadiye Pinar Ay (Department of Public Health, School of Medicine, Marmara University) for assistance in statistical analyses. This work was partly supported by a grant (SBAG-111S161) from the Scientific and Technological Research Council of Turkey and by grants (SAG-C-YLP-110412-0067 and SAG-C-YLP-210311-0048) from the Research Foundation of Marmara University (BAPKO).
Footnotes
Associate Editor: Anamaria Aranha Camargo
References
- Akkiprik M, Feng Y, Wang H, Chen K, Hu L, Sahin A, Krishnamurthy S, Ozer A, Hao X, Zhang W. Multifunctional roles of insulin-like growth factor binding protein 5 in breast cancer. Breast Cancer Res. 2008;10:212. doi: 10.1186/bcr2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie J, Allan GJ, Lochrie JD, Flint D. Insulin-like growth factor binding protein-5 (IGFBP5): A critical member of the IGF axis. Biochem J. 2006;395:1–19. doi: 10.1042/BJ20060086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker MA, Hou X, Harrington SC, Weroha SJ, Gonzalez SE, Jacob KA, Carboni JM, Gottardis MM, Haluska P. IGFBP ratio confers resistance to IGF targeting and correlates with increased invasion and poor outcome in breast tumors. Clin Cancer Res. 2012;18:1808–1817. doi: 10.1158/1078-0432.CCR-11-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buechli ME, Lamarre J, Koch TG. MicroRNA-140 expression during chondrogenic differentiation of equine cord blood-derived mesenchymal stromal cells. Stem Cells Dev. 2013;22:1288–1296. doi: 10.1089/scd.2012.0411. [DOI] [PubMed] [Google Scholar]
- Butt AJ, Dickson KA, Jambazov S, Baxter RC. Enhancement of tumor necrosis factor-alpha-induced growth inhibition by insulin-like growth factor-binding protein-5 (IGFBP-5), but not IGFBP-3 in human breast cancer cells. Endocrinology. 2005;146:3113–3122. doi: 10.1210/en.2004-1408. [DOI] [PubMed] [Google Scholar]
- Chen J, Feilotter HE, Paré GC, Zhang X, Pemberton JG, Garady C, Lai D, Yang X, Tron VA. MicroRNA-193b represses cell proliferation and regulates cyclin D1 in melanoma. Pathology. 2010;176:2520–2529. doi: 10.2353/ajpath.2010.091061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eroles P, Bosch A, Pérez-Fidalgo JA, Lluch A. Molecular biology in breast cancer: Intrinsic subtypes and signaling pathways. Cancer Treat Rev. 2011;6:698–707. doi: 10.1016/j.ctrv.2011.11.005. [DOI] [PubMed] [Google Scholar]
- Foekens J, Peters HA, Look MP, Portengen H, Schmitt M, Kramer MD, Brünner N, Jänicke F, Meijer-van Gelder ME, Henzen-Logmans SC, et al. The urokinase system of plasminogen activation and prognosis in 2780 breast cancer patients. Cancer Res. 2000;60:636–643. [PubMed] [Google Scholar]
- Gao XN, Lin J, Gao L, Li YH, Wang LL, Yu L. MicroRNA-193b regulates c-Kit proto-oncogene and represses cell proliferation in acute myeloid leukemia. Leuk Res. 2011;35:1226–1232. doi: 10.1016/j.leukres.2011.06.010. [DOI] [PubMed] [Google Scholar]
- Güllü G, Karabulut S, Akkiprik M. Functional roles and clinical values of insulin-like growth factor binding protein-5 in different types of cancers. Chin J Cancer. 2012;6:266–280. doi: 10.5732/cjc.011.10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Li S, Liu J, Ni B. MicroRNA-193b modulates proliferation, migration, and invasion of non-small cell lung cancer cells. Acta Biochim Biophys Sin (Shanghai) 2012;44:424–430. doi: 10.1093/abbs/gms018. [DOI] [PubMed] [Google Scholar]
- Hung PS, Kao SY, Shih YH, Chiou SH, Liu CJ, Chang KW, Lin SC. Insulin-like growth factor binding protein-5 (IGFBP-5) suppresses the tumourigenesis of head and neck squamous cell carcinoma. J Pathol. 2008;214:368–376. doi: 10.1002/path.2280. [DOI] [PubMed] [Google Scholar]
- Johnson SK, Haun RS. Insulin-like growth factor binding protein-5 influences pancreatic cancer cell growth. World J Gastroenterol. 2009;15:3355–3366. doi: 10.3748/wjg.15.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandwala HM, McCutcheon IE, Flyvbjerg A, Friend KE. The effects of insulin-like growth factors on tumori-genesis and neoplastic growth. Endocr Rev. 2000;21:215–244. doi: 10.1210/edrv.21.3.0399. [DOI] [PubMed] [Google Scholar]
- Kricker JA, Hyde CE, Van Lonkhuyzen DR, Hollier BG, Shooter GK, Leavesley DI, Herington AC, Upton Z. Mechanistic investigations into interactions between IGF-I and IGFBPs and their impact on facilitating cell migration on vitronectin. Growth Factors. 2010;28:359–369. doi: 10.3109/08977194.2010.494603. [DOI] [PubMed] [Google Scholar]
- Leivonen SK, Mäkelä R, Ostling P, Kohonen P, Haapa-Paananen S, Kleivi K, Enerly E, Aakula A, Hellström K, Sahlberg N, et al. Protein lysate microarray analysis to identify microRNAs regulating estrogen receptor signaling in breast cancer cell lines. Oncogene. 2009;28:3926–3936. doi: 10.1038/onc.2009.241. [DOI] [PubMed] [Google Scholar]
- Li Q, Yao Y, Eades G, Liu Z, Zhang Y, Zhou Q. Downregulation of miR-140 promotes cancer stem cell formation in basal-like early stage breast cancer. Oncogene. 2013;20:2589–2600. doi: 10.1038/onc.2013.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Cao X, Li X, Zhang W, Feng Y. Expression level of insulin-like growth factor binding protein 5 mRNA is a prognostic factor for breast cancer. Cancer Sci. 2007;98:1592–1596. doi: 10.1111/j.1349-7006.2007.00565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita K, Zhang Z, Ando Y, Toyama T, Hamaguchi M, Kobayashi S, Hayashi S, Fujii Y, Iwase H, Yamashita H. Prognostic significance of insulin-like growth factor binding protein (IGFBP)-4 and IGFBP-5 expression in breast cancer. Jpn J Clin Oncol. 2007;37:575–582. doi: 10.1093/jjco/hym066. [DOI] [PubMed] [Google Scholar]
- Mohan S, Baylink DJ. IGF-binding proteins are multi-functional and act via IGF-dependent and independent mechanisms. J Endocrinol. 2002;175:19–31. doi: 10.1677/joe.0.1750019. [DOI] [PubMed] [Google Scholar]
- Noh H, Hong S, Dong Z, Pan ZK, Jing Q, Huang S. Impaired microRNA processing facilitates breast cancer cell invasion by upregulating urokinase-type plasminogen activator expression. Genes Cancer. 2011;2:140–150. doi: 10.1177/1947601911408888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmero EI, de Campos SG, Campos M, de Souza NC, Guerreiro ID, Carvalho AL, Marques MM. Mechanisms and role of microRNA deregulation in cancer onset and progression. Genet Mol Biol. 2011;34:363–370. doi: 10.1590/S1415-47572011000300001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif G, Hum D, Pelletier JP, Duval N, Martel-Pelletier J. Regulation of the IGFBP-5 and MMP-13 genes by the microRNAs miR-140 and miR-27a in human osteoarthritic chondrocytes. BMC Musculoskelet Disord. 2009;10:148. doi: 10.1186/1471-2474-10-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentinis B, Baserga R. IGF-I receptor signalling in transformation and differentiation. Mol Pathol. 2001;54:133–137. doi: 10.1136/mp.54.3.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Liu S, Fu H, Li S, Tie Y, Zhu J, Xing R, Jin Y, Sun Z, Zheng X. MicroRNA-193b regulates proliferation, migration and invasion in human hepatocellular carcinoma cells. Eur J Cancer. 2010;15:2828–2836. doi: 10.1016/j.ejca.2010.06.127. [DOI] [PubMed] [Google Scholar]
- Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;11:2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Fang F, Chang R, Yang L. MicroRNA-140-5p suppresses tumor growth and metastasis by targeting transforming growth factor β receptor 1 and fibroblast growth factor 9 in hepatocellular carcinoma. Hepatology. 2013;58:205–217. doi: 10.1002/hep.26315. [DOI] [PubMed] [Google Scholar]
- Yoshimoto N, Toyama T, Takahashi S, Sugiura H, Endo Y, Iwasa M, Fujii Y, Yamashita H. Distinct expressions of microRNAs that directly target estrogen receptor α in human breast cancer. Breast Cancer Res Tr. 2011;130:331–339. doi: 10.1007/s10549-011-1672-2. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Eades G, Yao Y, Li Q, Zhou Q. Estrogen receptor α signaling regulates breast tumor-initiating cells by down-regulating miR-140 which targets the transcription factor SOX2. J Biol Chem. 2012;287:41514–41522. doi: 10.1074/jbc.M112.404871. [DOI] [PMC free article] [PubMed] [Google Scholar]