Abstract
Defects in centrosome, centrosomal-associated and spindle-associated proteins are the most frequent cause of Primary Microcephaly (PM) and Microcephalic Primordial Dwarfism (MPD) syndromes in humans. Mitotic progression and segregation defects, microtubule spindle abnormalities and impaired DNA damage-induced G2-M cell cycle checkpoint proficiency have been documented in cell lines from these patients. This suggests that impaired mitotic entry, progression and exit strongly contribute to PM and MPD. Considering the vast protein networks involved in coordinating this cell cycle stage, the list of potential target genes that could underlie novel developmental disorders is large. One such complex network, with a direct microtubule-mediated physical connection to the centrosome, is the kinetochore. This centromeric-associated structure nucleates microtubule attachments onto mitotic chromosomes. Here, we described novel compound heterozygous variants in CENPE in two siblings who exhibit a profound MPD associated with developmental delay, simplified gyri and other isolated abnormalities. CENPE encodes centromere-associated protein E (CENP-E), a core kinetochore component functioning to mediate chromosome congression initially of misaligned chromosomes and in subsequent spindle microtubule capture during mitosis. Firstly, we present a comprehensive clinical description of these patients. Then, using patient cells we document abnormalities in spindle microtubule organisation, mitotic progression and segregation, before modeling the cellular pathogenicity of these variants in an independent cell system. Our cellular analysis shows that a pathogenic defect in CENP-E, a kinetochore-core protein, largely phenocopies PCNT-mutated Microcephalic Osteodysplastic Primordial Dwarfism type II patient cells. PCNT encodes a centrosome-associated protein. These results highlight a common underlying pathomechanism. Our findings provide the first evidence for a kinetochore-based route to MPD in humans.
Keywords: Microcephaly, growth restriction, centromere, kinetochore, CENP-E, spindle microtubules
Introduction
Microcephalic primordial dwarfism (MPD) is the collective term for a group of clinically overlapping syndromes whose underlying genetic basis has started to emerge within the last decade or so. Specific examples include Seckel Syndrome, Microcephalic Primordial Dwarfism-Type II (MOPDii) and Meier-Gorlin Syndrome (Gorlin et al. 1975; Hall et al. 2004; Majewski et al. 1982b; Seckel 1960). For these specific disorders, causative defects have been identified in central components of the ATR-dependent DNA damage response (DDR), in a centrosome-associated protein and in components of the DNA replication licensing machinery respectively (Bicknell et al. 2011a; Bicknell et al. 2011b; Griffith et al. 2008; Guernsey et al. 2011; O’Driscoll et al. 2003; Ogi et al. 2012; Qvist et al. 2011; Rauch et al. 2008). Defects in DNA double strand break repair and other genome stability pathways can also cause MPD (Buck et al. 2006; Ijspeert et al. 2013; Murray et al. 2014; O’Driscoll et al. 2001; Shaheen et al. 2014; Shamseldin et al. 2012).
Primary Microcephaly (PM) with and without overt short stature is commonly caused by defects in centrosome and spindle pole associated proteins (Mahmood et al. 2011; Poirier et al. 2013; Thornton and Woods 2009). These PM and MPD-associated defects collectively underscore the requirement for optimal cell cycle progression and checkpoint control for normal human development, including growth and neurogenesis. Currently, mutations in genes encoding core centrosome and centrosome-associated proteins represent the most frequent causative defect identified so far in MPDs. The centrosome is an important microtubule-organizing centre essential for coordinating G2-M progression, normal mitotic cell division and even cilia formation. Interestingly, defects in the ATR-dependent DDR and in DNA replication licensing machinery are also associated with centrosome abnormalities, and vice versa, suggestive of a multifactorial but interconnected underlying pathobiology of MPDs (Alderton 2006; Alderton et al. 2004; Griffith et al. 2008; Stiff et al. 2013).
The centromeric kinetochore of mitotic chromosomes is the attachment site for spindle microtubules, ensuring their amphitelic attachments onto all of the chromosomes thereby enabling their alignment at metaphase (Hori and Fukagawa 2012; Santaguida and Musacchio 2009). This facilitates accurate and timely segregation. Unattached kinetochores activate the mitotic spindle assembly checkpoint (SAC) (Cleveland et al. 2003). SAC activation delays mitosis until the kinetochores of each chromosome has properly attached to spindle microtubules (Musacchio 2011; Musacchio and Salmon 2007). Congenital defects in the SAC gene BUB1B which encodes the SAC kinase BubR1 cause Mosaic Variegated Aneuploidy (MVA); an MPD associated with aneuploidy and elevated cancer incidence (Hanks 2004; Shinya Matsuura et al. 2006).
In contrast to the centrosome-spindle pole, pathogenic defects in core kinetochore components are currently notably under-represented as a cause of MPD. Using an exome sequencing strategy, we describe novel compound heterozygous variants in CENPE in two siblings characterized by a profound MPD with severe developmental delay, simplified gyri and various isolated abnormalities. CENP-E (centromere-associated protein-E) is a large (>300KD) kinetochore-associated kinesin-like motor protein required for spindle microtubule capture and attachment at the kinetochore (Abrieu et al. 2000; Yao et al. 2000). Unsurprisingly, Cenpe deletion in mice is early embryonic lethal (Putkey et al. 2002). Conditional Cenpe deletion in mouse embryonic fibroblasts and an adult regenerating liver system resulted in cells with profound mitotic defects including chromosome misalignment and segregation failure (Putkey et al. 2002; Weaver et al. 2003). Firstly, we detail the clinical presentation and progression of both siblings. Then, using patient-derived lymphoblastoid cell lines (LCLs) we catalogue a series of mitotic abnormalities including aberrant spindle microtubule organization, delayed mitotic progression and elevated levels of binucleate cells. The latter phenotype in particular is suggestive of an impaired ability to exit mitosis effectively. We also find that the nuclei of these binucleates are often of unequal size indicative of impaired chromosome segregation during mitosis and subsequent cytokinesis failure. To further characterize and consolidate the cellular pathogenicity of the CENPE variants we have identified, we model each individually and in combination using Flip-In technology (Invitrogen) coupled with siRNA mediated knockdown of the endogenous CENP-E in an independent cell system.
Interestingly, we find that many of these cellular phenotypes observed in the patient LCLs are also observed in LCLs from a patient with a pathogenic defect in PCNT; the centrosome associated protein implicated in MOPDii (Griffith et al. 2008; Rauch et al. 2008). Therefore, using MPD patient-derived material we show that a novel kinetochore-associated defect in CENP-E shares overlapping phenotypes of abnormal spindle microtubule structure and mitotic progression to that of a pathogenic defect in a centrosome–associated protein.
Methods
DNA extraction
Genomic DNA was extracted from peripheral blood samples using either the Puregene kit®, Magnapure® or Autogen® systems following the manufacturers’ recommendations.
Whole exome sequencing and analysis
We performed whole exome sequencing (WES) of peripheral blood DNA from subject LR05-054a1. We used the Nimbelgen whole exome capture kit, and sequence was generated on an Illumina GAII machine. Sequence was aligned to hg19 using BWA 0.6.2 and single nucleotide variants and indels were called using GATK 2.3.9 UnifiedGenotyper. Mean coverage was calculated using GATK 2.3.9 Depth Of Coverage Walker. Annotation of variants, including identification of variants present in dbSNP, 1000Genomes, and the NHLBI Exome Variant Server (EVS), was performed with Annovar (Wang et al. 2010). Pedigree-specific nonsynonymous variants, including potential compound heterozygotes, were identified using SOLVE-Brain. Variants in genes associated with known primary microcephaly and Seckel-related syndromes, as well as other known developmental brain disorders were identified using SOLVE to filter against the contents of the Developmental Brain Disorders Database 1.2 (DBDB). Final annotation of biological function and pathway membership of candidate genes with nonsynonymous compound heterozygous variants was performed using Lynx (Sulakhe et al. 2014).
Weblinks:
Sanger sequencing
PCR amplification was performed with 50ng of genomic DNA using Taq DNA polymerase (Applied Biosystems®). Exons 23 and 29 of CENPE (NM_001813.2) were targeted for mutation analysis by Sanger sequencing using standard methods.
Cell lines
EBV transformed lymphoblastoid cell lines (LCLs) were made from both patients (LR05-054a1 and LR05-054a2) using lymphocytes fractionated from whole blood. Wild-type (WT; AG09387) cells were obtained from Coriell Cell Repository (New Jersey, USA). PCNT-mutated MOPDii LCLs were from patient CV1559 and carry a homozygous mutation in PCNT resulting in p.E220X; Family 1 from Griffith et al (Griffith et al. 2008).
Antibodies
Anti-53BP1 was the mouse monoclonal from Bethyl. Anti-CENP-E was mouse monoclonal [1H12] from Abcam was used for patient LCL indirect immunofluorescence whilst anti-CENP-E (AKIN04) from Cytoskeleton was used for Western blotting. Anti-BUBR1 was a mouse monoclonal (ab4637) was from Abcam. Anti-α-tubulin was a mouse monoclonal (clone B512) from Sigma-Aldrich and anti-β-tubulin was a rabbit polyclonal (H-235) from Santa Cruz. Anti-PARP (MCS 1522G) was from AbD Serotec and anti-p85-PARP (cleaved-PARP: G7341) was obtained from Promega
Indirect immunofluorescence
Following various treatments (see below) LCLs were pelleted, swollen in 75mM KCL (10mins), fixed in 4% paraformaldehyde (10mins) prior to cytospinning onto poly-L-lysine coated slides. LCLs were then permeabilized (0.1% Triton X-100 in 5% BSA-PBS for 2mins) and blocked (5% BSA-PBS for 10mins) prior to incubation with primary and secondary antibodies diluted in 5%BSA-PBS. Slides were counter-stained with 4’,6-diamidino-2-phenylindole (DAPI) and mounted in Vectashield anti-fade medium. Images were captured using SimplePCI software on the Zeiss Axioplan platform.
LCL treatments
HU-induced 53BP1 foci
LCLs were treated with 5mM HU for 2hrs prior to harvesting and processing for indirect immunofluorescence staining using anti-53BP1.
Microtubule spindle stabilization
Spindles were stabilized by treating exponentially growing LCLs with Taxol (10µM) and the proteasome inhibitor MG132 (10µM) for 1hrs in complete medium at 37°C before shifting the cells to packed ice for 1.5hrs. LCLs were then pelleted and processed for indirect immunofluorescence using anti-α-tubulin.
BubR1 phosphorylation
Exponentially growing LCLs were treated with 0.2µg/ml colcemid for 2hrs prior to pelleting and whole cell extracts made using urea buffer (9M urea, 50mM Tris-HCL at pH7.5 and 10mM 2-mercaptoethanol) and sonication (15sec at 30% amplitude using a micro-tip; Sigma-Aldrich)
Colcemid block and release into cytochalasin B
LCLs were treated with colcemid (0.2µg/ml for 4hrs), pelleted, washed in complete medium before release into complete medium supplemented with cytochalasin B (5µg/ml). Cells were incubated for 24hrs and processed for indirect immunofluorescence using anti-β-tubulin.
G2-M checkpoint analysis
LCLs were irradiated with 3Gy IR in complete medium or 7J/m2 in PBS before seeding into complete medium supplemented with 0.2µg/ml colcemid and incubated for 4hrs. LCLs were pelleted, swollen in 75mM KCL (10mins) and fixed in 4% paraformaldehyde (10mins) prior to cytospinning onto poly-L-lysine coated slides. Cells were counter-stained with DAPI and mounted in Vectashield anti-fade prior to analysis on the Zeiss Axioplan system.
Flp-In system and variant modeling
The Flp-In (Invitrogen) system employed here for CENPE in DLD-1 cells has been described previously (Kim et al. 2010). The cDNA and siRNA for CENPE used here are identical to those of Kim et al (Kim et al. 2010). Lipofectamine RNAi MAX (Invitrogen) was used for CENPE siRNA (5’-CCACUAGAGUUGAAAGAUA-3’) with doxycycline used at 1µg/ml for induction of siRNA-resistant CENPE construct. For the live cell imaging an RFP-histone H2B expressing DLD-1 line was used to visualize chromosomes, as described in Kim et al (Kim et al. 2010).
Results
Clinical presentation and progression of siblings LR05-054a1 and LR05-054a2
The features of both siblings are summarized in Table 1. This family is a non-consanguineous family of European ancestry. The first affected, a boy (LR05-054a1), was born at 34 weeks of gestation to 33-year-old mother, who had a history of one miscarriage. Pregnancy was complicated by intrauterine growth restriction detected at 6 and 10 weeks of gestation; biparietal diameter, head circumference, femoral length, abdominal circumference and estimated fetal weight were all less than 10th percentile for gestational age. At 34 weeks of gestation, prenatal ultrasound was performed due to maternal hypertension and showed a severely small fetus for gestational age with decreased fetal activity, so delivery was induced at that time. At birth, LR05-054a1 exhibited severe congenital microcephaly, with a birth OFC of 24.5 cm (−5 SD), growth deficiency with a weight of 1.5 Kg (−3 to −4 SD) and length 40.5 cm (−2 to −3 SD). Facial features included a sloping forehead, prominent nose, and mild micrognathia. This boy had a single seizure at seven months of age which consisted of generalized body stiffening, extension of the arms and legs with breath holding. Electroencephalogram was abnormal showing generalized rhythmic fast activity followed by generalized slowing, and multifocal sharp waves. He had a few additional seizures between seven and ten months of age that were subsequently well controlled on phenobarbital.
Table 1.
A summary of the clinical and neuroimaging features of the two siblings.
| Features | LR05-054a1 | LR05-054a2 |
|---|---|---|
| Gender | Male | Female |
| Age last assessed | 8 years | 3 years |
| Growth at birth | ||
| OFC (SD) | −5 | nd |
| Weight (SD) | −3 to −4 | −3 |
| Length (SD) | −2 to −3 | −3 |
| Postnatal age | 5 years | 3 years |
| OFC (SD) | −9 | −7 |
| Weight (SD) | −5 | −3 to −4 |
| Length (SD) | −7 | −2 to −3 |
| Facial features | Apparent MIC, prominent nose, sloping forehead, relatively large ears, mild micrognathia |
Apparent MIC, low sloping forehead, round face, proportionately large ears and nose |
| Extremities | Mildly small hands and feet | Small hands and feet (– 2 SD below mean) |
| Cardiac abnormalities | Congenital restrictive cardiomyopathy |
None |
| Seizures | Tonic seizures, onset 7 months | None |
| Tone | Mild lower extremity spasticity | Mildly increased |
| Development | Severe DEV delay, nonverbal | Less severe DEV delay, 65% language delay at 1 year 4 months |
| Other medical problems | GER, mild congenital subglottic stenosis |
Otitis media, tight heel cords |
| Neuroimaging features | Simplified gyral pattern, thin cortex, mildly disproportionate cerebellar hypoplasia |
nd |
| Outcome | Deceased at 8 years (pneumonia) |
Alive |
DEV, developmental; GER, gastroesophageal reflux; MIC, microcephaly; nd, no data; OFC, occipitofrontal circumference; SD, standard deviation
At four months of age, he had a viral upper respiratory illness and, because of worsening respiratory symptoms, was admitted and underwent a cardiac echocardiogram which showed abnormal tricuspid and mitral valve inflow patterns with increased early and late atrial (e/a) ratios, suggestive of diastolic dysfunction, and decreased deceleration times varying with the respiratory cycles, indicative of increased flow reversal in the pulmonary veins with atrial contraction. Additional echo findings include trivial tricuspid and mitral regurgitation, biatrial enlargement with paradoxial septal wall motion suggesting elevated right ventricular systolic pressure, a trabeculated and hypertrophied apex of the left ventricle but with normal dimensions and good systolic function. Overall, these features were consistent with congenital restrictive cardiomyopathy, for which he was treated with digoxin, Lasix and aspirin. Other medical complications include poor weight gain, partly due to the underlying syndrome and partly due to gastroesophageal reflux, hyperopia requiring glasses, and mild congenital subglottic stenosis of no clinical significance.
On last examination at 5 years, LR05-054a1 exhibited the following growth measurements: weight 9.14 kg (−5 SD), length 83.7 cm (−7 SD), and OFC 37.0 cm (−9 SD). His facial appearance was significant for striking microcephaly with a very low forehead, round face, prominent nose, mild micrognathia and decreased subcutaneous fat (Fig.1 a–c). His head appeared disproportionately smaller than his body whilst his nose and ears disproportionately large. His hands and feet were mildly small. Even though LR05-054a1 did not exhibit the nasal preponderance with downward curved nasal tip classically associated with Seckel syndrome, his overall features appeared compatible with profound MPD of this type (Hall et al. 2004; Majewski et al. 1982a; Majewski et al. 1982b). On neurological exam, he was alert but did not follow commands or communicate. Eye movements were full, pupils normal and facial movements symmetric. Motor exam showed mild spasticity in the hip adductors and ankles. Deep tendon reflexes were normal (2+) and symmetric. Developmentally, this child made very slow but steady progress albeit remaining severely handicapped. He began to pull himself to stand only briefly at 3.5 years of age, and crawled at 4 years. On assessment at 5 years, he had no communication skills besides vocalizing without signs, words or pointing. He used a rake and pincer grasp. He was fed a soft diet by mouth. His general health including his cardiac course deteriorated over time; he developed pericardial effusion on a few occasions requiring percutaneous drainage and also had several episodes of pneumonia, with progressive worsening of his oxygen saturation levels. A final episode of pneumonia at 8 years of age proved fatal.
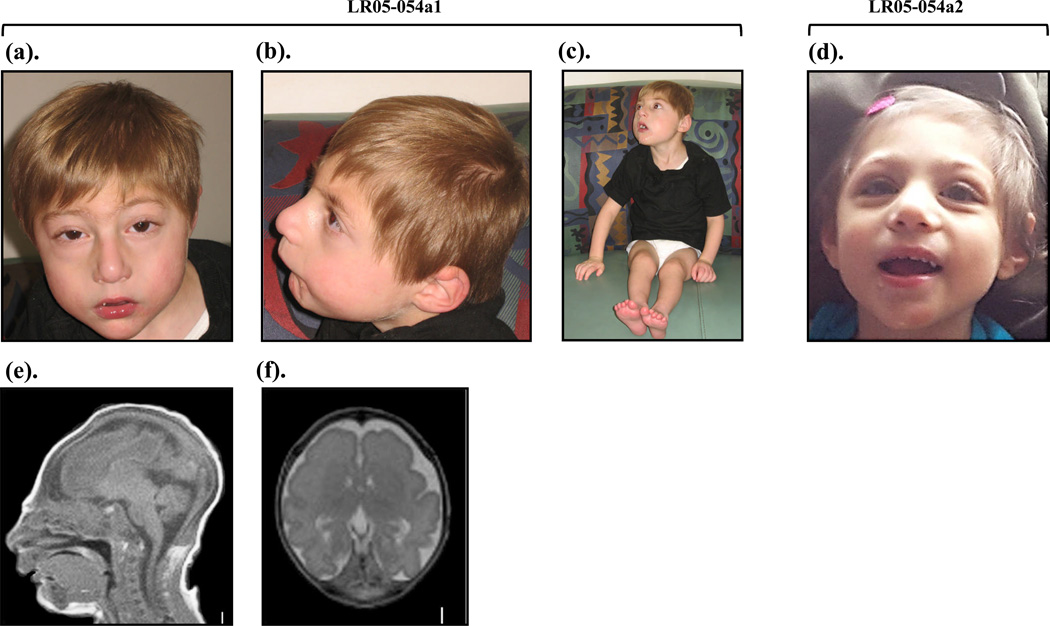
Figure 1. Images of the affected siblings LR05-054a1 and LR05-054a2.
(a–c). Patient LR05-054a1 at 16 months of age.
(d). Patient LR05-054a2 at 2 years of age showing microcephaly, low sloping forehead, proportionately large ears and nose, a prominent nose, micrognathia, and severe growth deficiency.
(e). T1-weighted mid-sagittal and (f). T2-weighted axial brain MRI images of LR05-054a1 at 17 days of life show severe microcephaly, with low-sloping forehead, severely simplified gyral pattern with virtually no gyri in the frontal region, mildly increased extra-axial space, and disproportionately small cerebellar vermis.
The pregnancy of the second affected child of the family, a girl (LR05-054a2), was also complicated by microcephaly and intrauterine growth restriction prenatally detected on ultrasound. She was born at 37 weeks of gestation with birth weight 1.76 Kg (−3 SD) and length 41.5 cm (−3 SD). She was noticed to have the same facial features as her brother’s at birth. She exhibited frequent episodes of otitis media requiring myringotomy tube placement, and minor foot deformities consisting of tight heel cords and medially deviated halluces, for which she underwent heel cord lengthening and tenotomy of great toes for better alignment. Unlike her brother, she did not have episodes of pneumonia or other severe illnesses requiring hospitalization. Echocardiogram and electrocardiogram were normal. She did not have seizures, and was described as a generally happy child.
On last exam at 3 years, her weight was 9.1 kg (−3 to −4 SD), length 83 cm (−2 to −3 SD), and OFC 37.5 cm (−7 SD). Her palms, fingers and toes measured less than −2 SD. Her overall appearance was significant for a small head, small round face with a low forehead and proportionately large ears and nose (Fig. 1d). The helix of the right ear was mildly under-folded, and great toes medially deviated. Neurological exam revealed an alert child with brief visual tracking and a strong cry. Tone was mildly increased and deep tendon reflexes minimally brisk (2++). Eye movements were full with intermittent nystagmus. Facial movements were symmetrical. Developmentally, this girl sat alone at 1–1.5 years, pulled to stand, cruised and climbed on furniture at 2–2.5 years, and crawled at 2.5 years. At 2.5 years, her language was limited to a few words such as “hi” and “bye”, along with mimicking speech sounds. She picked small objects but did not point or draw. She attended preschool at 3 years of age. She was able to chew solids with some feeding difficulties such as choking occasionally, requiring a softer diet.
Clinical work-up of siblings LR05-054a1 and LR05-054a2
Clinical workup performed on LR05-054a1 with inconclusive results includes a karyotype, a chromosomal microarray (Signature Genomics), chromosome breakage studies, metabolic tests and TORCH titers. A skeletal survey demonstrated apparent microcephaly with no evidence of craniosynostosis, subtle widening of the ribs and possible metaphyseal areas of sclerosis in the distal femurs and proximal tibias. The hands appeared osteopenic and metacarpals were relatively short. Overall, these findings were not characteristic of a specific bone dysplasia. Brain MRI scan of LR05-054a1 at 17 days of life showed extreme microcephaly with a low forehead and a diffuse severely simplified gyral pattern with virtually no gyri over the frontal lobe, a thin 2–3 mm cortex and immature white matter (Fig. 1e–f). Additional findings include partial agenesis of the corpus callosum and cerebellar hypoplasia that appeared disproportionate to the degree of microcephaly (Fig. 1e–f). Notably, neither sibling had a history of blood dyscrasias or other childhood cancers.
Molecular-genetic analysis of family LR05–054
Although LR05-054a1 did not present with the skeletal features characteristic of MOPDii (e.g. coxa vara, small iliac wings, V-shaped distal femoral epiphysis, metaphyseal flaring of radius and ulna), these features can take time to manifest (Hall et al. 2004). Since defects in the centrosome-associated protein pericentrin, encoded by PCNT, are currently one of the most commonly identified defects associated with profound MPD in humans (MOPDii) (Bober et al. 2012), we sequenced PCNT and did not find any pathogenic mutations in this family.
Whole exome sequencing (WES) was then performed using peripheral blood from subject LR05-054a1. Mean depth of coverage across the targeted exome was 99.5x. The mean depth of coverage across CENPE was 45.5x. SOLVE identified 4322 genes with two or more heterozygous nonsynonymous variants. However, most of these variants were in genes not likely contributory to the phenotype (DBDB, dbSNP and EVS). There were no novel variants detected in other known PM, MPD or Seckel syndrome genes.
Annotation of biological function and pathway membership in Lynx allowed us to identify compound heterozygous nonsynonymous variants in CENPE (c.2797G>A:p.D933N and c.4063A>G:p.K1355E) as putatively causative of the phenotype (Fig 2a). Although both of these variants were present in dbSNP and the EVS, it was with very low frequency (1/8597 for c.2797G>A:p.D933N and 3/8595 for c.4063A>G:p.K1355E), not ruling out their involvement in an autosomal recessive developmental disorder. Two additional CENPE variants (chr4: 104059542 encoding c.6269C>T:p.T2090M and chr4: 104066461 encoding c.4603T>C:p.F1535L) were identified, however as these were both found in high frequency in dbSNP and the EVS they were not considered to be pathogenic. We therefore proceeded with confirmatory studies for the CENPE c.2797G>A:p.D933N and c.4063A>G:p.K1355E variants. Subsequent Sanger sequencing confirmed that the parents (LR05-054F: father. LR05-054M: mother), were found to each carry one of these mutations (Fig 2a). Mutations status of both siblings was also confirmed by Sanger sequencing (Fig 2a). Both residues (D933 and K1355) are relatively highly conserved in mammals, although not in the mouse M. musculus (Fig. 2b). Patient-derived LCLs were prepared from both siblings for functional analysis. Sanger sequencing of CENPE genomic DNA from ten clinically overlapping individuals did not reveal any mutations (See Online Resource Table 1 for a summary).
Figure 2. Family LR05-054 have compound heterozygous variants in CENPE.
(a). Representative sequence chromatographs identifying mutations in CENPE (NM001813.2) showing the two missense mutations identified in both siblings (c.2797G>A, p.D933N and c.4063A>G, p.K1355E; a-b), as well as parental carrier status: p.D933N is paternally-inherited (LR05-054F; father) and p.K1355E is maternally-inherited (LR05-054M; mother).
(b). Multiple sequence alignments of the regions around amino acids 933 and 1355 were assembled using MUSCLE (version 3.6) via NCBI. Identical sequences are identified in grey, divergent in black and the target residues in red and by the red asterix. Both D933 and K1355 are relatively well conserved in mammals. Although interestingly, neither residue is conserved in mice, M. musculus. In fact, the mouse contains N933, one of the variants identified in our patients. It is notable that CENP-E protein identity between H. sapien and Cenpe of M.musculus is only 64.4% (for R.noregicus 56%). This is in marked contrast to chimpanzee P. troglodytes (98.8%), Rhesus monkey M.mulatta (95.8%) or cattle B.taraus (79%). In fact, the mouse (M. musculus) Cenpe is only 2471 amino acids in length compared to 2701 amino acids of the human CENP-E. Whilst both proteins possess the kinesin motor domain ATPase, only the human protein possesses the SMC (C0G1196) chromosome segregation ATPase and SMC_N(pFam02463) RecF/RecN/SMC N-terminal domain, highlighting the significant sequence and domain disparity between mouse and human herein.
(c). Schematic representation of a mitotic cell, showing the microtubule nucleating centrosome (purple), the microtubule spindles (blue), mitotic chromosomes (grey) and their centromeric kinetochores (green). Known genetic defects in centrosomal and centrosome-associated proteins are shown in purple and spindle pole components in blue. The dashed box indicates a magnified schematic of the kinetochore showing CENP-E capturing microtubules. CENP-E is characterized by a large 230nm flexible coiled-coil domain that can move about to enable microtubule capture by the globular head regions which are composed of microtubule interaction, kinesin motor and ATP binding domains. The CENPE mutations identified here encode residues within this coiled-coil.
LR05-054a1 mitotic LCLs exhibit reduced centromeric accumulation of CENP-E
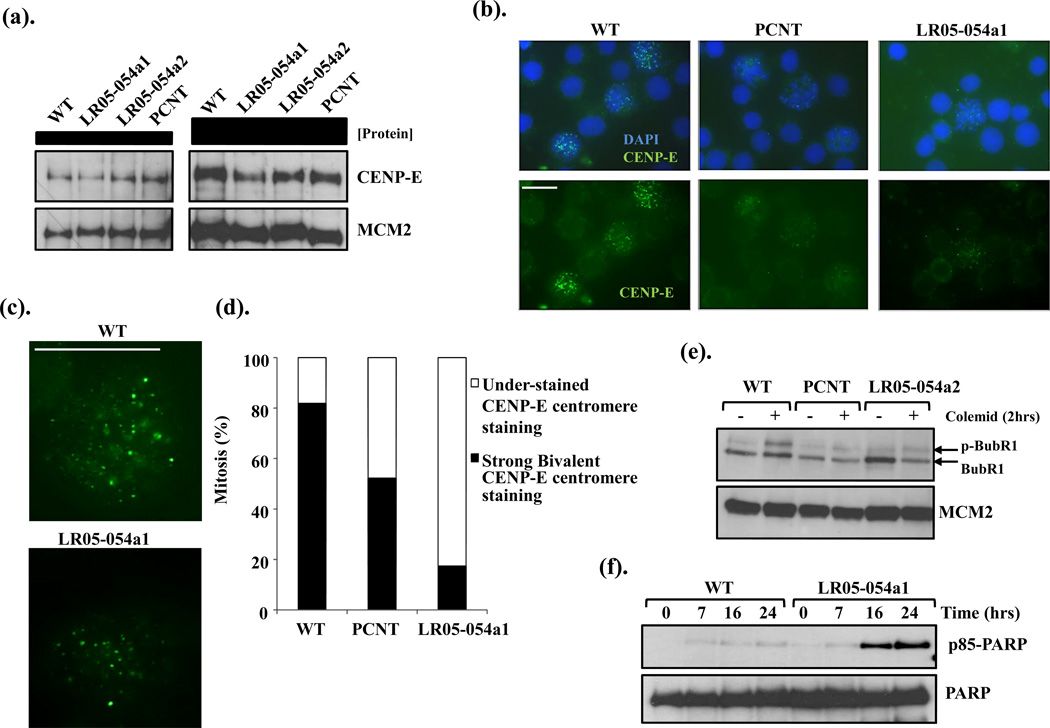
CENP-E is required for spindle microtubule capture to the kinetochore (Fig.2c). Its expression typically peaks during G2-M and is then rapidly degraded in telophase (Brown et al. 1994). CENP-E expression by Western blotting was comparable in whole cell extracts from asynchronous wild-type (WT) LCLs, to those LCLs from both siblings (LR05-054a1 and LR05-054a2) and PCNT-mutated MOPDii, suggesting the variants did not adversely impact upon CENP-E expression or stability (Fig. 3a). As expected, the majority of WT mitotic cells exhibited a strong bivalent CENP-E signal at chromosome centromeres (Fig. 3b–d). This was in stark contrast to LCLs from LR05-054a1 where approximately 80% of mitotic cells exhibited a barely detectable CENP-E signal (Fig. 3b–d). Interestingly, PCNT-defective MOPDii LCLs also exhibited a reduced intensity of bivalent CENP-E signal in about 50% of mitotic cells, highlighting a phenotypic similarity between CENP-E mutant cells and those of MOPDii (Fig. 3d).
Figure 3. CENPE- and PCNT-mutated patient LCLs exhibit reduced kinetochore localization of CENP-E and reduced BubR1 phosphorylation.
(a). CENP-E expression is comparable in whole cell extracts (WCE) from wild-type (LCLs) and LCLs from both siblings (LR05-054a1 and LR05-054a2) and from PCNT-mutated MOPDii LCLs. The left-hand panels show 25µg WCE and the right-hand panel 50µg of WCE.
(b). Indirect immunofluoresence (IF) showing CENP-E focalization onto kinetochores of mitotic cells in wild-type (WT), PCNT-mutated MOPDii (PCNT) and CENPE-mutated patient derived LCLs (LR05-054a1). Cells were treated with Taxol (10µM) and MG132 (10µM) for 1hrs followed by 1.5hrs on ice to stabilize spindle microtubules. Both PCNT and LR05-054a1 mitotic cells exhibit a clear under-representation of defined CENP-E foci. All images were acquired under identical exposure conditions (Scale bar; 10µm).
(c). Close-up image of CENP-E focalization in a mitotic cell from wild-type (WT) and CENPE-mutated patient derived LCLs (LR05-054a1). It has been estimated that only approximately 50 CENP-E dimers occupy each normal human kinetochore (Brown et al. 1994). (Scale bar; 10µm).
(d). The distribution of mitotic cells exhibiting strong bivalent CENP-E foci and those under-stained for CENP-E foci in wild-type (WT), PCNT-mutated MOPDii (PCNT) and CENPE-mutated patient derived LCLs (LR05-054a1). Between 30-50 mitotic cells were scored for each cell line under identical conditions as (A). Strong bivalent CENP-E staining is under-represented in PCNT and LR05-054a1 LCLs compared to WT.
(e). LCLs, from wild-type (WT), PCNT-mutated MOPDii (PCNT) and the female CENPE-mutated patient sib (LR05-054a2) were treated with colcemid (0.2µg/ml) for 2hrs and phosphorylation status of BubR1 was determined by electrophoretic shift following Western blotting. In contrast to WT, colcemid treatment did not induce significant BubR1 retardation (phosphorylation) in PCNT or LR05-054a2 LCLs under these conditions. Interestingly, following a more prolonged treatment with colcemid (>4hrs), BubR1 retardation is evident in both these LCLs suggestive of a leaky/hypomorphic defect. The blot was re-probed for MCM2 expression as a loading control.
(f). LCLs were treated with 2.5µM Taxol and incubated for the times indicated. Apoptosis induction and levels were assessed by blotting for cleaved form of PARP; p85-PARP. The p85-PARP was detected at 16hrs post-treatment in LR05-054a1 LCLs and further increasing at 24hrs. Under these conditions no apoptosis was evident in the WT LCLs.
LR05-054a2 LCLs exhibit impaired BubR1 Phosphorylation
Phosphorylation of the kinetochore associated kinase BubR1 occurs at unattached kinetochores as part of the Spindle Assembly Checkpoint (SAC). This has been proopsed to represent autophosphorylation that is dependent upon CENP-E (Guo et al. 2012; Mao et al. 2005; Tanudji et al. 2004). Although other have argued that phosphorylation must be mediated by other kinases as BubR1 itself has been proposed to be a pseudokinase without intrinsic kinase activity (Suijkerbuijk et al. 2012). Regardless, mitotic-BubR1 phosphorylation can be monitored as an overt electrophoretic retardation following treatment with spindle poisons (Tanudji et al. 2004). As an indirect assay of CENP-E functional proficiency, we treated LCLs with colcemid for 2hrs and examined the electrophoretic mobility of BubR1 by Western blotting. Colcemid treatment of WT LCLs resulted in the expected retardation in BubR1 mobility, in contrast to LR05-054a2 LCLs (Fig. 3e). Interestingly PCNT-MOPDii LCLs, similar to LR05-054a2, failed to induce a shift in BubR1 mobility suggestive of impaired CENPE-dependent BubR1 phosphorylation under these conditions (Fig. 3e). These results are consistent with the reduced CENP-E localization to mitotic centromeres observed in both these LCLs (Fig. 3d).
LR05-054a1 LCLs exhibited elevated sensitivity to Taxol
Prolonged treatment of LR05-054a1 LCLs for up to 24hrs with the spindle poison Taxol was associated with elevated levels of apoptosis compared to WT LCLs, as revealed by elevated p85-PARP expression levels. These data demonstrate that these patient cells are sensitive to treatment with a microtubule spindle stabilizing agent.
LR05-054a1 LCLs exhibit altered spindle dynamics
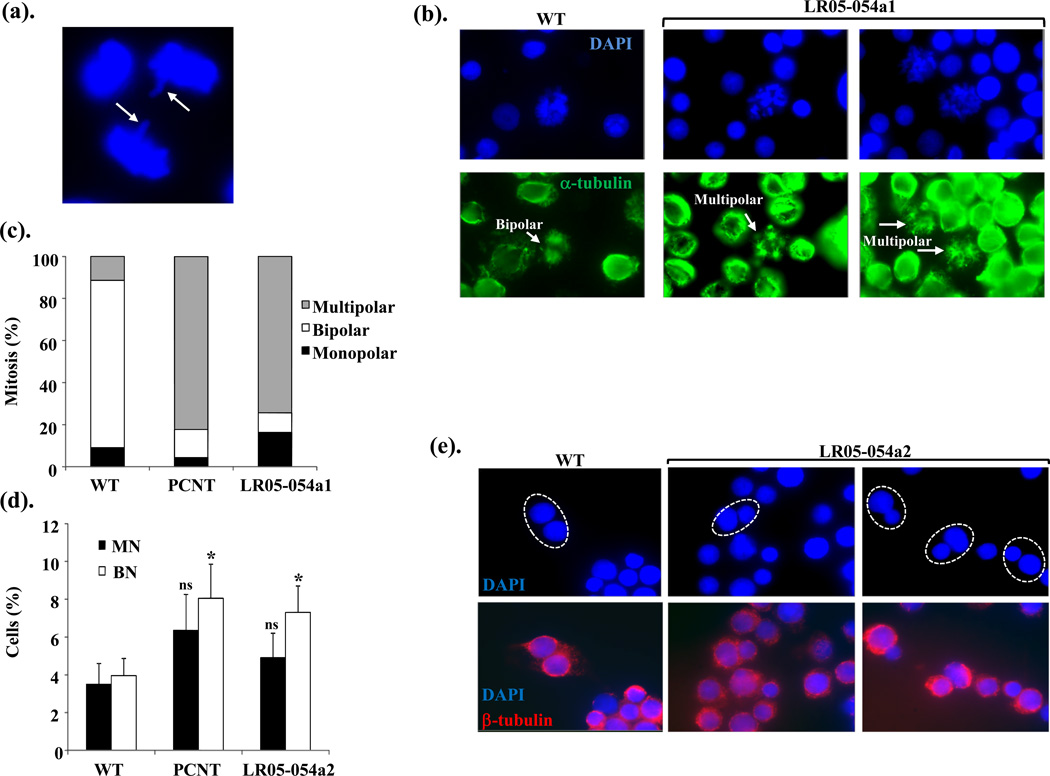
Through its role in chromosome congression and spindle capture at the kinetochore, CENP-E enables chromosomal alignment prior to effective segregation (Fig. 2c). Occasionally, lagging chromosomes were evident in mitotic cells from untreated asynchronous LR05-054a1 cells, but the frequency of this was quite variable (Fig. 4a). Nevertheless, these were not seen in over 100 mitotic cells analyzed from WT LCLs. We next examined the structure of the microtubule spindle network following stabilization with a short pulse of Taxol via indirect immunofluorescence with antibodies against α-tubulin. Most (>80%) of WT mitotic cells exhibited a bipolar spindle (Fig. 4b–c). In direct contrast, approximately 80% of mitotic cells from PCNT-MOPDii (PCNT) and LR05-054a1 exhibited multipolar spindles suggestive of massively disorganized spindle network in these cells (Fig. 4b–c). Abnormal spindle structure has been reported for PCNT-MOPDii previously (Rauch et al. 2008). Interestingly, again we observe similar cellular phenotypes between a centrosomal defect (PCNT) and that of CENPE-mutated LR05-054a1 LCLs.
Figure 4. CENPE-mutated patient LCLs exhibit abnormal spindle microtubule organization and evidence of aberrant segregation and delayed progression through mitosis.
(a). An image of a lagging chromosome in an anaphase cells from an asynchronous culture of LR05-054a1.
(b). The lower panels show IF images of α-tubulin staining microtubule network (green) from wild-type (WT) and LR05-054a1 LCLs following treatment with Taxol (10µM) and MG132 (10µM) for 1hrs then 1.5hrs on ice to stabilize spindle microtubules. The upper panels are the corresponding DAPI-staining nuclei. A normal bipolar spindle is seen in WT mitotic cell whilst multipolar spindles are frequently seen in mitotics from LR05-054a1.
(c). The distribution of spindle abnormalities noted in mitotic cells in wild-type (WT), PCNT-mutated MOPDii (PCNT) and CENPE-mutated patient derived LCLs (LR05-054a1). Between 40–50 mitotic cells were scored for each cell line. Multipolar spindles are over-represented in mitotic cells from PCNT and LR05-054a1 LCLs compared to WT.
(d). The spontaneous levels of micronuclei (MN) are not statistically significantly elevated (ns; not significant Student t-test) in PCNT-mutated MOPDii (PCNT) or CENPE-mutated patient derived LCLs (LR05-054a2) compared to WT. In contrast, binucleates (BN) levels are approximately 2-fold higher in these LCLs compared to WT potentially suggestive of elevated rates of cytokinesis failure (* p<0.05 Student t-test).
(e). DAPI (upper panel) and their corresponding IF-mediated β-tubulin images (lower panel) of binucleates show that the binucleates (circled) from LR05-054a2 LCLs frequently exhibit nuclei of different sizes. The β-tubulin staining facilitates the identification of true binucleate cells. Differing sized nuclei within a binucleate are indicative of unequal segregation during mitosis.
LR05-054a2 LCLs exhibit elevated levels of binucleates with features consistent with altered chromosome segregation
CENP-E deficiency is associated with elevated frequency of polar chromosomes and mis-segregation (Guo et al. 2012; Putkey et al. 2002; Tanudji et al. 2004; Weaver et al. 2003). We did not observe a significantly elevated spontaneous frequency of micronuclei in PCNT or CENP-E LCLs (LR05-054a2), although we did detect an approximately two-fold increase in the level of bi-nucleated cells here, compared to WT (Fig. 4d). Binucleates are the product of impaired and/or delayed cytokinesis which can be a consequence of a failure to coordinately transit mitosis effectively. Strikingly, approximately 50% of binucleates detected in the LR05-054a2 LCLs exhibited nuclei of unequal size, suggestive of unequal segregation during mitosis and impaired cytokinesis (Fig. 4e). We did not observe this phenotype in the binucleates from the PCNT-MOPDii LCLs.
Expression of the D933N and K1355E variants of CENP-E induce mitotic spindle abnormalities and polar chromosomes
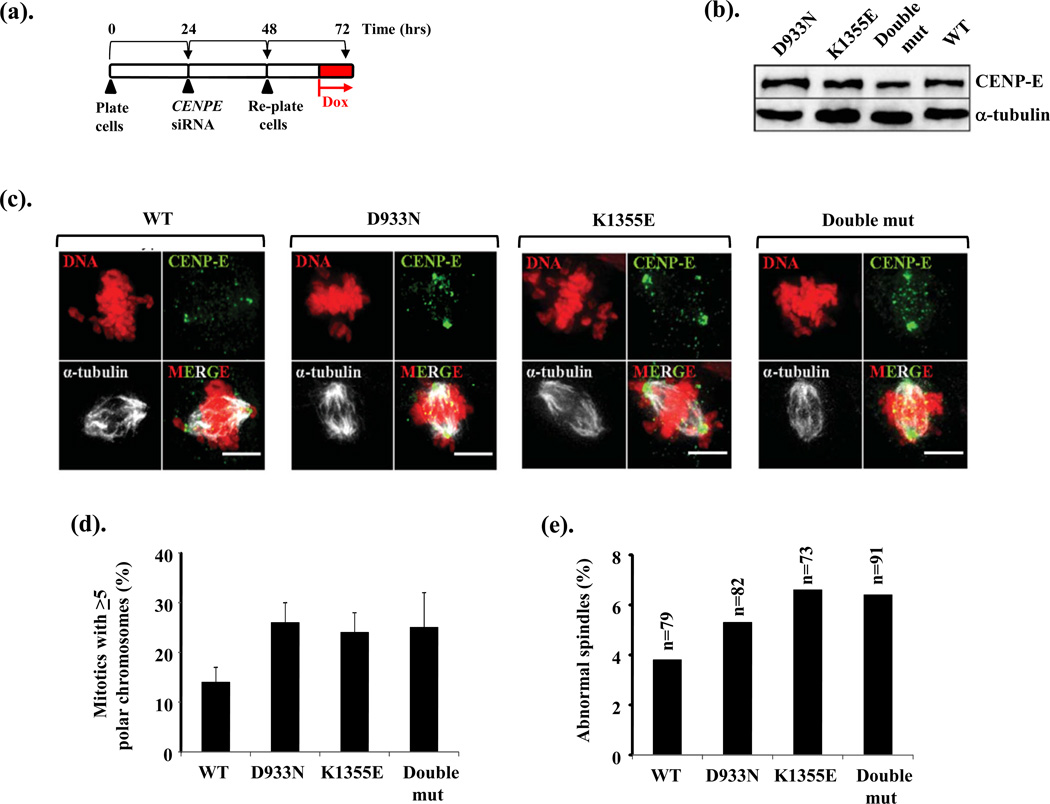
We used Flp-In (Invitrogen) recombination to introduce siRNA-resistant p.D933N and p.K1355E variants of CENPE, individually and in-cis (Double mut; double mutant), into DLD-1 cells under doxycycline (Dox) conditional regulation, as described in detail by Kim et al (Kim et al. 2010). Following siRNA-mediated silencing of the endogenous CENPE, Dox treatment (8hrs) resulted in strong expression of these variants and cells were fixed for immunofluorescence imaging. This allowed us to interrogate the impacts upon mitotic spindle structure and chromosome segregation when each variant was expressed individually and when engineered together into CENP-E. The induction scheme used is shown in Fig 5a and comparable CENP-E expression levels shown in Fig5b. In this independent model cell system, we found that p.D933N and p.K1355E individually or when in-cis, caused an elevated frequency of mitotic cells with polar chromosomes (Fig 5c–d), along with elevated levels of mitotic cells with abnormal spindles (monopolar and multipolar mitotic spindles; Fig 5c and 5e). These findings independently show that both these patient-derived CENP-E variants can adversely impact upon normal mitotic spindle formation and chromosome segregation, phenotypes consistent with our observations derived from patient LCLs.
Figure 5. Expression of the D933N and K1355E variants of CENP-E results in mitotic spindle abnormalities and polar chromosomes.
(a). DLD-1 cells engineered by Flp-In recombination for the conditional (doxycycline-induced) expression of siRNA-resistant CENPE and CENPE patient variants, were plated and 24hrs later subjected to CENPE siRNA targeting endogenous CENP-E only. After 40hrs approx. the cells were treated with doxycycline (Dox; 1µg/ml) for 8hrs to induce expression of the siRNA-resistant CENPE variants. Cells were then fixed and processed for immunofluorescence.
(b). Expression levels of the various CENP-E variants including a construct with both variants engineered in-cis (Double mut; double mutant) were compared to that of wild-type (WT) CENP-E following 8hrs in doxycycline.
(c). Immunofluoresence images following 8hrs treatment with doxycycline for wild-type (WT), D933N alone, K1355E alone and the double mutant (Double mut) with both D933N and K1355E in-cis co-stained with anti-CENP-E and anti-α-tubulin. Polar chromosomes not aligned properly stain strongly for CENP-E under these conditions as seen in D933N alone, K1355E alone and the double mutant. (Scale bar; 5µm).
(d). The frequency of mitotic cells with >5 unaligned polar chromosomes is elevated following expression of D933N alone, K1355E alone or following expression of both variants in-cis (Double mut). Between 70–90 mitotic cells were evaluated in total for each construct.
(e). Expression of D933N alone, K1355E alone or both variants in-cis (Double mut) resulted in an increased frequency of mitotic cells with abnormal spindle structure (monopolar plus multipolar spindles) compared to expression of wild-type (WT) CENP-E in this system (n= the number of mitotic cells scored).
LR05-054a2 LCLs exhibit impaired mitotic progression
To examine the proficiency of mitotic progression in patient LCLs in more detail we performed a block-release-block experiment using colemid-coupled with cytochalasin B treatments. Specifically, LCLs were briefly treated with colcemid (4hrs) to synchronize a sub-population of cells in mitosis. The colcemid was removed by washing and cells then released into cytochalasin B, a cytokinesis inhibitor, for 24hrs. The frequency of cytochalasin B-induced binucleate cells was compared to untreated cells for each of the WT, PCNT-MOPDii and LR05-054a2 LCLs. The ratio of binucleate cells; that is the number of binucleates following colcemid block and release into cytochalasin B, over the level of binucleates in untreated (Unt) LCLs, reflects the progression through mitosis following release from the colcemid block. We observed a decreased ratio of binucleate cells following this block-release-block procedure in PCNT and CENPE (LR05-054a2)-mutated LCLs compared to WT consistent with a delay in progression through mitosis upon release from colcemid (Fig 6a).
Figure 6. CENPE-mutated patient LCLs exhibit delayed mitotic progression which is also observed upon conditional expression of CENP-E carrying D933N and K1355E in-cis.
(a). The ratio of binucleates following colcemid block and release into cytochalasin B, compared to untreated (Unt) cells is lower in PCNT-mutated MOPDii (PCNT) or CENPE-mutated patient derived LCLs (LR05-054a2), compared to WT, consistent with a slower progression through mitosis under these conditions in these patient cells.
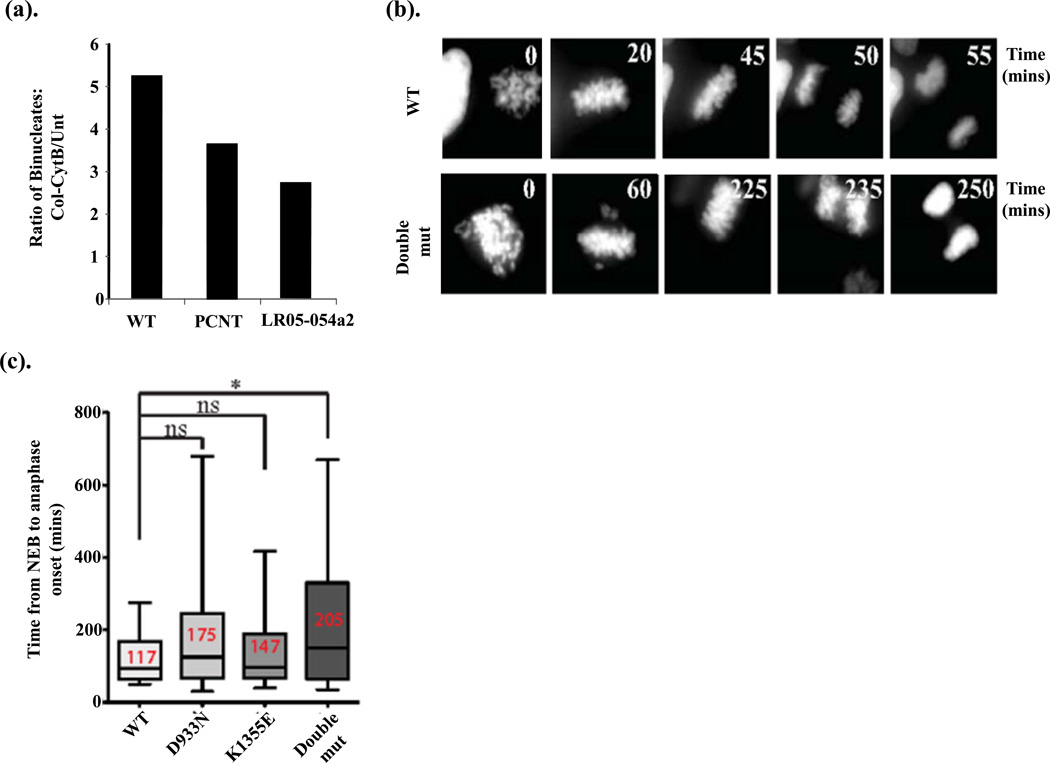
(b). DLD-1 cells engineered by Flp-In recombination for the conditional (doxycycline-induced) expression of siRNA-resistant CENPE and CENPE patient variants, were plated and 24hrs later subjected to CENPE siRNA targeting endogenous CENP-E only as described in Fig 5a. These cells also contain a RFP-tagged histone H2B to enable real-time analysis of chromosome dynamics during mitosis by live cell imaging. Approximately 40hrs post-siRNA the cells were treated with doxycycline (Dox; 1mg/ml) to induce expression of the siRNA-resistant CENPE variants and cells analyzed by live cell imaging. Still images from the live cell movies following mitosis when wild-type (WT) CENP-E was expressed compared to the double mutant (Double mut) expressing D933N and K1355E in-cis. The time (in minutes) is indicated in the various panels showing onset of the transition from prophase-metaphase-anaphase-telophase under each conditional expression condition.
(c). The time from the onset of nuclear envelope breakdown (NEB) to anaphase was calculated for each CENP-E variant when expressed individually or in-cis (Double mut; double mutant) and represented in box-whisker plots. The average time of onset of NEB to anaphase is shown in red for each variant. Expression of D933N alone, or K1355E alone, did not result in a statistically significant delay compared to expression of wild-type (WT) CENPE (ns; not significant by Student t-test), although the average transition for each variant was slightly longer compared to WT. *Expression of both variants in-cis did result in a statistically significant delay from NEB to anaphase onset compared to WT (p <0.05 Student t-test).
Expression of the D933N and K1355E variants of CENP-E are associated with delayed mitotic progression
Using our Flp-In (Invitrogen) CENPE system in DLD-1 cells engineered to stably co-express RFP-histone H2B to enable chromosome visualization, we employed live cell imaging to determine the kinetics of mitotic progression (Fig 6b–c). Whilst we found that expression of p.D933N alone or expression of p.K1355E alone, on average, was associated with a slower progression through mitosis compared to WT CENP-E, these delays were not statistically significant (Fig 6c). The mean mitotic transit time for WT CENP-E was 117mins compared to 175mins for D933N and 147mins for K1355E. But, when both variants were engineered in-cis into CENP-E, this resulted in a statistically significant delay in mitosis (205mins) compared to WT CENP-E (117mins) (Fig 6b–c). These findings show that expression of a CENPE cDNA engineered to carry both patient-derived variants (p.D933N and p.K1355E) in-cis, resulted in a significantly delayed mitosis. These findings are consistent with the block-release-block findings using LR05-054a2 LCLs (Fig 6a).
LR05-054a1 LCLs exhibit a functional ATR-dependent DNA damage response
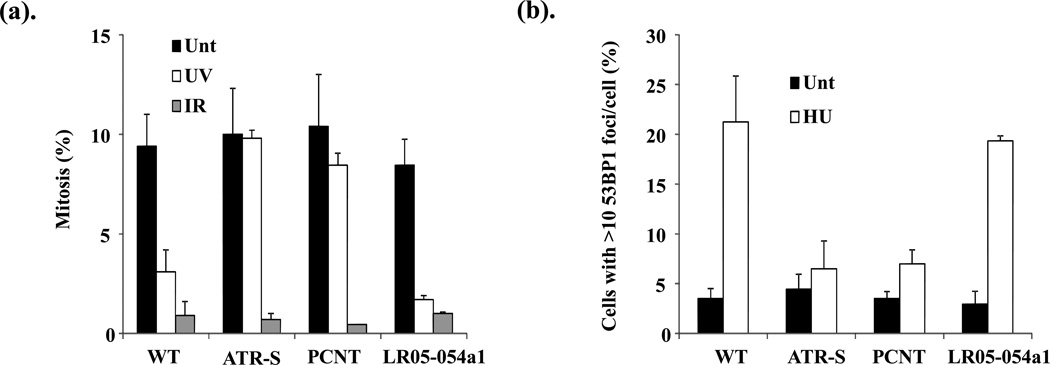
Seckel syndrome and MOPDii patient-derived LCLs exhibit defects in the ATR-dependent DNA damage response (DDR), including impaired UV-induced G2-M cell cycle checkpoint activation and compromised ATR-DDR dependent signaling (Alderton et al. 2004; Griffith et al. 2008; O’Driscoll et al. 2003; Ogi et al. 2012; Qvist et al. 2011). To determine whether LCLs from patient LR05-054a1 exhibited an impaired ATR-dependent DDR, we treated the cells with different forms of radiation; UV to activate the ATR-DDR and IR to activate the ATM-DDR, and then assessed mitotic index (MI). G2-M cell cycle checkpoint activation is indicated by a decrease in MI due to cell cycle arrest in G2 phase following radiation. Similar to WT LCLs, LR05-054a1 exhibited a decrease in MI following UV indicative of a functional ATR-dependent checkpoint (Fig. 7a). This is in contrast to LCLs derived from a patient mutated in ATR (ATR-S; ATR-Seckel syndrome) or in PCNT-mutated MOPDii LCLs (PCNT). All patient-derived LCLs demonstrated a drop in MI following IR indicative of a functional ATM-dependent G2-M checkpoint (Fig. 7a).
Figure 7. CENPE-mutated patient LCLs exhibit a functional ATR-dependent DNA damage response in contrast to PCNT-MOPDii and ATR-Seckel syndrome patient cells.
(a). LCLs from wild-type (WT), ATR-mutated Seckel syndrome (ATR-S), PCNT-mutated MOPDii (PCNT) and the CENPE-mutated male sib (LR05-054a1) were either un-irradiated (Unt), irradiated with ultraviolet light (UV) or ionizing radiation (IR) and their mitotic index (MI: % mitosis) enumerated at 4hrs. UV-irradiation specifically activates the ATR-dependent G2-M cell cycle checkpoint whilst IR activates the ATM-dependent checkpoint. Both WT and LR05-054a1 LCLs demonstrate a robust decrease in MI under both conditions. This is in contrast to LCLs from ATR or PCNT-mutated patients who exhibit a specific defect in the ATR-mediated G2-M cell cycle checkpoint arrest, as demonstrated previously .
(b). Hydroxyurea (HU)-induced 53BP1 foci formation is an ATR and CHK1-dependent process. Cells were either untreated (Unt) or treated with 5mM HU and 53BP1 foci positive cells enumerated after 2hrs via IF. Both WT and LR05-054a1 LCLs demonstrate a similarly increased frequency of cells with 53BP1 foci under these conditions, in contrast to LCLs from ATR or PCNT-mutated patients.
Following stalling of DNA replication forks with hydroxyurea (HU), 53BP1 forms microscopically detectable foci in an ATR and CHK1 dependent manner (Sengupta et al. 2004). Seckel syndrome and PCNT-mutated MOPDii patient-derived LCLs exhibit impaired HU-induced 53BP1 foci formation indicative of an impaired ATR-DDR (Griffith et al. 2008; Ogi et al. 2012). Consistent with the normal UV-induced G2-M checkpoint activation in LR05-054a1, these cells were proficient in HU-induced 53BP1 foci formation, in contrast to ATR-S and PCNT-defective MOPDii LCLs (Fig. 7b). Collectively, these results suggest that LR05-054a1 does not exhibit an impaired ATR-DDR.
Discussion
The developing embryonic neuroepithelium is exquisitely sensitive to perturbations in cell cycle kinetics as this system is characterized by a temporally restricted rapid expansion of stem cells (Rakic 1995). It is also sensitive to perturbations in spindle orientation, as alterations in the cleavage plane can disrupt symmetric-asymmetric division, migration and differentiation (Fish et al. 2006). Furthermore, defects in microtubule dynamics and microtubule interacting proteins are associated with a range of neuronal migration disorders (e.g. lissencephaly, pachygyria, polymicrogyria) coupled with microcephaly (Jaglin and Chelly 2009; Poirier et al. 2013).
Collectively, disruption of centrosome-spindle processes can result in an overlapping set of human pathologies ranging from PM, PM with short stature to overt MPD (Al-Dosari et al. 2010; Bond J et al. 2005; Bond J 2002; Guernsey et al. 2010; Kalay et al. 2011; Sir et al. 2011). Whilst pathogenic defects have been identified in centrosome, microtubule spindle and spindle-associated components, defects in core kinetochore components underlying MPD with cortical malformation have not yet been reported. Recently, a rare homozygous mutation in CASC5 was reported in three consanguineous families from the same isolated geographical region; one of whom was the original Moroccan family used to define the MCPH4 locus (Genin et al. 2012; Jamieson et al. 1999). CASC5 is a component of the KMN complex (KNL1-Mis12 complex-Ndc80 complex), a kinetochore localized network implicated in several functions including microtubule stabilization and SAC silencing (Bolanos-Garcia et al. 2009; Kiyomitsu et al. 2011; Kiyomitsu et al. 2007). In contrast to the CENP-E patients described here, the CASC5-mutated individuals were characterized by PM (age-related OFC ranging from −4 to −7 SD), normal stature, normal motor milestones without evidence of epilepsy or neurological deficits and an otherwise unremarkable medical history with the oldest patients in their thirties (Genin et al. 2012; Jamieson et al. 1999). Interestingly, detailed analysis did not yield any spontaneous mitotic and/or cell cycle defect or evidence of genomic instability in cells from these patients, again in contrast to the CENP-E defect described here (Genin et al. 2012). The authors postulated that this may perhaps be due to the restriction of such anticipated cellular phenotypes to the neuronal system (Genin et al. 2012).
Different defects even within the same centrosomal proteins have previously been found to exhibit a spectrum of growth and neurodevelopmental phenotypes, the basis of which is unclear (Al-Dosari et al. 2010; Bond J et al. 2005; Guernsey et al. 2010; Kalay et al. 2011). An important implication of our findings is that hypomorphic defects in other core kinetochore and kinetochore-associated factors (aside from CENP-E), may also underlie MPD in humans. This should be considered when designing targeted sequencing strategies and/or analyzing WES datasets. Our work also illustrates a useful platform to model the functional impacts of candidate gene variants (Flip-In recombination of siRNA-resistant variant carrying cDNA). This is particularly relevant in our situation as one of the patient variant changes reflected the amino acid at equivalent position in mouse Cenpe. No doubt this variant would likely have been excluded based on standard filtering approaches if used in isolation. Modeling variant pathogenicity in an independent cell system when possible represents a powerful approach when used in combination with patient cell-based functional analysis.
Another important aspect of our findings is that they provide a wider clinical picture for how, a hypomorphic defect in a core kinetochore component presents. Nevertheless, with regard to the congenital heart defect (CHD) observed here specifically in LR05-054a1, we cannot be absolutely sure that impaired CENP-E is a causative and/or a contributing factor. Interestingly, CHDs have been reported in MVA patients, although specifically in those with CEP57 mutations (CEP57 encodes a centrosome-associated protein), rather than BUB1B. But, the underlying pathomechanism of CHDs in this context has not been described (Snape et al. 2011). As part of our exome analysis of LR05-054a1, we examined 78 genes with known associations with CHDs for potential pathogenic variants but did not find any strong candidates (Online Resource Table 4 & 5).
The functional analysis of patient LCLs outlined here also demonstrates that defects in spatially distinct proteins that play different roles in mitosis can manifest with similar cellular phenotypes (Online Resource Tables 3). We find that impaired PCNT-function mimics impaired CENP-E function with respect to CENP-E focalization, CENP-E-dependent BubR1 phosphorylation, mitotic spindle organization and mitotic progression. The basis of this is unclear but may be related to the abnormal spindles characteristic of PCNT-MOPDii cells (Rauch et al. 2008). Nevertheless, we also did observe some divergence in cellular phenotypes between CENP-E and PCNT (Fig. 4e). Additionally, we did not detect an impaired ATR-dependent DDR in CENP-E LCLs, in contrast to PCNT-MOPDii LCLs (Fig 7a–b). This is likely due to the fact that the centrosome appears to be an important site of localized modification and/or interaction for many DDR factors, including CHK1, a direct substrate of ATR (Kramer 2004; Tibelius et al. 2009; Zhang et al. 2007).
At 2701 amino acids in length human CENP-E is one of the largest fibrous extensions emanating from the kinetochore. Structurally, CENP-E is a dimeric kinesin with a large 230nm flexible α-helical coiled-coil region (D336-A2471) separating a pair of the head domains that constitute the kinesin motor, ATP binding and microtubule interacting regions (M1-K327), from the C-terminal kinetochore binding domain (Q2472-Q2663) (Fig. 2c)(Kim et al. 2008). Interestingly, defects in other kinesin-family proteins, with roles in microtubule dynamics and trafficking have been identified in patients with overt cortical structural abnormalities including MIC (Poirier et al. 2013). Both CENPE mutations identified here localize to the α-helical coiled-coil region of CENP-E. A noted feature of the structure of CENP-E is the inherent flexibility of the coiled-coil region which is thought to be essential for searching out microtubules (Fig. 2c)(Kim et al. 2008). It is difficult to accurately postulate how together both of the amino acid changes identified here (D933N and K1355E) could impact on CENP-E function. But, the K1355E does represent a change in both charge and size of the side chain (Online Resource Table 2). Our findings provide the basis for further modeling and analysis of these changes at the structural level, as conditional expression of either variant in a model cell system does appear to adversely impact upon aspects of normal mitotic progression (Fig 5&Fig 6).
In summary, we identify rare variants in CENPE in two siblings with a profound MPD, structural brain malformations and isolated abnormalities. We catalogue various abnormalities in mitotic spindle organization, mitotic progression and segregation in LCLs from these patients, as well as using a model cell system following conditional expression of each CENPE variant, alone and together in-cis. In demonstrating significant phenotypic overlap between CENPE-mutated LCLs and those from a PCNT-mutated MOPDii patient, our findings are consistent with a model wherein aberrant mitotic spindle structure, chromosome mis-segregation and delayed mitotic progression represent a significant underlying pathomechanism for MPD in humans. Our findings demonstrate the first example of a core kinetochore defect in contributing to this pathomechanism. This has implications for our understanding the molecular basis of normal brain and body development and potential for novel genetic defect identification.
Supplementary Material
Acknowledgements
We wish to thank the family and the referring physicians for their contribution to this study. The O’Driscoll laboratory is funded by Cancer Research UK, Leukaemia Lymphoma Research (UK) and the Medical Research Council (UK). The Dobyns laboratory is funded by the US National Institutes of Health under NINDS grant NS058721. The Paciorkowski laboratory is funded by the US National Institutes of Health under NINDS grant NS078054. Thanks also to the Center for Integrated Research Computing at the University of Rochester Medical Center for the computational resources for data analysis. The Cleveland laboratory is supported by the Ludwig Cancer Institute and NIH (R01-GM29513). B.V is a Human Frontiers Science Program postdoctoral fellow.
Footnotes
Conflict of Interest Statements
The authors declare they have no conflict of interest.
References
- Abrieu A, Kahana JA, Wood KW, Cleveland DW. CENP-E as an Essential Component of the Mitotic Checkpoint In Vitro. Cell. 2000;102:817–826. doi: 10.1016/s0092-8674(00)00070-2. doi: http://dx.doi.org/10.1016/S0092-8674(00)00070-2. [DOI] [PubMed] [Google Scholar]
- Al-Dosari MS, Shaheen R, Colak D, Alkuraya FS. Novel CENPJ mutation causes Seckel syndrome. Journal of Medical Genetics. 2010;47:411–414. doi: 10.1136/jmg.2009.076646. [DOI] [PubMed] [Google Scholar]
- Alderton GK, Galbiati L, Griffith E, Surinya KH, Neitzel H, Jackson AP, Jeggo PA, O’Driscoll M. Regulation of mitotic entry by microcephalin and its overlap with ATR signalling. Nature Cell Biology. 2006;8:725–733. doi: 10.1038/ncb1431. [DOI] [PubMed] [Google Scholar]
- Alderton GK, Joenje H, Varon R, Borglum AD, Jeggo PA, O’Driscoll M. Seckel syndrome exhibits cellular features demonstrating defects in the ATR signalling pathway. Human Molecular Genetics. 2004;13:3127–3138. doi: 10.1093/hmg/ddh335. [DOI] [PubMed] [Google Scholar]
- Bicknell LS, Bongers EMHF, Leitch A, Brown S, Schoots J, Harley ME, Aftimos S, Al-Aama JY, Bober M, Brown PAJ, van Bokhoven H, Dean J, Edrees AY, Feingold M, Fryer A, Hoefsloot LH, Kau N, Knoers NVAM, MacKenzie J, Opitz JM, Sarda P, Ross A, Temple IK, Toutain A, Wise CA, Wright M, Jackson AP. Mutations in the pre-replication complex cause Meier-Gorlin syndrome. Nat Genet. 2011a;43:356–359. doi: 10.1038/ng.775. doi: http://www.nature.com/ng/journal/v43/n4/abs/ng.775.html#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell LS, Walker S, Klingseisen A, Stiff T, Leitch A, Kerzendorfer C, Martin C-A, Yeyati P, Al Sanna N, Bober M, Johnson D, Wise C, Jackson AP, O’Driscoll M, Jeggo PA. Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier-Gorlin syndrome. Nat Genet. 2011b;43:350–355. doi: 10.1038/ng.776. doi: http://www.nature.com/ng/journal/v43/n4/abs/ng.776.html#supplementary-information. [DOI] [PubMed] [Google Scholar]
- Bober MB, Niiler T, Duker AL, Murray JE, Ketterer T, Harley ME, Alvi S, Flora C, Rustad C, Bongers EMHF, Bicknell LS, Wise C, Jackson AP. Growth in individuals with Majewski osteodysplastic primordial dwarfism type II caused by pericentrin mutations. American Journal of Medical Genetics Part A. 2012;158A:2719–2725. doi: 10.1002/ajmg.a.35447. [DOI] [PubMed] [Google Scholar]
- Bolanos-Garcia VM, Kiyomitsu T, D’Arcy S, Chirgadze DY, Grossmann JG, Matak-Vinkovic D, Venkitaraman AR, Yanagida M, Robinson CV, Blundell TL. The Crystal Structure of the N-Terminal Region of BUB1 Provides Insight into the Mechanism of BUB1 Recruitment to Kinetochores. Structure. 2009;17:105–116. doi: 10.1016/j.str.2008.10.015. doi: http://dx.doi.org/10.1016/j.str.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J, Roberts E, Springell K, Lizarraga SB, Scott S, Higgins J, Hampshire DJ, Morrison EE, Leal GF, Silva EO, Costa SM, Baralle D, Raponi M, Karbani G, Rashid Y, Jafri H, Bennett C, Corry P, Walsh CACGW. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet. 2005;37:353–355. doi: 10.1038/ng1539. [DOI] [PubMed] [Google Scholar]
- Bond JRE, Mochida GH, Hampshire DJ, Scott S, Askham JM, Springell K, Mahadevan M, Crow YJ, Markham AF, Walsh CA, Woods CG. ASPM is a major determinant of cerebral cortical size. Nat Genet. 2002;32:316–320. doi: 10.1038/ng995. [DOI] [PubMed] [Google Scholar]
- Brown KD, Coulson RM, Yen TJ, Cleveland DW. Cyclin-like accumulation and loss of the putative kinetochore motor CENP-E results from coupling continuous synthesis with specific degradation at the end of mitosis. The Journal of Cell Biology. 1994;125:1303–1312. doi: 10.1083/jcb.125.6.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche M-C, Sanal O, Plebani A, Stephan J-L, Hufnagel M, le Deist F, Fischer A, Durandy A, de Villartay J-P, Revy P. Cernunnos, a Novel Nonhomologous End-Joining Factor, Is Mutated in Human Immunodeficiency with Microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF. Centromeres and Kinetochores: From Epigenetics to Mitotic Checkpoint Signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. doi: http://dx.doi.org/10.1016/S0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- Fish JL, Kosodo Y, Enard W, Pääbo S, Huttner WB. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proceedings of the National Academy of Sciences. 2006;103:10438–10443. doi: 10.1073/pnas.0604066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genin A, Desir J, Lambert N, Biervliet M, Van Der Aa N, Pierquin G, Killian A, Tosi M, Urbina M, Lefort A, Libert F, Pirson I, Abramowicz M. Kinetochore KMN network gene CASC5 mutated in primary microcephaly. Human Molecular Genetics. 2012;21:5306–5317. doi: 10.1093/hmg/dds386. [DOI] [PubMed] [Google Scholar]
- Gorlin R, Cervenka J, Moller K, Horrobin M, Witkop CJ. Malformation syndromes. A selected miscellany. Birth Defects Orig Artic Ser. 1975;11:39–50. [PubMed] [Google Scholar]
- Griffith E, Walker S, Martin C-A, Vagnarelli P, Stiff T, Vernay B, Sanna NA, Saggar A, Hamel B, Earnshaw WC, Jeggo PA, Jackson AP, O’Driscoll M. Mutations in pericentrin cause Seckel syndrome with defective ATR-dependent DNA damage signaling. Nat Genet. 2008;40:232–236. doi: 10.1038/ng.2007.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guernsey DL, Jiang H, Hussin J, Arnold M, Bouyakdan K, Perry S, Babineau-Sturk T, Beis J, Dumas N, Evans SC, Ferguson M, Matsuoka M, Macgillivray C, Nightingale M, Patry L, Rideout AL, Thomas A, Orr A, Hoffmann I, Michaud JL, Awadalla P, Meek DC, Ludman M, Samuels ME. Mutations in Centrosomal Protein CEP152 in Primary Microcephaly Families Linked to MCPH4. The American Journal of Human Genetics. 2010;87:40–51. doi: 10.1016/j.ajhg.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guernsey DL, Matsuoka M, Jiang H, Evans S, Macgillivray C, Nightingale M, Perry S, Ferguson M, LeBlanc M, Paquette J, Patry L, Rideout AL, Thomas A, Orr A, McMaster CR, Michaud JL, Deal C, Langlois S, Superneau DW, Parkash S, Ludman M, Skidmore DL, Samuels ME. Mutations in origin recognition complex gene ORC4 cause Meier-Gorlin syndrome. Nat Genet. 2011;43:360–364. doi: 10.1038/ng.777. doi: http://www.nature.com/ng/journal/v43/n4/abs/ng.777.html#supplementary-information. [DOI] [PubMed] [Google Scholar]
- Guo Y, Kim C, Ahmad S, Zhang J, Mao Y. CENP-E-dependent BubR1 autophosphorylation enhances chromosome alignment and the mitotic checkpoint. The Journal of Cell Biology. 2012;198:205–217. doi: 10.1083/jcb.201202152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JG, Flora C, Scott CI, Jr, Pauli RM, Tanaka KI. Majewski osteodysplastic primordial dwarfism type II (MOPD II): Natural history and clinical findings. American Journal of Medical Genetics Part A. 2004;130A:55–72. doi: 10.1002/ajmg.a.30203. [DOI] [PubMed] [Google Scholar]
- Hanks S, Coleman K, Reid S, Plaja A, Firth H, Fitzpatrick D, Kidd A, Mehes K, Nash R, Robin N, Shannon N, Tolmie J, Swansbury J, Irrthum A, Douglas J, Rahman N. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet. 2004;36:1159–1161. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- Hori T, Fukagawa T. Establishment of the vertebrate kinetochore. Chromosome Res. 2012;20:547–561. doi: 10.1007/s10577-012-9289-9. [DOI] [PubMed] [Google Scholar]
- Ijspeert H, Warris A, van der Flier M, Reisli I, Keles S, Chishimba S, van Dongen JJM, van Gent DC, van der Burg M. Clinical Spectrum of LIG4 Deficiency Is Broadened with Severe Dysmaturity, Primordial Dwarfism, and Neurological Abnormalities. Human Mutation. 2013;34:1611–1614. doi: 10.1002/humu.22436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglin XH, Chelly J. Tubulin-related cortical dysgeneses: microtubule dysfunction underlying neuronal migration defects. Trends Genet. 2009;25:555–566. doi: 10.1016/j.tig.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Jamieson CR, Govaerts C, Abramowicz MJ. Primary Autosomal Recessive Microcephaly: Homozygosity Mapping of MCPH4 to Chromosome 15. The American Journal of Human Genetics. 1999;65:1465–1469. doi: 10.1086/302640. doi: http://dx.doi.org/10.1086/302640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalay E, Yigit G, Aslan Y, Brown KE, Pohl E, Bicknell LS, Kayserili H, Li Y, Tuysuz B, Nurnberg G, Kiess W, Koegl M, Baessmann I, Buruk K, Toraman B, Kayipmaz S, Kul S, Ikbal M, Turner DJ, Taylor MS, Aerts J, Scott C, Milstein K, Dollfus H, Wieczorek D, Brunner HG, Hurles M, Jackson AP, Rauch A, Nurnberg P, Karaguzel A, Wollnik B. CEP152 is a genome maintenance protein disrupted in Seckel syndrome. Nat Genet. 2011;43:23–26. doi: 10.1038/ng.725. doi: http://www.nature.com/ng/journal/v43/n1/abs/ng.725.html#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Heuser JE, Waterman CM, Cleveland DW. CENP-E combines a slow, processive motor and a flexible coiled coil to produce an essential motile kinetochore tether. The Journal of Cell Biology. 2008;181:411–419. doi: 10.1083/jcb.200802189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Holland AJ, Lan W, Cleveland DW. Aurora Kinases and Protein Phosphatase 1 Mediate Chromosome Congression through Regulation of CENP-E. Cell. 2010;142:444–455. doi: 10.1016/j.cell.2010.06.039. doi: http://dx.doi.org/10.1016/j.cell.2010.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T, Murakami H, Yanagida M. Protein Interaction Domain Mapping of Human Kinetochore Protein Blinkin Reveals a Consensus Motif for Binding of Spindle Assembly Checkpoint Proteins Bub1 and BubR1. Molecular and Cellular Biology. 2011;31:998–1011. doi: 10.1128/MCB.00815-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T, Obuse C, Yanagida M. Human Blinkin/AF15q14 Is Required for Chromosome Alignment and the Mitotic Checkpoint through Direct Interaction with Bub1 and BubR1. Developmental Cell. 2007;13:663–676. doi: 10.1016/j.devcel.2007.09.005. doi: http://dx.doi.org/10.1016/j.devcel.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Kramer AM, Lukas N, Syljuasen C, Wilkinson RG, Nigg CJ, Bartek EA, Lukas JJ. Centrosome-associated Chk1 prevents premature activation of cyclin-B-Cdk1 kinase. Nat Cell Biol. 2004;6:884–891. doi: 10.1038/ncb1165. [DOI] [PubMed] [Google Scholar]
- Mahmood S, Ahmad W, Hassan M. Autosomal recessive primary microcephaly (MCPH): clinical manifestations, genetic heterogeneity and mutation continuum. Orphanet Journal of Rare Diseases. 2011;6:39. doi: 10.1186/1750-1172-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski F, Goecke T, Opitz JM. Studies of microcephalic primordial dwarfism I: Approach to a delineation of the seckel syndrome. American Journal of Medical Genetics. 1982a;12:7–21. doi: 10.1002/ajmg.1320120103. [DOI] [PubMed] [Google Scholar]
- Majewski F, Ranke M, Schinzel A, Opitz JM. Studies of microcephalic primordial dwarfism II: The osteodysplastic type II of primordial dwarfism. American Journal of Medical Genetics. 1982b;12:23–35. doi: 10.1002/ajmg.1320120104. [DOI] [PubMed] [Google Scholar]
- Mao Y, Desai A, Cleveland DW. Microtubule capture by CENP-E silences BubR1-dependent mitotic checkpoint signaling. The Journal of Cell Biology. 2005;170:873–880. doi: 10.1083/jcb.200505040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JE, Bicknell LS, Yigit G, Duker AL, van Kogelenberg M, Haghayegh S, Wieczorek D, Kayserili H, Albert MH, Wise CA, Brandon J, Kleefstra T, Warris A, van der Flier M, Bamforth JS, Doonanco K, Adès L, Ma A, Field M, Johnson D, Shackley F, Firth H, Woods CG, Nürnberg P, Gatti RA, Hurles M, Bober MB, Wollnik B, Jackson AP. Extreme Growth Failure is a Common Presentation of Ligase IV Deficiency. Human Mutation. 2014;35:76–85. doi: 10.1002/humu.22461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A. Spindle assembly checkpoint: the third decade. Philosophical Transactions of the Royal Society B: Biological Sciences. 2011;366:3595–3604. doi: 10.1098/rstb.2011.0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- O’Driscoll M, Cerosaletti KM, Girard P-M, Dai Y, Stumm M, Kysela B, Hirsch B, Gennery A, Palmer SE, Seidel J, Gatti RA, Varon R, Oettinger MA, Sperling K, Jeggo PA, Concannon P. DNA Ligase IV mutations identified in patients exhibiting development delay and immunodeficiency. Molecular Cell. 2001;8:1175–1185. doi: 10.1016/s1097-2765(01)00408-7. [DOI] [PubMed] [Google Scholar]
- O’Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nature Genetics. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- Ogi T, Walker S, Stiff T, Hobson E, Limsirichaikul S, Prescott K, Suri M, Byrd PJ, Matsuse M, Mitsutake N, Nakazawa Y, Vasudevan P, Barrow M, Stewart GS, Taylor AMR, O’Driscoll M, Jeggo PA. Identification of the first ATRIP-deficient patient and novel mutations in ATR define a clinical spectrum for ATR-ATRIP Seckel syndrome. PLoS Genet. 2012;8:e1002945. doi: 10.1371/journal.pgen.1002945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier K, Lebrun N, Broix L, Tian G, Saillour Y, Boscheron C, Parrini E, Valence S, Pierre BS, Oger M, Lacombe D, Genevieve D, Fontana E, Darra F, Cances C, Barth M, Bonneau D, Bernadina BD, N’Guyen S, Gitiaux C, Parent P, des Portes V, Pedespan JM, Legrez V, Castelnau-Ptakine L, Nitschke P, Hieu T, Masson C, Zelenika D, Andrieux A, Francis F, Guerrini R, Cowan NJ, Bahi-Buisson N, Chelly J. Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nat Genet. 2013;45:639–647. doi: 10.1038/ng.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- http://www.nature.com/ng/journal/v45/n6/abs/ng.2613.html#supplementary-information.
- Putkey FR, Cramer T, Morphew MK, Silk AD, Johnson RS, McIntosh JR, Cleveland DW. Unstable Kinetochore-Microtubule Capture and Chromosomal Instability Following Deletion of CENP-E. Developmental Cell. 2002;3:351–365. doi: 10.1016/s1534-5807(02)00255-1. doi: http://dx.doi.org/10.1016/S1534-5807(02)00255-1. [DOI] [PubMed] [Google Scholar]
- Qvist P, Huertas P, Jimeno S, Nyegaard M, Hassan MJ, Jackson SP, Børglum AD. CtIP Mutations Cause Seckel and Jawad Syndromes. PLoS Genet. 2011;7:e1002310. doi: 10.1371/journal.pgen.1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. A small step for the cell, a giant leap for mankind: a hypothesis of neocortical expansion during evolution. Trends in Neurosciences. 1995;18:383–388. doi: 10.1016/0166-2236(95)93934-p. [DOI] [PubMed] [Google Scholar]
- Rauch A, Thiel CT, Schindler D, Wick U, Crow YJ, Ekici AB, van Essen AJ, Goecke TO, Al-Gazali L, Chrzanowska KH, Zweier C, Brunner HG, Becker K, Curry CJ, Dallapiccola B, Devriendt K, Dorfler A, Kinning E, Megarbane A, Meinecke P, Semple RK, Spranger S, Toutain A, Trembath RC, Voss E, Wilson L, Hennekam R, de Zegher F, Dorr H-G, Reis A. Mutations in the Pericentrin (PCNT) Gene Cause Primordial Dwarfism. Science. 2008;319:816–819. doi: 10.1126/science.1151174. [DOI] [PubMed] [Google Scholar]
- Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. doi: http://www.nature.com/emboj/journal/v28/n17/suppinfo/emboj2009173a_S1.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seckel HPG. Thomas CT, editor. Bird-headed dwarfs: Studies in developmental anthropolgy incuding human proportions. Bird-headed dwarfs: Studies in developmental anthropology including human proportions. S.Karger, Basel. 1960 [Google Scholar]
- Sengupta S, Robles AI, Linke SP, Sinogeeva NI, Zhang R, Pedeux R, Ward IM, Celeste A, Nussenzweig A, Chen J, Halazonetis TD, Harris CC. Functional interaction between BLM helicase and 53BP1 in a Chk1-mediated pathway during S-phase arrest. J Cell Biol. 2004;166:801–813. doi: 10.1083/jcb.200405128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen R, Faqeih E, Ansari S, Abdel-Salam G, Al-Hassnan ZN, Al-Shidi T, Alomar R, Sogaty S, Alkuraya FS. Genomic analysis of primordial dwarfism reveals novel disease genes. Genome Research. 2014 doi: 10.1101/gr.160572.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamseldin HE, Elfaki M, Alkuraya FS. Exome sequencing reveals a novel Fanconi group defined by XRCC2 mutation. Journal of Medical Genetics. 2012;49:184–186. doi: 10.1136/jmedgenet-2011-100585. [DOI] [PubMed] [Google Scholar]
- Shinya Matsuura, Matsumoto Y, Morishima K-i, Izumi H, Matsumoto H, Ito E, Tsutsui K, Kobayashi J, Tauchi H, Kajiwara Y, Hama S, Kurisu K, Tahara H, Oshimura M, T KK, Ikeuchi a, Kajii T. Monoallelic BUB1B mutations and defective mitotic-spindle checkpoint in seven families with premature chromatid separation (PCS) syndrome. American Journal of Medical Genetics Part A. 2006;140A:358–367. doi: 10.1002/ajmg.a.31069. [DOI] [PubMed] [Google Scholar]
- Sir J-H, Barr AR, Nicholas AK, Carvalho OP, Khurshid M, Sossick A, Reichelt S, D’Santos C, Woods CG, Gergely F. A primary microcephaly protein complex forms a ring around parental centrioles. Nat Genet. 2011;43:1147–1153. doi: 10.1038/ng.971. doi: http://www.nature.com/ng/journal/v43/n11/abs/ng.971.html#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snape K, Hanks S, Ruark E, Barros-Nunez P, Elliott A, Murray A, Lane AH, Shannon N, Callier P, Chitayat D, Clayton-Smith J, FitzPatrick DR, Gisselsson D, Jacquemont S, Asakura-Hay K, Micale MA, Tolmie J, Turnpenny PD, Wright M, Douglas J, Rahman N. Mutations in CEP57 cause mosaic variegated aneuploidy syndrome. Nat Genet. 2011;43:527–529. doi: 10.1038/ng.822. doi: http://www.nature.com/ng/journal/v43/n6/abs/ng.822.html#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiff T, Alagoz M, Alcantara A, Outwin E, Brunner HG, Bongers EMHF, O’Driscoll M, Jeggo PA. Deficiency in Origin Licensing proteins impairs cilia formation: implications for the aetiology of Meier-Gorlin syndrome. PLoS Genet In Press. 2013 doi: 10.1371/journal.pgen.1003360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suijkerbuijk Saskia JE, van Dam Teunis JP, Karagöz GE, von Castelmur E, Hubner Nina C, Duarte Afonso MS, Vleugel M, Perrakis A, Rüdiger Stefan GD, Snel B, Kops Geert JPL. The Vertebrate Mitotic Checkpoint Protein BUBR1 Is an Unusual Pseudokinase. Developmental Cell. 2012;22:1321–1329. doi: 10.1016/j.devcel.2012.03.009. doi: http://dx.doi.org/10.1016/j.devcel.2012.03.009. [DOI] [PubMed] [Google Scholar]
- Sulakhe D, Balasubramanian S, Xie B, Feng B, Taylor A, Wang S, Berrocal E, Dave U, Xu J, Börnigen D, Gilliam TC, Maltsev N. Lynx: a database and knowledge extraction engine for integrative medicine. Nucleic Acids Research. 2014;42:D1007–D1012. doi: 10.1093/nar/gkt1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanudji M, Shoemaker J, L’Italien L, Russell L, Chin G, Schebye XM. Gene Silencing of CENP-E by Small Interfering RNA in HeLa Cells Leads to Missegregation of Chromosomes after a Mitotic Delay. Molecular Biology of the Cell. 2004;15:3771–3781. doi: 10.1091/mbc.E03-07-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton GK, Woods CG. Primary microcephaly: do all roads lead to Rome? Trends in Genetics. 2009;25:501–510. doi: 10.1016/j.tig.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibelius A, Marhold J, Zentgraf H, Heilig CE, Neitzel H, Ducommun B, Rauch A, Ho AD, Bartek J, Krämer A. Microcephalin and pericentrin regulate mitotic entry via centrosome-associated Chk1. The Journal of Cell Biology. 2009;185:1149–1157. doi: 10.1083/jcb.200810159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Research. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BAA, Bonday ZQ, Putkey FR, Kops GJPL, Silk AD, Cleveland DW. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. The Journal of Cell Biology. 2003;162:551–563. doi: 10.1083/jcb.200303167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Abrieu A, Zheng Y, Sullivan KF, Cleveland DW. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat Cell Biol. 2000;2:484–491. doi: 10.1038/35019518. [DOI] [PubMed] [Google Scholar]
- Zhang S, Hemmerich P, Grosse F. Centrosomal localization of DNA damage checkpoint proteins. J Cell Biochem. 2007;101:451–465. doi: 10.1002/jcb.21195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.