Summary
An intercentrosomal linker keeps the cell’s two centrosomes joined together until it is dissolved at the onset of mitosis. A second connection keeps daughter centrioles engaged to their mothers until they lose their orthogonal arrangement at the end of mitosis. Centriole disengagement is required to license centrioles for duplication. We show that the intercentrosomal linker protein Cep68 is degraded in prometaphase through the SCFβTrCP (Skp1-Cul1-F-box protein) ubiquitin ligase complex. Cep68 degradation is initiated by PLK1 phosphorylation of Cep68 on Ser332, allowing recognition by βTrCP. We also found that Cep68 forms a complex with Cep215/Cdk5Rap2 and PCNT/Pericentrin, two PCM (pericentriolar material) proteins involved in centriole engagement. Cep68 requires two different pools of Cep215. We propose that Cep68 degradation allows Cep215 removal from the peripheral PCM preventing centriole separation following disengagement, whereas PCNT cleavage mediates Cep215 removal from the core of the PCM to inhibit centriole disengagement and duplication.
Introduction
Cells begin G1 phase with two centrosomes, each containing one centriole. During S phase, they produce a procentriole (or daughter centriole), which grows from the lateral surface of the preexisting centriole. From G1 to late G2, the cell’s two centrosomes are joined by a flexible, loosely-organized, and dynamic intercentrosomal linker, allowing them to function as a single microtubule organizing center (MTOC)1–4. The intercentrosomal linker proteins Cep68, Rootletin, LRCC45, and Centlein localize to rootlets emanating from the proximal end of the parental centrioles through the tethering protein c-Nap1 (Refs.5–8). Depletion of any of these proteins results in loss of intercentrosome cohesion and premature centrosome separation in interphase5, 6, 9.
At the onset of mitosis, the intercentrosomal linker is disassembled, suggesting that centrosome separation requires the dissolution of the linker. For example, c-Nap1 and Rootletin dissociate from centrosomes during early mitosis in response to phosphorylation by the NEK2 kinase5, 10, 11. Cep68 is also removed from mitotic centrosomes6, but the mechanism of its removal is not known. Following linker disassembly, the microtubule-dependent motor protein Eg5 facilitates centrosome migration to the opposite poles of the cell11–15.
In addition to the intercentrosomal linker between parental centrioles, each newly formed daughter centriole remains connected, or “engaged,” to its parent in an orthogonal arrangement through a poorly understood mechanism. Centriole engagement prevents the assembly of additional daughter centrioles16, until, in late mitosis, disengagement (the loss of the connection between the mother and daughter centrioles and loss of their orthogonal orientation) licenses centrioles for duplication in the subsequent S-phase17–19. Following disengagement, the two centrioles separate and move apart from each other, sometimes considerable distances20. Two temporally separate stages of centriole disengagement have been described18. The first stage occurs in early mitosis and requires PLK1 activity18. The second stage of disengagement occurs in late mitosis, when centriole pairs finally lose their orthogonal connection, and involves Separase-dependent proteolytic cleavage of PCNT (also known as Pericentrin)21, 22. Centriolar Cohesin is a second putative Separase substrate, although its cleavage may play a role in meiotic, rather than mitotic, disengagement18, 23–25.
Super-resolution microscopy studies have illustrated that the pericentriolar material (PCM) is organized into higher-order domains in both interphase and mitosis26–30. In interphase, PCNT localizes in a toroid around the proximal end of the mother centriole wall, where it acts as a scaffold for recruitment of outer matrix components of the PCM, including Cep215 (also known as Cdk5RAP2)26, 27, 29, 31, 32. In G2/early mitosis, PLK1 promotes PCM maturation, or expansion, into a matrix-like structure that mediates microtubule nucleation. PLK1 is required for PCNT and Cep215 recruitment31, 33–35. Intriguingly, it has been suggested that PCM integrity contributes to centriole engagement25, 36. Accordingly, the PCM disassembles in late mitosis, coincident with centriole disengagement37.
In mammals, there are 69 distinct F-box proteins, each serving as substrate-specificity components of the multi-subunit Skp1-Cul1-F-box protein (SCF) family of ubiquitin ligase complexes38. SCF complexes have been shown to ensure that centrosome duplication only occurs once per cell cycle39–49, but whether SCFs regulate other aspects of the centrosome cycle (e.g., PCM disassembly, centriole disengagement and licensing, etc.) is not known.
We identified the centrosome cohesion protein Cep68 as a target of PLK1 and SCFβTrCP and an interactor of both Cep215 and PCNT. The biological significance of these findings is presented herein.
Results
Cep68 is degraded in prometaphase in a βTrCP-dependent manner
Centrosome cohesion is maintained until late G2/M, when the two centrosomes separate to promote the formation of the bipolar mitotic spindle. Centrosome cohesion proteins, including Cep68, are lost from mitotic centrosomes (Refs.5, 6, 50 and Figure S1A), consistent with the idea that linker proteins must be removed from centrosomes to allow their separation. To establish if the decrease in the levels of centrosomal Cep68 during mitosis is due to proteolysis, Cep68 levels were analyzed in synchronized HeLa cells by immunoblotting. Whereas Cep68 levels were high in S and G2 cells, Cep68 was absent in prometaphase cells (Figure 1A–B). Specifically, Cep68 downregulation occurred between prophase and prometaphase, as judged by immunofluorescence staining to detect endogenous Cep68 in cells co-stained with DAPI and an antibody to Lamin B (Figure 1C). MG132 (a proteasome inhibitor) and MLN4924 [a cullin-RING ligase (CRL) inhibitor] prevented the decrease in Cep68 levels (Figure 1B), suggesting that one of approximately 200 mammalian CRL complexes51, 52 is responsible for Cep68 degradation. Cep68 levels remained low throughout mitosis, and progressively increased through the next G1 phase, peaking in G2 (Figure S1B–C).
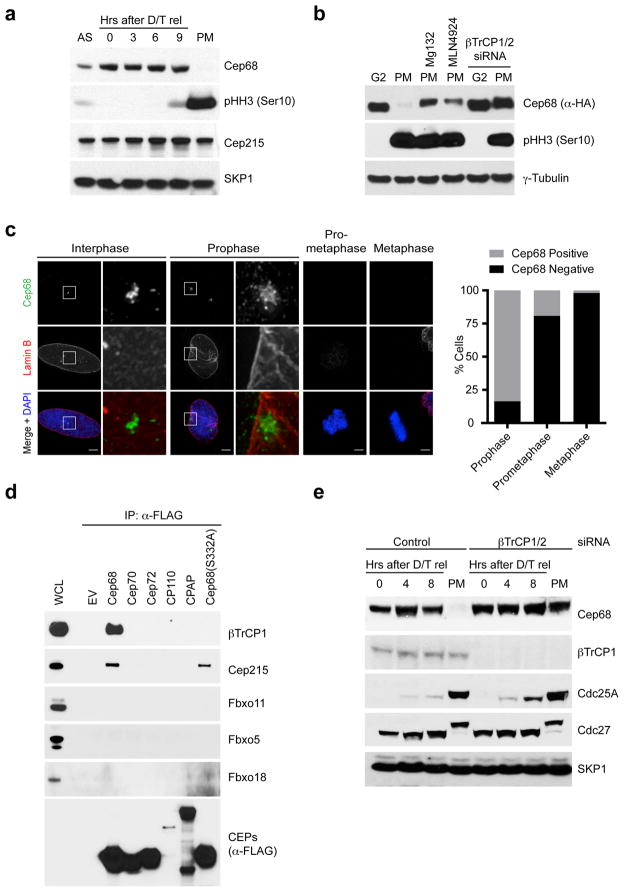
Figure 1. Cep68 is degraded in prometaphase in a βTrCP-dependent manner.
a) Cep68 protein levels are downregulated in prometaphase. HeLa cells were synchronized by double-thymidine arrest before release into fresh media. Seven hours after release, nocodazole was added to the media to accumulate cells in prometaphase, which were collected by mitotic shake-off. Cells were collected at the indicated times and lysed for immunoblotting as indicated. AS, asynchronous. PM, prometaphase.
b) Cep68 is degraded in prometaphase by a cullin-RING ligase. HeLa cells stably expressing Flag-HA-Cep68 were released from double-thymidine arrest. Seven hours after release, nocodazole was added to the media to accumulate cells in prometaphase. Where indicated, MG132 (a proteasome inhibitor) or MLN4924 (a cullin-RING ligase inhibitor) were also added to cells seven hours after release. Where indicated, cells were transfected with βTrCP siRNA. Cells were harvested either in G2 phase (eight hours after release from double-thymidine arrest) or prometaphase (twelve hours after release).
c) Cep68 degradation occurs between prophase and prometaphase. U-2OS cells were fixed and stained with DAPI and antibodies recognizing Cep68 (green) and Lamin B (red). The areas in the white boxes are shown at higher magnification directly beside the corresponding image. The graph indicates the percentage of cells that were either Cep68 positive or negative in each stage of mitosis. Prophase: n=110; Prometaphase: n=130; Metaphase: n=102 (from one experiment).
d) Cep68 interacts specifically with βTrCP1. HEK293T cells were transfected with either empty vector (EV) or FLAG-tagged centrosomal proteins (CEPs). Cell lysates were immunoprecipitated with anti-FLAG resin, and immunoprecipitates were probed with the indicated antibodies. WCL, whole cell lysate.
e) Cep68 is stabilized in prometaphase by βTrCP depletion. HeLa cells were transfected with βTrCP siRNA. Cells were synchronized by double-thymidine arrest and released into fresh media for the indicated times, before processing as in Figure 1A.
Analysis of immunopurified FLAG-tagged Cep68 by Multidimensional Protein Identification Technology (MudPIT) revealed that Cep68 interacted with βTrCP2, in addition to Cul1 and Skp1 (Figure S1D), suggesting that SCFβTrCP might regulate Cep68 degradation. Mammalian cells express two paralogs of βTrCP [βTrCP1 (also known as FBXW1) and βTrCP2 (also known as FBXW11)], which are biochemically indistinguishable (and we use βTrCP when referring to both). We screened several additional F-box proteins for binding to Cep68 and found that only endogenous βTrCP1 interacted specifically with Cep68 (Figure 1D). Furthermore, endogenous βTrCP1 also interacted with PLK4, and, to a lesser extent, PLK1, but not other centrosome proteins (Figures 1D and S1E). Lastly, depletion of βTrCP using distinct siRNAs resulted in the stabilization of both endogenous and exogenous Cep68 in mitosis (Figures 1B, 1E, S1C, and S1F). In contrast, depletion of Cdc20, the activator of the mitotic ubiquitin ligase APC/C (Anaphase Promoting Complex/Cyclosome), did not stabilize Cep68 in mitosis (Figure S1F).
PLK1 phosphorylates Cep68 on S332 and is required for Cep68 degradation
We mapped the βTrCP-binding domain in Cep68 using a series of Cep68 deletion constructs, establishing a 100 amino acid region required for binding (aa 300–400) (Figure S2A–B). βTrCP typically recognizes a consensus DpSGXX(X)pS degron sequence in substrates, where both serine residues are phosphorylated53. Cep68 does not possess a classical DSGXX(X)S motif, but it has a partial match within the region of interaction (DSGVDLD; aa 331-337 in human), which is also conserved in other species (Figure S2C). Deletion of these seven amino acids prevented both binding to βTrCP1 (Figure S2D) and Cep68 downregulation in prometaphase (Figure 2A). Substituting Serine 332 to Alanine prevented both binding to βTrCP1 (Figures 1D and S2D) and downregulation of Cep68 during mitosis (Figure 2A), demonstrating that this one amino acid is critical for Cep68 degradation. Cep68(S332A) still exhibited a migration shift, suggesting that it is phosphorylated on other sites during mitosis. Mutation of Aspartic acid 337 to Alanine also reduced Cep68 binding to βTrCP1 significantly (Figure S2D), suggesting that this residue also contributes to the degron, possibly by mimicking the second phosphorylated Serine residue found within the consensus degron.
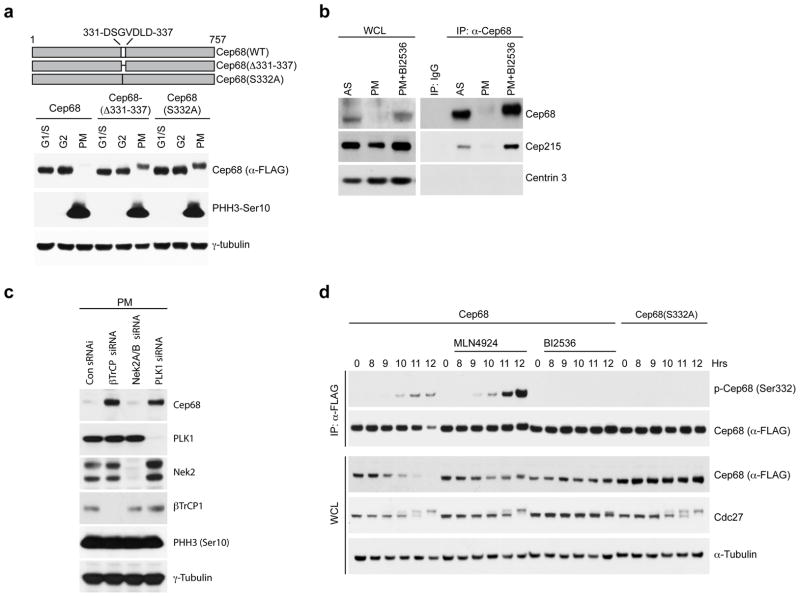
Figure 2. PLK1 phosphorylates Cep68 on Ser332 to promote βTrCP-mediated Cep68 degradation.
a) Ser332 is necessary for Cep68 degradation in prometaphase. HeLa cells expressing inducible FLAG-tagged Cep68, Cep68(Δ331-337), or Cep68(S332A) were synchronized by double-thymidine arrest. Cells were released and harvested at the G1/S transition, in G2 phase (8 hours after release), or in prometaphase (PM). Cell lysates were immunoblotted as indicated.
b) PLK1 inhibition prevents the degradation of endogenous Cep68. HeLa cells were synchronized by double-thymidine arrest, treated with nocodazole seven hours after release, and incubated for a further five hours to allow cells to enter prometaphase (PM). Where indicated, cells were treated with PLK1 inhibitor (BI2536). Cep68 was immunoprecipitated from cell lysates and immunoblotted as indicated. Cep68 interacts with Cep215 both in asynchronous cells and when Cep68 is stabilized by PLK1 inhibition.
c) PLK1 depletion, but not Nek2 depletion, prevents the degradation of Cep68. HeLa cells expressing inducible FLAG-Cep68 were transfected with siRNAs to βTrCP, Nek2, or PLK1. Cells in prometaphase were harvested and imunoblotted as indicated.
d) PLK1 inhibition prevents phosphorylation of Cep68 on Ser332 in vivo. HeLa cells expressing either inducible FLAG-Cep68 or FLAG-Cep68(S332A) were released from a double-thymidine arrest. Seven hours after release, cells were treated with nocodazole and, where indicated, either BI2536 (a PLK1 inhibitor) or MLN4924 (a CRL inhibitor). Cells were then harvested at the indicated time points. Cep68 or Cep68(S332A) were immunoprecipitated from cell lysates using an anti-FLAG resin. Whole cell lysates (WCL) and immunoprecipitates were immunoblotted as indicated.
To identify the mitotic enzymes promoting Cep68 degradation, we tested a panel of small-molecule inhibitors for their ability to prevent downregulation of Cep68 in prometaphase. Whereas an Eg5 inhibitor (Monastrol) and an Aurora kinase inhibitor (VX680) were ineffective at preventing Cep68 degradation, a PLK1 inhibitor (BI2536) completely prevented Cep68 downregulation (Figures 2B, 2D, S2E, S2G), suggesting that PLK1 promotes Cep68 degradation. RNAi-mediated depletion of PLK1, but not NEK2, inhibited the degradation of Cep68 (Figure 2C). PLK1 was able to phosphorylate Cep68 on Ser332 in vitro, as detected by a phospho-specific antibody raised against a peptide containing phospho-Serine at position 332 (Figure S2F). In vivo, the phospho-specific Ser332 antibody recognized wild type Cep68 in prometaphase cells, particularly when Cep68 degradation was prevented by MLN4924 (Figures 2D and S2G). In contrast, when Cep68 was stabilized by inhibiting PLK1 with BI2536, the phospho-specific Ser332 antibody was unable to recognize Cep68 (Figures 2D and S2G). Finally, the phospho-specific Ser332 antibody did not recognize Cep68(S332A) under any of the above conditions (Figures 2D and S2G). We were also able to detect phosphorylation of Ser332 using mass spectrometry, and this modification was not detected after treatment with BI2536 (Table 1). Interestingly, PLK1 interacted with Cep68 in cells treated with BI2536, but not in untreated cells (Figure S2H), suggesting that, when inhibited, PLK1 is trapped in complex with Cep68. Collectively, these results demonstrate that PLK1 phosphorylates Cep68 on S332 in vivo, and this event is required for binding to βTrCP in prometaphase.
Table 1. Identification of Cep68 phosphorylation sites in vivo by mass spectrometry.
FLAG-tagged Cep68 was immunoprecipitated from HEK293T cells that were either untreated or treated with BI2536 (a PLK1 inhibitor) for 16 hours. Note that PLK1 inhibition results in prometaphase arrest. The table lists the number of spectra matching phosphorylated peptides as a fraction of total spectra detected for peptides bearing each residue and their corresponding modification percentage. Ser332 was identified as a phosphorylated residue in untreated cells, but not in cells treated with BI2536.
| Residue and modification | Untreated | PLK inhibition | Surrounding Sequence |
|---|---|---|---|
| T106 | 1/664 | 0/972 |
|
| S109 | 17/664 | 17/972 |
|
| S126 | 1/128 | 3/286 |
|
| S136 | 0/128 | 4/286 |
|
| S186 | 0/279 | 1/482 |
|
| S249 | 0/61 | 8/113 |
|
| S268 | 3/50 | 6/40 |
|
| Y273 | 0/50 | 8/40 |
|
| S285 | 0/50 | 5/40 |
|
| S332 | 1/147 | 0/215 |
|
| S340 | 0/147 | 2/215 |
|
| S342 | 9/147 | 12/215 |
|
| S354 | 3/203 | 6/188 |
|
| S374 | 0/144 | 1/89 |
|
| S435 | 2/419 | 8/437 |
|
| S472 | 4/37 | 2/47 |
|
| S478 | 23/37 | 32/47 |
|
Cep68 degradation is not required for c-Nap1 and Rootletin dissociation or centrosome separation at G2/M
Similar to endogenous Cep68, exogenous wild type Cep68 and Cep68(S332A) localized to the centrosome during interphase (3A–B). Cep68 degradation was complete by prometaphase, whereas Cep68(S332A) was present at the centrosome throughout mitosis (Figures 3C and 5A–B). Notably, both endogenous and exogenous Cep68 were detected on G2-phase and prophase centrosomes that were already separated (Figures 3B–C and 4), indicating that Cep68 degradation occurs after - and is therefore not required for - centrosome separation. Cells expressing non-degradable Cep68(S332A) successfully formed a bi-oriented spindle (Figures 3C and S3B). Moreover, cells expressing Cep68(S332A) accumulated centrosomal γ-tubulin (Figure 3C) and PCNT (see later Figure 5A) in metaphase, suggesting that centrosome maturation occurs in the presence of the non-degradable Cep68(S332A) mutant. Similar to cells expressing wild-type Cep68, the majority of cells expressing Cep68(S332A) already exhibited centrosome separation in prophase and prometaphase (Figure S3C), indicating that Cep68(S332A) did not slow the kinetics of centrosome separation. Furthermore, cells expressing non-degradable Cep68 progressed through mitosis with the same kinetics as cells expressing wild-type Cep68, as determined by time-lapse microscopy (Figure S3D).
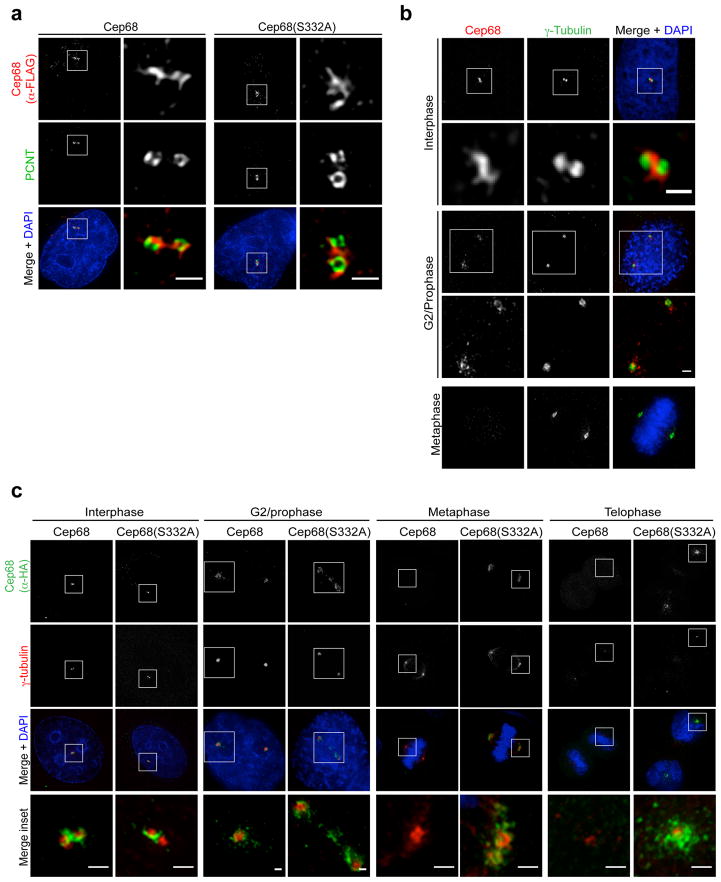
Figure 3. Cep68 degradation is not required for centrosome separation in mitosis.
a) Cep68(S332A) localizes to the intercentrosomal linker in interphase cells. Cells expressing inducible FLAG-tagged Cep68 or Cep68(S332A) were fixed and analyzed by immunofluorescence using anti-FLAG (red) and anti-PCNT (green) antibodies. The areas in the white boxes are shown at higher magnification directly beside the corresponding image. Scale bars represent 1 μm.
b) Centrosome separation occurs before the degradation of endogenous Cep68. HeLa cells were fixed and analyzed by immunofluorescence using antibodies recognizing Cep68 (red) and γ-tubulin (green). The areas in the white boxes are shown at higher magnification directly below the corresponding image. Scale bars represent 1 μm.
c) Centrosome separation and γ-tubulin recruitment occur normally in cells expressing Cep68(S332A). Cells expressing HA-tagged Cep68 or Cep68(S332A) were fixed and stained with anti-HA (green) and anti-γ–tubulin (red) antibodies. Cells in interphase, G2 phase, metaphase, and telophase were analyzed by immunofluorescence. The areas in the white boxes are shown at higher magnification directly below the corresponding image. Scale bars represent 1 μm. This experiment was reproduced three times. For each experiment in this figure, approximately 30 cells per condition were analysed.
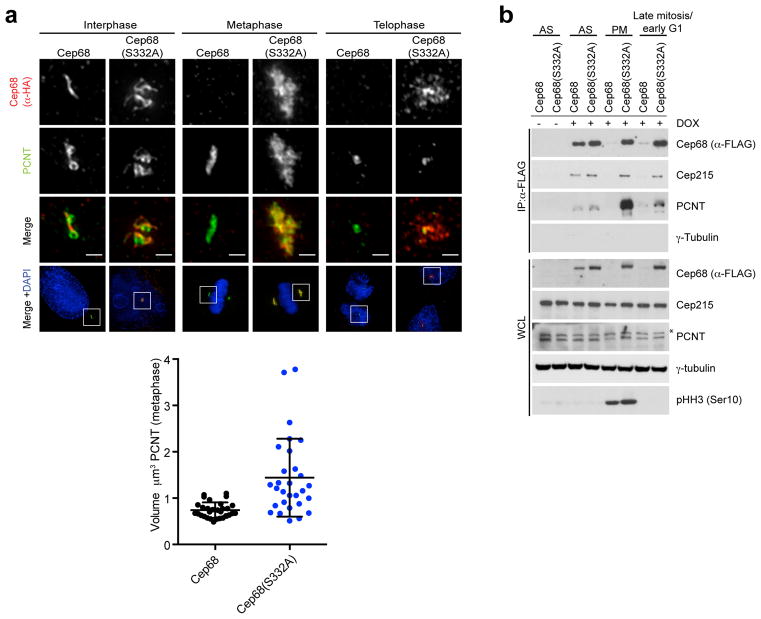
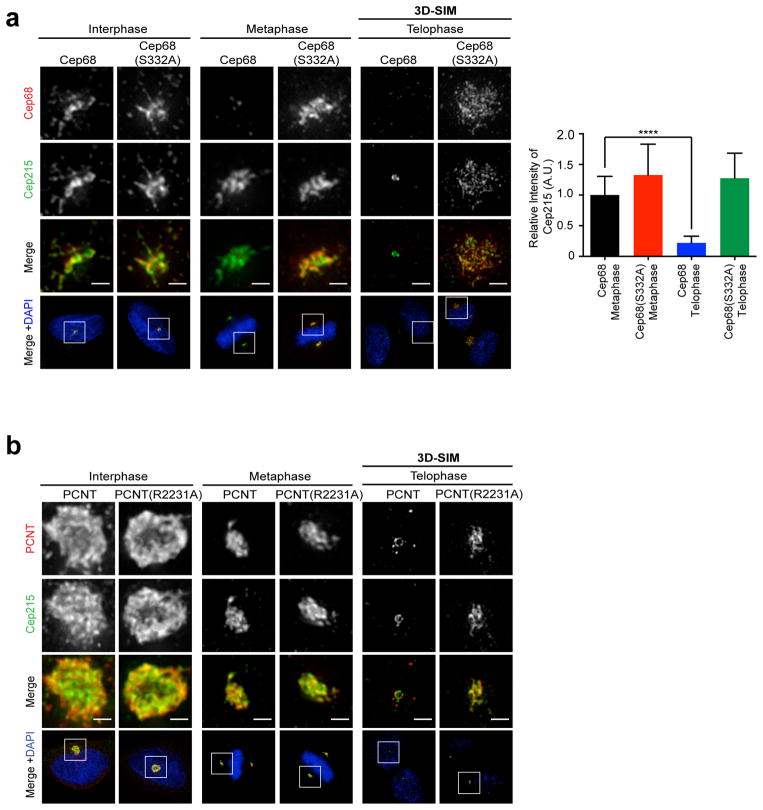
Figure 5. Cep68 stabilization increases the amount of PCNT at metaphase centrosomes, but does not affect its removal at the end of mitosis.
a) Cep68(S332A) expression results in increased levels of PCNT on metaphase centrosomes. Cells expressing pBABE-HA-tagged Cep68 or Cep68(S332A) were fixed and stained with anti-HA and anti-PCNT antibodies. PCNT levels decrease in telophase centrosomes, as previously reported21, regardless of the expression of Cep68. The areas in the white boxes are shown at higher magnification directly above the corresponding image. This phenomenon was observed in >3 experiments. The graph on the right shows the volume of PCNT-positive material in metaphase cells expressing Cep68 or Cep68(S332A) measured using Volocity 3D image analysis software. Bars represent the mean ± standard deviation (S.D.). Cep68: n=32 centrosomes; Cep68(S332A):n=33 centrosomes. P< 0.0001.
b) The biochemical interaction between stable Cep68 and PCNT increases in metaphase, and decreases at the end of mitosis. In contrast, the interaction between stable Cep68 and Cep215 persists throughout mitosis. Cells expressing doxycycline (DOX)-inducible FLAG-tagged Cep68 or Cep68(S332A) were collected from asynchronous conditions (AS), prometaphase (PM), or late mitosis/early G1 phase (the latter obtained by releasing cells from nocodazole arrest for two hours). Cep68 or Cep68(S332A) were immunoprecipitated from cell lysates with anti-FLAG resin, and whole cell lysates (WCL) and immunoprecipitates were immunoblotted as indicated. Total Cep215 levels do not change following release from mitosis. The asterisk denotes a non-specific band.
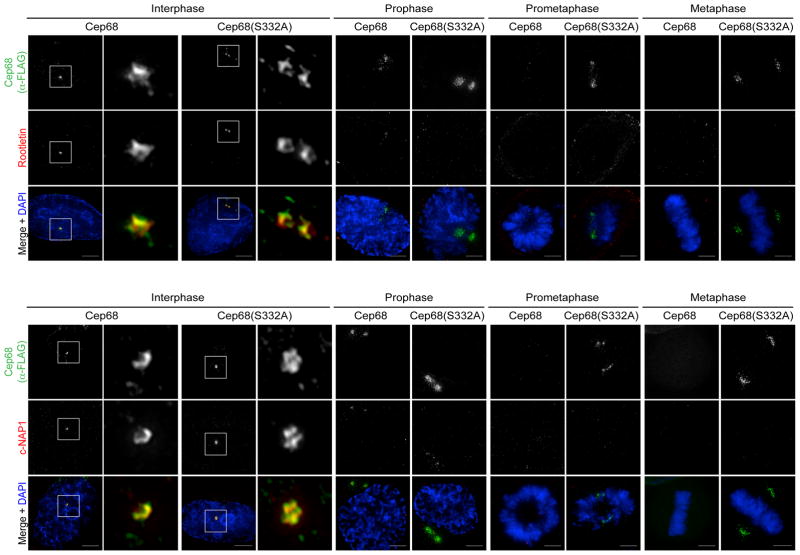
Figure 4. Cep68 degradation is not required for the displacement of Rootletin or c-Nap1 from mitotic centrosomes.
Cells expressing inducible FLAG-Cep68 or FLAG-Cep68(S332A) were stained with an anti-FLAG antibody (green) and either an anti-Rootletin or an anti-c-Nap1 antibody (red). Cells in interphase and mitosis were analyzed. The areas in the white boxes are shown at higher magnification directly beside the corresponding image. Scale bar represents 5 μm. This experiment was independently reproduced two times and each time approximately 30 cells per condition were analysed.
Cep68 is required for the localization of Rootletin during interphase (Ref6 and Figure S4A), and Rootletin is removed, along with c-NAP1, in a NEK2-dependent manner upon mitotic entry5, 7, 10, 11. We found that the displacement of these intercentrosomal linker proteins occurred normally in cells expressing non-degradable Cep68(S332A) (Figure 4). Their removal started in prophase and was complete by prometaphase, with similar kinetics in cells expressing either Cep68 or Cep68(S332A). Thus, Cep68 degradation does not influence the dissociation of intercentrosomal linker proteins, c-Nap1 or Rootletin.
The Eg5 motor protein can overcome the holding forces of the intercentrosomal linker, allowing centrosome separation to occur in the absence of NEK2 activity11. To determine whether Cep68 degradation similarly becomes a limiting step for centrosome separation when Eg5 activity is inhibited, we monitored the distance between centrosomes in cells treated with a low dose of an Eg5 inhibitor (monastrol)11. We found that the average distance between centrosomes was the same in cells expressing Cep68 or Cep68(S332A) (Figure S3E), indicating that degradation of the Cep68-linker does not affect centrosome disjunction in mitosis. We also monitored the kinetics of mitotic progression in cells stably expressing Cep68 or Cep68(S332A) with and without low doses of monastrol. We found that, whereas monastrol inhibited the kinetics of mitotic progression, cells expressing Cep68 or Cep68(S332A) passed through mitosis with similar kinetics (Figure S3D), suggesting that Cep68 degradation does not facilitate Eg5-independent bipolar spindle assembly. Overall, we concluded that Cep68 degradation does not facilitate and is not required for centrosome separation and bipolar spindle assembly.
Cep68 degradation and PCNT cleavage are required for the removal of different populations of Cep215 from centrosomes during late mitosis
We hypothesized that the degradation of Cep68 might result in the concomitant removal of its interacting proteins from mitotic centrosomes. Using immunopurification followed by MudPIT analysis, we identified Cep215 and PCNT as two interactors of Cep68 (Figure S1D). We confirmed that Cep68 co-immunoprecipitated with endogenous Cep215 and PCNT (Figures 1D, 2B, 5A, 5B, S2B), so we investigated whether Cep68 acts as a scaffold protein to localize these interacting proteins, as well as intercentrosomal linker proteins, during interphase. We silenced Cep68 expression and analyzed the localizations of Cep215, PCNT, Rootletin, and Cep135. In agreement with the role of Cep68 in Rootletin recruitment and intercentrosome cohesion6, depletion of Cep68 resulted in the loss of Rootletin from centrosomes (Figure S4A). We also observed that, in interphase, Cep215 levels were significantly reduced at the intercentrosomal linker in Cep68 depleted cells (Figures S4A), although not as severely as in PCNT depleted cells6, 32, 33, 54. Although Cep215 was largely displaced from the intercentrosomal linker after Cep68 depletion, total levels of Cep215 remained unchanged (Figure S4B). Finally, the localizations of PCNT and Cep135 were not affected by Cep68 depletion (Figure S4A).
Next, we tested whether the stabilization of Cep68 affected the temporal localization of these proteins in mitosis. The aberrant retention of Cep68(S332A) on metaphase centrosomes resulted in increased PCNT recruitment to the pericentriolar material (PCM), increasing the volume of PCNT-positive material from an average of 0.74 μm3 to 1.4 μm3 during mitosis (Figure 5A). Moreover, whereas Cep68 and Cep68(S332A) faintly interacted with PCNT in interphase cells, non-degradable Cep68(S332A) showed a much more robust interaction with PCNT in mitosis (Figure 5B). Despite this increased recruitment and binding, by telophase, PCNT was barely detected on centrosomes in cells engineered to express either Cep68(S332A) or wild type Cep68 (Figure 5A) and, reflecting this, the biochemical interaction decreased as well (Figure 5B).
In contrast, in the presence of the non-degradable Cep68(S332A) mutant, a strong Cep215 signal was visible in telophase, colocalizing with Cep68 at diffuse granules and fibers emanating from the PCM (Figure 6A). When Cep68 was degraded, Cep215 centrosomal localization was significantly reduced in telophase cells (Figure 6A), in agreement with previous reports36. We found that the Cep68(S332A) stable mutant co-immunoprecipitated Cep215 both in prometaphase and in late mitosis (Figure 5B), unlike PCNT, in support of the retention of Cep215 in mitosis observed by immunofluorescence. Similarly, when Cep68 was stabilized in prometaphase using the PLK1 inhibitor BI2536, Cep68 still interacted with Cep215 (Figure 2B). The interaction between Cep215 and Cep68 was not mediated by PCNT since siRNA depletion of PCNT did not affect this interaction (Figure S5A). Likewise, the mitotic interaction between PCNT and Cep68(S332A) did not require Cep215 (Figure S5A). Cep215 protein levels remained constant throughout mitosis, as judged by immunoblot analysis (Figures 2B and 5B), indicating that the inability to detect Cep215 by immunofluorescence is due to its dissociation from centrosomes in late mitosis rather than its degradation. These results indicate that the degradation of Cep68 is a prerequisite for the removal of Cep215 from the PCM in late mitosis.
Figure 6. Retention of Cep68 or PCNT in late mitosis prevents the removal of Cep215.
a) Expression of non-degradable Cep68(S332A) results in the retention of Cep215 on centrosomes in late mitosis. Cells expressing Cep68 or Cep68(S332A) were fixed and stained with antibodies recognizing Cep68 (red) and anti-Cep215 (green) antibodies. Cells in interphase and metaphase expressing HA-tagged Cep68 constructs were analyzed by immunofluorescence. The localization of Cep215 in telophase in cells expressing FLAG-tagged Cep68 constructs was analyzed by three-dimensional structured-illumination microscopy (3D-SIM). Scale bars represent 1 μm. The graph below shows the relative intensity of Cep215 quantified in metaphase and telophase cells expressing HA-tagged Cep68 or Cep68(S332A). The bars represent the mean ± standard deviation (S.D.). Cep68 metaphase: n=30 centrosomes; Cep68(S332A) metaphase: n=26 centrosomes; Cep68 telophase: n=30 centrosomes; Cep68(S332A) telophase: n=32 centrosomes. ****P < 0.0001.
b) Expression of non-cleavable PCNT(R2231A) in late mitosis prevents Cep215 removal. HeLa cells were transiently transfected with FLAG-PCNT or FLAG-PCNT(R2231A). Cells were fixed and stained with antibodies to FLAG (red) and Cep215 (green). The localization of Cep215 in telophase cells expressing FLAG-tagged PCNT constructs was analyzed by 3D-SIM. The areas in the white boxes are shown at higher magnification directly above the corresponding image. Scale bars represent 1 μm. This experiment was reproduced three times.
In prometaphase and metaphase cells (when Cep68 is absent), Cep215 is still present on centrosomes, implying that a mechanism other than Cep68 degradation is required for Cep215 removal. PCNT also interacts with Cep215 and is required for Cep215 localization in interphase and mitosis6, 32, 33, 54. We tested whether PCNT retention in late mitosis, like that of Cep68(S332A), can result in the retention of Cep215. We transiently transfected cells with PCNT or non-cleavable PCNT(R2231A). Overexpression of PCNT or PCNT(R2231A) increased the size of the PCM in around 50% of cells (Figure 6B), as described previously16, 26. In all cases, Cep215 co-localized with PCNT and PCNT(R2231A). Like endogenous PCNT21, wild-type exogenous PCNT was reduced in late mitosis in ~ 70% of cells. In contrast, we found that PCNT(R2231A) remained on centrosomes in late mitosis in ~ 80% of cells, until its levels eventually dropped in cells undergoing cytokinesis (Figure S5B). Moreover, we found that Cep215 colocalized with PCNT in all cases where PCNT was present in late mitosis (Figure 6B). Therefore, the aberrant retention of either PCNT or Cep68 in late mitosis is sufficient to retain Cep215. Notably, the pattern of Cep215 retention caused by stabilization of Cep68 differed from that caused by expression of non-cleavable PCNT. Cep68(S332A) recruited Cep215 to diffuse granules and fibers radiating away from the centrosome (Figure 6A), whereas PCNT(R2231A) recruited Cep215 to a dense, compact lattice, in proximity to the proximal end of the centrioles [Figure 6B and Ref.26]. These findings, together with the fact that Cep68-Cep215 and PCNT-Cep215 interactions are independent from each other (Figures S5A and 5A), suggest the existence of two distinct pools of Cep215.
Cep215 removal promotes centriole disengagement and subsequent centriole separation
Cep215 maintains centrosome cohesion and centriole engagement in interphase6, 36, and we found that Cep215 removal in late mitosis requires Cep68 degradation and PCNT removal (Figure 6A,B). Therefore we investigated the role of Cep68 degradation in centriole disengagement and/or centriole cohesion after disengagement.
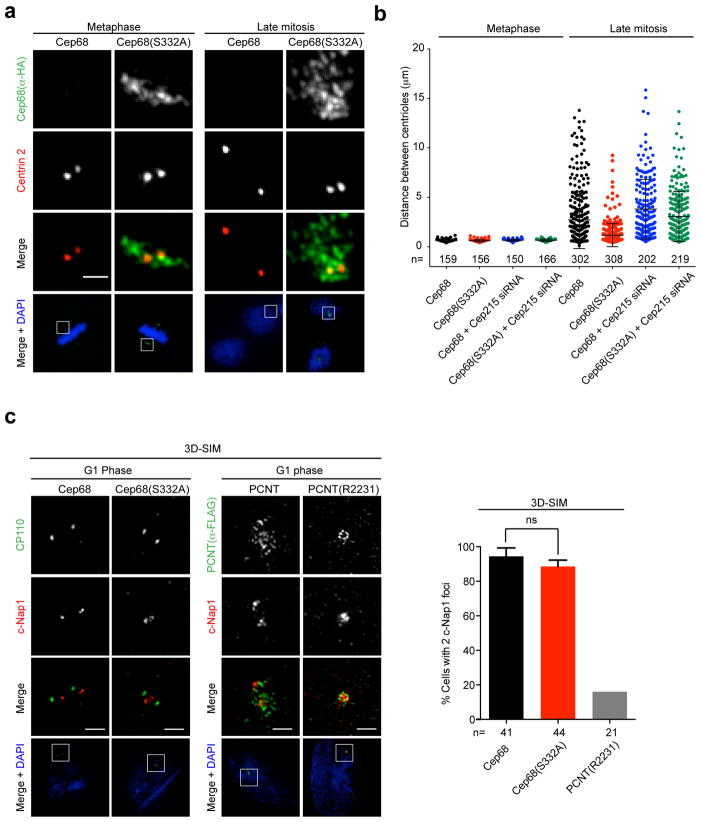
We found that centrioles separated from each other in late mitotic cells expressing wild-type Cep68, displaying widely variable distances (mean 3.2 μm; median 2.1 μm) (Figures 7A,B). In contrast, centrioles in cells expressing non-degradable Cep68(S332A) remained in closer proximity to each other in late mitosis (mean 1.4 μm; median 1 μm) (Figures 7A,B). Cep215 depletion by siRNA reversed the phenotype caused by retention of Cep68(S332A) in late mitosis (Figures 7B). These results suggest that Cep68(S332A) and Cep215 prevent either centriole disengagement and/or centriole separation in late mitosis.
Figure 7. Cep68 degradation promotes centriole separation in a Cep215-dependent manner.
a) The distance between mother and daughter centrioles in late mitosis is restricted in cells expressing non-degradable Cep68(S332A). Cells expressing HA-Cep68 or HA-Cep68(S332A) were synchronized by double-thymidine arrest and released into fresh media. Cells in metaphase or cytokinesis were analyzed by immunofluorescence using antibodies recognizing HA (green) or Centrin 2 (red). The areas in the white boxes are shown at higher magnification directly above the corresponding image. Scale bars represent 1 μm.
b) Measurement of the distance between mother and daughter centrioles in late mitosis. Cells expressing inducible FLAG-tagged Cep68 or Cep68(S332A) were transfected with Cep215 siRNA. Cells were synchronized by double-thymidine arrest and released into fresh media. Cells in metaphase or cytokinesis were analyzed by immunofluorescence. The distance between centrioles (CP110 foci) was measured using the SoftWorx measure tool and is displayed as a scatter plot. Bars represent the mean distance ± standard deviation (S.D.). The n value is pooled from at least three independent experiments.
c) Cep68 degradation is not required for centriole disengagement. In contrast, non-cleavable PCNT(R2231A) prevents centriole disengagement. Left panels: Cells expressing Cep68 or Cep68(S332A) were synchronized by double-thymidine arrest and released into fresh media for 14 hrs. Cells in G1 phase were stained with for c-Nap1 (red) and CP110 (green) and analyzed by three-dimensional structured-illumination microscopy (3D-SIM). c-Nap1 can be resolved into two populations in cells expressing Cep68 and Cep68(S332A). Scale bars represent 1 μm. Right panels: Cells expressing wild-type FLAG-PCNT or non-cleavable FLAG-PCNT(R2231A) were synchronized by double thymidine arrest and released into fresh media until they entered G1 phase. Cells were stained with antibodies to FLAG (green) and c-Nap1 (red) and analyzed by 3D-SIM. The graph on the right shows the quantification of cells containing 2 c-Nap1 dots in cells expressing Cep68 or Cep68(S332A). > 10 cells per experiment were analysed per experiment. Bars represent the mean of three experiments ± standard deviation (S.D.). Non-cleavable PCNT served as the positive control in two out of the three experiments and is shown without error bars. ns= non-significant.
In immunofluorescence microscopy, c-Nap1 appears as one mark at the proximal end of the parent centriole in engaged centrosomes that is resolved into two populations when centrioles disengage and lose their orthogonal arrangement18. In contrast, distal centriole proteins, like Centrin 2, are detected as two separate signals in both engaged and disengaged configurations.
Thus, the appearance of c-Nap1 and Centrin 2 foci in a 2:2 ratio indicates successful centriole disengagement, whereas a 1:2 ratio indicates that centrioles are engaged. We observed that, compared with cells expressing wild type Cep68, there were significantly more cells containing c-Nap1 and Centrin 2 foci in a 1:2 ratio in the presence of non-degradable Cep68(S332A) (Figure S6). One caveat to this assay is that, using standard light microscopy, it is difficult to distinguish between engaged centriole pairs and closely associated centrioles that are disengaged. Therefore, we used three-dimensional structured-illumination microscopy (3D-SIM) to analyze centriole orientation in cells expressing Cep68(S332A). We observed that in the presence of non-degradable Cep68(S332A), at the resolution provided by 3D-SIM, in contrast to our results using standard light microscopy, we could distinguish two c-Nap1 populations in most cells (Figure 7C). As expected, the majority of cells expressing wild-type Cep68 displayed centrioles configurations that indicated successful disengagement and separation. In contrast, even by 3D-SIM, expression of non-cleavable PCNT prevented the resolution of c-Nap1 into two populations (Figure 7C), corresponding with the established role of PCNT cleavage in centriole disengagement21, 22. Therefore, we concluded that expression of non-degradable Cep68(S332A) does, in fact, not interfere with the centriole disengagement process, but does affect the ability of centrioles to separate after this step.
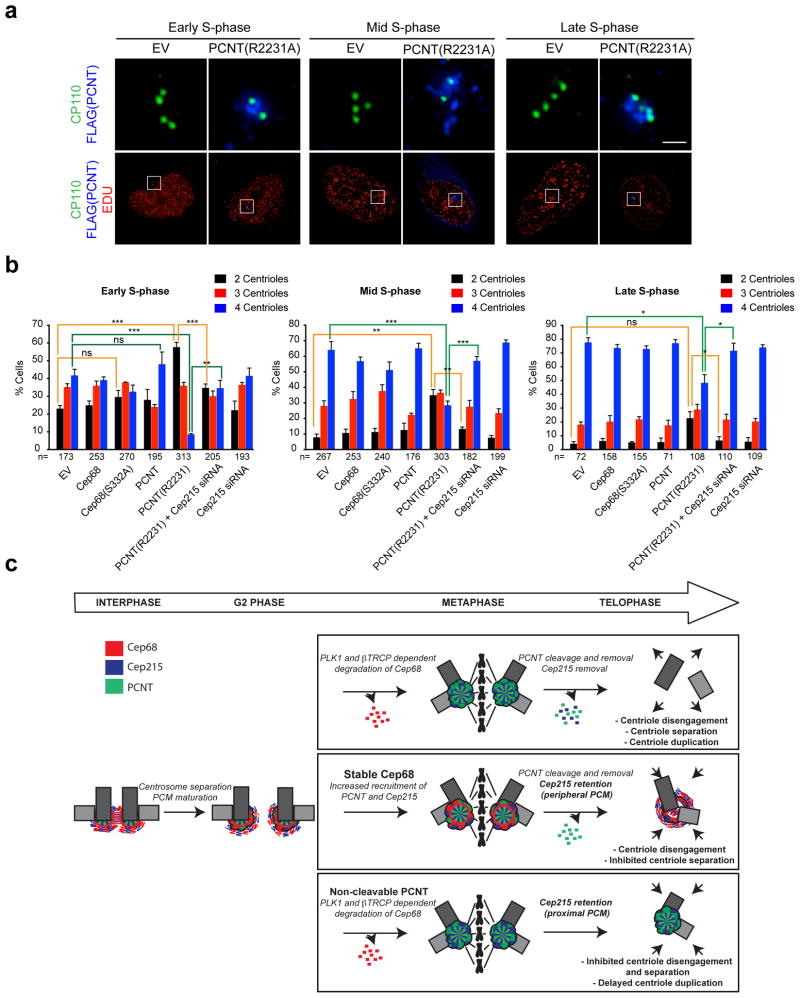
To study whether Cep215 retention by PCNT(R2231A) contributes to its role in centriole disengagement and duplication21, 22, we looked at centriole duplication in S-phase. We labeled cells with Edu to identify cells undergoing DNA replication and counted the number of centrioles in cells in different conditions. We categorized S-phase into different stages (early S-phase, mid S-phase or late S-phase) based on the incorporation pattern of Edu55 (Figure 8A). We found that centriole duplication occurred normally in cells expressing Cep68(S332A) (Figure 8B). In contrast, as previously shown21, 22, expression of non-cleavable PCNT prevented centriole duplication in early and mid S-phase, although centrioles eventually duplicated by late S-phase. Importantly, Cep215 depletion by siRNA could reverse the phenotype caused by PCNT(R2231A) (Figure 8B). Collectively, our results show that that PCNT(R2231A) inhibits centriole disengagement and licensing in a Cep215-dependent manner. In contrast, the stabilization of Cep68 during mitosis inhibits centriole separation in a Cep215-dependent manner.
Figure 8. Cep215 mediates the effects of non-cleavable PCNT(R2231A) on centriole duplication.
a) Representative Edu incorporation patterns for early S-phase, mid S-phase, and late S-phase cells. HeLa cells were incubated with Edu for one hour prior to fixation to identify cells that were actively undergoing DNA replication, then stained using the Click-it Edu imaging system (red) together with antibodies to CP110 (green) and FLAG (blue). The distinct stages of S-phase were identified according to the Edu incorporation pattern55. Cells were considered to be in early S-phase if they contained numerous small Edu foci, distributed evenly throughout the nucleus. Cells were considered to be in mid S-phase if the Edu was incorporated at perinuclear or perinucleolar sites. Late S-phase was scored if cells contained very large Edu-positive foci. The areas in the white boxes are shown at higher magnification directly above the corresponding image. Scale bar represents 1 μm.
b) Cep215 depletion rescues the centriole duplication delay caused by expression of non-cleavable PCNT(R2231A). HeLa cells expressing doxycycline-inducible FLAG-tagged Cep68 or Cep68(S332A) were induced for 48 hours with doxycycline. Where indicated, cells were transfected with Cep215 siRNA at the time of transient transfection of FLAG-tagged PCNT(R2231A). Centriole numbers (CP110 foci) were scored. The average of three independent experiments is shown ± standard error of the mean (SEM). The n value is pooled over the three experiments. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
c) Model. Top panel: Cep68 is degraded in a βTrCP- and PLK1-dependent manner in early mitosis. PCNT is cleaved by Separase in anaphase21, 22, after which the C-terminus of PCNT (which contains the Cep215-binding region) is degraded via the N-end rule22. Cep215 is subsequently removed from the centrosome. Middle panel: Stabilization of Cep68 at the centrosome in mitosis is not required for centrosome separation and bipolar spindle assembly. Instead, Cep68 stabilization in late mitosis results in the retention of Cep215, which prevents centriole separation following disengagement. Bottom panel: Expression of non-cleavable PCNT results in the retention of a different population of Cep215, which prevents centriole disengagement and subsequent licensing.
Discussion
In this study, we have shown that PLK1 triggers the βTrCP-dependent degradation of Cep68 in prometaphase. We identified PCNT and Cep215, both products of genes involved in microcephaly syndromes56, 57, as interactors of Cep68. PCNT and Cep215 play integral roles in PCM maturation and engagement22, 31, 33, 36, 58, 59 and interact with each other32, 54, 60, sharing a similar localization pattern in mitosis (i.e. increased in prophase and metaphase, and reduced by telophase, when the PCM disassembles and centriole disengagement occurs). Indeed, PCNT controls the localization of Cep215 in interphase and early mitosis6, 26, 29, 32, as well as late mitosis (shown here). Our study suggests that there are two pathways required for Cep215 removal from late mitotic centrosomes. One pathway involves the βTrCP- and PLK1-mediated degradation of Cep68 in prometaphase; the other the cleavage of PCNT at Arginine 2231 by Separase upon mitotic exit21, 22, after which the C-terminus of PCNT (which contains the Cep215-binding region) is subject to N-end rule mediated degradation22, 32. Moreover, our results are consistent with the notion that there are two pools of Cep215, a peripheral, granular PCM pool (interacting with Cep68), and proximal, dense centriolar pool (interacting with PCNT). When Cep215 is retained on the peripheral PCM by its interaction with Cep68(S332A), centriole separation following disengagement is inhibited. When Cep215 is retained at the core of the PCM by its interaction with non-cleavable PCNT, centriole disengagement and duplication are delayed (see model in Figure 8C).
Several different models of disengagement have been proposed. In one version, the orthogonal configuration of engaged centrioles is maintained by “glue proteins,” such as Cohesin23 and other, unidentified factors that may form a linker between mother and daughter centrioles. Although not mutually exclusive, an alternative model suggests a function for the PCM in trapping the mother-daughter centriole pair in their orthogonal configuration21, 22, 25, sterically inhibiting the disengagement process. We confirmed that lack of PCNT cleavage and removal inhibits centriole disengagement and licensing and further show that these events depend on the retention of Cep215 by PCNT. Our results suggest that PCM remodeling facilitates the processes of centriole disengagement and separation.
Interestingly, other centrosomal proteins (including CP110, PLK4, cyclin E, and Sas6) are also degraded via SCF ligases (SCFcyclin F, SCFβTrCP, SCFFbxw7 and SCFFbxw5, respectively)39–48 to limit centriole duplication to once per cell cycle. In contrast, SCFβTrCP-mediated degradation of Cep68 degradation does not inhibit centrosome overduplication and, instead, prevents Cep215 removal and centriole separation after centriole disengagement. As Separase is activated by APC/C, our study also highlights the crosstalk between SCF and APC/C ubiquitin ligase complexes in regulating PCM disassembly.
Materials and Methods
Antibodies
A rabbit polyclonal anti-Cep68 antibody was generated against GST-Cep681-355 (YenZym Antibodies, South San Francisco, CA) and affinity purified (used at 1:1000 dilution). A rabbit polyclonal antibody against phospho-S332-Cep68 (PADPVLQDpSGVDLD) was generated and affinity purified by YenZym Antibodies (used at 1:1000). The following commercial rabbit antibodies were used: pHH3-Ser10 (1:1000; D2C8; Cell signaling), γ-tubulin (1:1000; T5192; Sigma), FLAG (1:1000 (IF), 1:5000 (WB); F7425; Sigma), βTrCP1 (1:500; D13F10; Cell Signaling), Cep215 (Cdk5RAP2) (1:1000; A300-554A; Bethyl), Fbxo11 (1:1000; NB100-59825; Novus Biologicals), Fbxo5 (1:500; Invitrogen), Rootletin (CROCC) (1:100; Ab2; Sigma), Centrin 2 (1:100; N-17; Santa Cruz), PCNT (1:500; ab99342; Abcam), CP110 (1:100; 12780-1-AP; Protein Tech), Cdc20 (1:500; p55 CDC (H-175) sc-8358; Santa Cruz), and Cep135 (1:500; A302-250A; Bethyl). Antibodies to Fbxo18 (1:500) and Skp1 (1:5000) were generated in the Pagano laboratory61. The goat pericentrin 2 (N20) antibody was from Santa Cruz (1:1000 (IF)). The following mouse antibodies were used: HA.11 (Covance) (1:1000), γ-tubulin (1:1000; T5326; Sigma), Cdc25A (1:500; F6; Santa Cruz), GST (1:1000; 13-6700; Invitrogen), PLK1 (1:1000; F-8, Santa Cruz), Cdc27 (1:1000; AF3.1; Sigma), α-Tubulin (1:1000; T6199; Sigma), Nek2 (1:200; 610593; BD Transduction Laboratories), Lamin B (1:1000; sc-365962; Santa Cruz), Centrin 3 (1:100; SS12; Santa Cruz), cNAP-1 (1:100: BD Transduction Labs) and c-NAP-1 (1:50; F3; Santa Cruz).
Plasmids
Cep68 isoform 1 (KIAA0582) from the Kazusa DNA Research Institute or Cep68 isoform 2 (BC004873.2) were amplified by PCR and subcloned into a variety of vectors: a modified pcDNA3 vector containing a FLAG tag (FLAG-pcDNA3), a modified pBABE construct containing 2×FLAG and 2×HA tags (FLAG-HA-pBABE), pGEX-4T-2 (Amersham), and pcDNA-FRT-FLAG-dest. Cep68 truncation mutants and phosphosite-mutants were generated by site-directed mutagenesis (QuikChange, Agilent Technologies). Cep68 isoform 2 is used in figures 1b, 1d, 2a, s1b, s1c, s2b, s2d, s2e, s2f, and s2g. Both isoforms contain the βTrCP-degron and behave identically in our experiments. The FLAG-PCNTB-wt-MYC and FLAG-PCNTB-R2231A-myc in pLVX-IRES-PURO have been described previously22.
Cell culture procedures
HeLa and U-2 OS cells were propagated in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum (FBS). Stable cell lines expressing FLAG-HA-Cep68 or HA-Cep68 constructs (from a pBABE backbone) were generated by retroviral infection, as reported previously62. Cells expressing doxycycline-inducible FLAG-Cep68 and FLAG-Cep68(S332A) were generated using Flippase (Flp) recombination target (FRT)/Flp-mediated recombination technology in HeLa-T-rex Flp-in cells, described previously63. Cep68 was induced with 0.5 μg/ml doxycycline (Sigma). HEK293T cells were transfected with DNA using polyethylenimine (Polysciences). Cells were synchronized at G1/S using the double-thymidine block method. The following drugs were used: Nocodazole (Sigma; 100 ng/ml), BI2536 (Selleck; 100 nM), VX680 (Selleck; 300 nM), Monastrol (Tocris; 50 μM), Mg132 (Peptides International; 10 μM), and MLN4924 (Active Biochem; 0.5 μM).
siRNA oligonucleotides
The following siRNAs from Dharmacon were used: GUGGAAUUUGUGGAACAUC (βTrCP1/2, sequence 1), L-003463-00 and L-003490-00 (βTrCP1 and βTrCP2, sequence 2), ON-TARGETplus SMARTPOOL Cdc20 (L-015377-00-0005), GGAAAGUGAUGACGAGUAU (Cep68), and GCAAGGAUCUGAAUUUGUU (Cep215). The siRNA duplexes were transfected into cells using RNAiMax (Invitrogen) according to the manufacturer’s instructions.
Biochemical Methods, Purifications, and Mass-Spectometry
Cell lysis, immunoprecipitation, and immunoblotting have been previously described41. For purification and MudPIT analysis, HEK293T cells were transiently transfected with FLAG-Cep68. Briefly, the method follows that of MacCoss, M. J. et al with modifications64. TCA-precipitated proteins were urea-denatured, reduced, alkylated, and digested with endoproteinase Lys-C (Roche) followed by digestion with modified trypsin (Roche). Samples were loaded onto triple-phase 100 μm fused silica microcapillary columns and placed in-line with an LTQ Velos-Orbitrap Elite hybrid mass spectrometer equipped with a nano-LC electrospray ionization source (Thermo Fisher Scientific), coupled with a quaternary Agilent 1260 series HPLC. Each differently-digested sample was analyzed using fully automated 10-step chromatography run as described in Florens et al.65. Full MS spectra were recorded on the peptides over a 400 to 1,600 m/z range in the Orbitrap-elite at 60K resolution, followed by fragmentation in the ion trap (at 35% collision energy) on the first to tenth most intense ions selected from the full MS spectrum. Dynamic exclusion was enabled for 90 seconds66, with an exclusion window of 0.03 to 1.03. Mass spectrometer scan functions and HPLC solvent gradients were controlled by the Xcalibur data system (Thermo Fisher Scientific). For data analysis, RAW files were extracted into .ms2 file format67 using RawDistiller v. 1.0 (Ref.67), in-house developed software. RawDistiller D(g, 6) settings were used to abstract MS1 scan profiles68. Tandem mass (MS/MS) spectra were interpreted using SEQUEST (Ref.69) with a peptide mass tolerance of 10 ppm and of +/− 0.5 amu for fragment ions. The data was searched against a database of 58614 sequences, consisting of 29147 H. sapiens non-redundant proteins (downloaded from NCBI on 2011-8-16), 177 usual contaminants (such as human keratins, IgGs, and proteolytic enzymes), and, to estimate false discovery rates, 29307 randomized amino acid sequences derived from each non-redundant protein entry.
Kinase assay
GST-Cep68 or GST-Cep68-S332A were purified from Escherichia coli (BL-21) and incubated with purified PLK1 (Sigma) in a kinase reaction buffer (50 mM Tris-HCl pH 7.5, 10 mM MgCl2, 0.6 mM DTT, 0.01% Triton X-100, and 2 mM ATP) at 30°C for one hour.
Immunofluorescence Microscopy
Cells were fixed with ice-cold methanol for 10 minutes, then blocked with 3% BSA in PBS prior to incubation with primary antibodies. Alexa-555, Alexa-647 or Alexa-488-conjugated secondary antibodies were from Life Technologies. Edu labeled DNA was detected using the Click-iT Edu Alexa Fluor 455 imaging kit from Life Technologies according to the manufacturer’s instructions. Slides were mounted in ProlongGold with DAPI (Invitrogen). Imaging was performed using a DeltaVision Elite inverted microscope system (Applied Precision), using a 100×/1.4NA Oil PSF Objective from Olympus. The system was equipped with a CoolSNAP HQ2 camera and SoftWorx imaging software version 5.0. Serial optical sections obtained 0.2-μm apart along the z-axis were processed using the SoftWorx deconvolution algorithm and projected into one picture using SoftWorx software (Applied Precision). Image J (http://rsb.info.nih.gov/ij) was used to measure fluorescence intensities from a defined area around the centrosome. Signal intensities were normalized against γ-tubulin. Background intensity was substracted from each measurement. Images from each data set were acquired on the same day using the same exposure times. 3D-SIM images were acquired using a DeltaVision OMX V4/Blaze system (GE Healthcare) fitted with an Olympus 100x/1.40 NA UPLSAPO oil objective and Photometrics Evolve EMCCD cameras. 405, 488 and 568 nm laser lines were used for excitation and the corresponding emission filter sets were 436/31, 528/48 and 609/37 nm respectively. Image stacks were acquired with an optical section spacing of 125 nm. SI reconstruction and Image Registration were performed using softWoRx v. 6.1 software, using Optical Transfer Functions (OTFs) generated from Point Spread Functions (PSFs) acquired using 100 nm (green and red) or 170 nm (blue) FluoSpheres and alignment parameters refined using 100 nm TetraSpeck beads (Invitrogen/Life Technologies).
Statistics
P values were derived from unpaired two-tailed t-tests using GraphPad Prism software. All immunoblot images and immunofluorescence images are representative of at least two independent experiments, except where specified in the figure legends.
Supplementary Material
Supplementary Figure 1. βTrCP targets Cep68 for degradation during mitosis.
a) Cep68 immunofluorescence in interphase and metaphase. U-2OS cells were transfected with control siRNA or Cep68 siRNA and analyzed 48 hours post-transfection. Cells were fixed and analyzed by immunofluorescence using Cep68 antibodies (red) and γ-tubulin antibodies (green). The areas in the white boxes are shown at higher magnification directly below the corresponding image. Cep68 is absent from mitotic centrosomes. Depletion of Cep68 by siRNA demonstrates that the Cep68 antibody specifically recognizes Cep68. Scale bars represent 1 μm.
b) Cep68 levels during a release from prometaphase. HeLa cells stably expressing FLAG-HA-Cep68 were synchronized in prometaphase with nocodazole. Round, prometaphase cells were collected by mitotic shake-off and released into freh media. Cells were collected at the indicated times and lysed for immunoblotting as indicated.
c) Cep68 degradation is mediated by βTrCP. HeLa cells stably expressing FLAG-HA-Cep68 were transfected with βTrCP siRNA. Cells were synchronized by double-thymidine arrest and released into fresh media for the indicated times, before processing as in Figure 1a.
d) Multidimensional Protein Identification Technology analysis of a FLAG-Cep68 immunopurification from HEK293T cells, listing the number of spectra, peptides, and the dNSAF (distributed normalized spectra abundance factor) for the indicated co-purifying proteins.
e) βTrCP interacts with Cep68. HEK293T cells were transfected with FLAG-tagged centrosomal proteins (CEPs). Cell lysates were immunoprecipitated with an anti-FLAG resin, and immunoprecipitates were probed with antibodies to βTrCP1. PLK1 and PLK4 are known βTrCP-interacting proteins. Asterisks denote expression of FLAG-tagged CEPs.
f) Different siRNA sequences targeting βTrCP prevent the downregulation of Cep68 in prometaphase, whereas siRNA targeting Cdc20 does not. HeLa cells were transfected with the indicated siRNA sequences. Prometaphase cells (PM) were harvested by mitotic shake-off after overnight nocodazole treatment. NS, non-synchronized. Please note that Cdc20 silencing synchronizes cells in PM, as shown by the phosphorylation state of Cdc27 (see lane 8).
Supplementary Figure 2. Mapping of the βTrCP degron in Cep68 and identification of PLK1 as the kinase phosphorylating Cep68 degron.
a) Schematic representation of Cep68 mutants used in mapping experiments. Cep68 mutants found to interact with βTrCP or Cep215 are indicated by the symbol (+). ‘−/+’ denotes reduced binding.
b) Mapping the βTrCP binding region in Cep68 with Cep68 truncation mutants. HEK293T cells were transfected with empty vector (EV), FLAG-tagged Cep68, or the indicated FLAG-tagged Cep68 constructs. Whole cell extracts were immunoprecipitated with an anti-FLAG resin, and immunoprecipitates were immunoblotted with the indicated antibodies.
c) Alignment of the βTrCP binding motif in Cep68 orthologs and previously reported βTrCP substrates. Critical amino acids required for βTrCP binding are highlighted in red.
d) Ser332 is required for binding to βTrCP1. HEK293T cells were transfected with empty vector (EV), FLAG-tagged Cep68, or the indicated FLAG-tagged Cep68 mutants. Cell lysates were immunoprecipitated with an anti-FLAG resin, and immunoprecipitates were probed with the indicated antibodies.
e) PLK1 inhibition prevents Cep68 degradation in prometaphase. HeLa cells expressing FLAG-HA-Cep68 were synchronized by double-thymidine arrest, as in Figure 1b. Where indicated, cells were treated with a PLK1 inhibitor (BI2536), an Aurora kinase inhibitor (VX680), or an Eg5 inhibitor (monastrol) for three hours prior to their collection and analysis by immunoblotting.
f) PLK1 phosphorylates Cep68 on Ser332 in vitro. Recombinant, purified GST-Cep68, GST-Cep68(S332A), or GST alone were incubated with ATP and increasing amounts of purified PLK1. Proteins were detected by immunoblotting as indicated.
g) PLK1 inhibition prevents the in vivo phosphorylation of Cep68 on Ser332. HeLa cells expressing either inducible FLAG-Cep68 or FLAG-Cep68(S332A) were released from a double-thymidine arrest. Seven hours after release, cells were treated with nocodazole and, where indicated, either BI2536 (a PLK1 inhibitor) or MLN4924 (a CRL inhibitor). Cells were then harvested at the indicated time points. Cep68 or Cep68(S332A) was immunoprecipitated from cell lysates using an anti-FLAG resin. Whole cell lysates (WCL) and immunoprecipitates were immunoblotted as indicated.
h) Analysis of Cep68 binding to PLK1 following BI2536 treatment by Multidimensional Protein Identification Technology. The table lists the total number of spectral counts and the dNSAF values for Cep68 and PLK1 in the untreated and the treated samples.
Supplementary Figure 3. Cep68 degradation does not promote centrosome separation and bipolar spindle assembly.
a) HA-Cep68 is expressed at near to physiological levels. HeLa cells or HeLa cells stably expressing pBABE-HA-tagged Cep68, Cep68(S332A), or Cep68(Δ331-337) were synchronized by a double-thymidine arrest and harvested at the G1/S transition, in G2 phase (eight hours after release), or in prometaphase (PM). Cell lysates were immunoblotted as shown.
b) Bipolar spindles assemble in the presence of Cep68(S332A). Cells expressing inducible FLAG-tagged Cep68 or Cep68(S332A) were fixed and stained with anti-FLAG and antiα–tubulin antibodies. Cells in metaphase were analyzed by immunofluorescence. This experiment was reproduced twice.
c) Expression of FLAG-Cep68(S332A) does not prevent centrosome separation in early mitosis. Cells expressing inducible FLAG-tagged Cep68 or Cep68(S332A) were fixed and analyzed by immunofluorescence. The percentage of cells with separated centrosomes in prophase or prometaphase was scored. ≥100 cells were counted for each condition from one experiment. n=cell number.
d) Time spent in mitosis for cells expressing Cep68 or Cep68(S332A). Cells expressing inducible FLAG-tagged Cep68 or Cep68(Δ331-338) were synchronized by double thymidine release. Live cell microscopy was used to calculate time in mitosis (judged from rounded cell morphology). Where indicated, monastrol (50 μm) was added to cells eight hours after release. Bars represent mean ± standard deviation (S.D.). n=30 cells per condition. ns=not significant. This experiment was performed once.
e) Centrosome disjunction is not inhibited by expression of Cep68(S332A). Cells expressing inducible FLAG-Cep68 or FLAG-Cep68(S332A) were arrested with monopolar spindles in prometaphase with 100 μm monastrol. The distance between γ-tubulin foci was measured using Softworx software. Cep68: n=28; Cep68(S332A): n=27. Bars represent mean ± standard deviation (S.D.). ns=not significant.
Supplementary Figure 4. Cep215 levels are reduced at the intercentrosomal linker in Cep68 depleted cells.
a) Cep68 depletion results in removal of Cep215 from the intercentrosomal linker in interphase. U-2OS cells were transfected for 72 hours with siRNAs targeting Cep68 or a non-targeting control sequence. Cells were fixed and stained with antibodies to γ-tubulin (green) and the specified centrosome cohesion factors (red). After Cep68 silencing, only a small amount of Cep215 is observed on interphase centrioles. The graph represents the relative intensity of Cep215 after Cep68 depletion. Bars represent mean ± standard deviation (S.D.). Control siRNA: n=10 cells; Cep68 siRNA: n=11 cells. ***P=0.0006.
b) Cep68 knockdown does not affect the total levels of Cep215. U-2OS cells were transfected with siRNAs targeting Cep68 or Cep215. Seventy-two hours post-transfection, cells were lysed for immunoblotting as indicated.
Supplementary Figure 5. The interaction between Cep215 and Cep68 is not mediated by PCNT.
a) Cep68 interacts with Cep215 and PCNT independently. Cells expressing doxycycline (DOX)-inducible FLAG-tagged Cep68 or Cep68(S332A) were transfected with the indicated siRNA sequences. Cep68 or Cep68(S332A) were immunoprecipitated from whole cell lysates (WCL) with anti-FLAG resin, and both immunoprecipitates and WCLs were immunoblotted with the indicated antibodies. Cep215 interacts with Cep68 when PCNT is depleted in interphase and mitosis. Likewise, PCNT interacts with Cep68(S332A) when Cep215 is depleted. AS, asynchronous. PM, prometaphase. The asterisk denotes a non-specific band.
b) PCNT and PCNT(R2231A) localization in cytokinesis. HeLa cells were transiently transfected with FLAG-tagged PCNT or PCNT(R2231A). Cells in cytokinesis were stained for immunofluorescence using antibodies recognizing FLAG (red) and CP110 (green). Scale bars represent 1 μm.
Supplementary Figure 6. Cep68 degradation is required for centriole separation after disengagement.
Quantification of cells containing 1 c-Nap1 dot in cells expressing Cep68(S332A) using conventional microscopy. HeLa cells expressing HA-tagged Cep68 or Cep68(S332A) were synchronized by double-thymidine arrest and allowed to progress into the next G1 phase (15 hours after release). Where indicated, cells were transfected with Cep215 siRNA during the first release from double-thymidine arrest. G1 cells were fixed and stained for c-Nap1 (green), centrin 2 (red), and Cep68 (α-HA) (blue) to determine their engagement status. The magnification bar represents 1 μm. The areas in the white boxes are shown at higher magnification directly above the corresponding image. The graph shows the quantification of disengagement. G1 cells with 1: 2 ratio of c-Nap1:Centrin 2 foci were scored as engaged. n≥100 cells for each experiment from three independent experiments were analysed. **P ≤ 0.01. Bars represent the mean ± standard deviation (S.D.) from n= three experiments.
Acknowledgments
The authors thank BD Dynlacht, JR Skaar, and MFB Tsou for critical reading of the manuscript; K Lee and S Kim for helpful advice; Y Deng at the NYU SOM Microscopy core for image processing assistance; A North (Rockefeller University) for use of the DeltaVision OMX V4/Blaze system, supported by award S10RR031855 from the National Center for Research Resources; K Rhee for the PCNT constructs, and I Hoffmann for the FLAG-CPAP construct. MP is grateful to TM Thor for continuous support. This work was funded by grants from the National Institutes of Health (R01-GM057587 and R37-CA076584) and New York State Health Department (NYSTEM-N11G-255) to MP and fellowships from the National Health and Medical Research Council of Australia and the Lymphoma Research Foundation to JP. AS, LF, and MPW are supported by the Stowers Institute for Medical Research. PVJ and MJKJ were supported by NIH grant R01 GM094972 and an award from the Mathers Foundation. MP is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Author Contributions. JP planned and performed most experiments and co-wrote the manuscript. MP coordinated the study, oversaw the results, and co-wrote the manuscript. AM helped with many biochemical experiments. MJKJ helped with several experiments and provided intellectual advice. PVP provided reagents and intellectual advice. AS, LF, and MW performed the mass spectrometry analyses of the purifications performed by JP. All authors discussed the results and commented on the manuscript.
References
- 1.Bornens M, Paintrand M, Berges J, Marty MC, Karsenti E. Structural and chemical characterization of isolated centrosomes. Cell Motil Cytoskeleton. 1987;8:238–249. doi: 10.1002/cm.970080305. [DOI] [PubMed] [Google Scholar]
- 2.Mardin BR, Schiebel E. Breaking the ties that bind: new advances in centrosome biology. J Cell Biol. 2012;197:11–18. doi: 10.1083/jcb.201108006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nigg EA, Stearns T. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol. 2011;13:1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paintrand M, Moudjou M, Delacroix H, Bornens M. Centrosome organization and centriole architecture: their sensitivity to divalent cations. J Struct Biol. 1992;108:107–128. doi: 10.1016/1047-8477(92)90011-x. [DOI] [PubMed] [Google Scholar]
- 5.Bahe S, Stierhof YD, Wilkinson CJ, Leiss F, Nigg EA. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J Cell Biol. 2005;171:27–33. doi: 10.1083/jcb.200504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graser S, Stierhof YD, Nigg EA. Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. J Cell Sci. 2007;120:4321–4331. doi: 10.1242/jcs.020248. [DOI] [PubMed] [Google Scholar]
- 7.Fang G, et al. Centlein mediates an interaction between C-Nap1 and Cep68 to maintain centrosome cohesion. J Cell Sci. 2014;127:1631–1639. doi: 10.1242/jcs.139451. [DOI] [PubMed] [Google Scholar]
- 8.He R, et al. LRRC45 is a centrosome linker component required for centrosome cohesion. Cell Rep. 2013;4:1100–1107. doi: 10.1016/j.celrep.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 9.Mayor T, Stierhof YD, Tanaka K, Fry AM, Nigg EA. The centrosomal protein C-Nap1 is required for cell cycle-regulated centrosome cohesion. J Cell Biol. 2000;151:837–846. doi: 10.1083/jcb.151.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fry AM, et al. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J Cell Biol. 1998;141:1563–1574. doi: 10.1083/jcb.141.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mardin BR, et al. Components of the Hippo pathway cooperate with Nek2 kinase to regulate centrosome disjunction. Nat Cell Biol. 2010;12:1166–1176. doi: 10.1038/ncb2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sawin KE, LeGuellec K, Philippe M, Mitchison TJ. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature. 1992;359:540–543. doi: 10.1038/359540a0. [DOI] [PubMed] [Google Scholar]
- 13.Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol. 2000;150:975–988. doi: 10.1083/jcb.150.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roof DM, Meluh PB, Rose MD. Multiple kinesin-related proteins in yeast mitosis. Cold Spring Harb Symp Quant Biol. 1991;56:693–703. doi: 10.1101/sqb.1991.056.01.078. [DOI] [PubMed] [Google Scholar]
- 15.Blangy A, et al. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83:1159–1169. doi: 10.1016/0092-8674(95)90142-6. [DOI] [PubMed] [Google Scholar]
- 16.Loncarek J, Hergert P, Magidson V, Khodjakov A. Control of daughter centriole formation by the pericentriolar material. Nat Cell Biol. 2008;10:322–328. doi: 10.1038/ncb1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006;442:947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- 18.Tsou MF, et al. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev Cell. 2009;17:344–354. doi: 10.1016/j.devcel.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loncarek J, Hergert P, Khodjakov A. Centriole reduplication during prolonged interphase requires procentriole maturation governed by Plk1. Curr Biol. 2010;20:1277–1282. doi: 10.1016/j.cub.2010.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J Cell Biol. 2000;149:317–330. doi: 10.1083/jcb.149.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuo K, et al. Kendrin is a novel substrate for separase involved in the licensing of centriole duplication. Curr Biol. 2012;22:915–921. doi: 10.1016/j.cub.2012.03.048. [DOI] [PubMed] [Google Scholar]
- 22.Lee K, Rhee K. Separase-dependent cleavage of pericentrin B is necessary and sufficient for centriole disengagement during mitosis. Cell Cycle. 2012;11:2476–2485. doi: 10.4161/cc.20878. [DOI] [PubMed] [Google Scholar]
- 23.Schockel L, Mockel M, Mayer B, Boos D, Stemmann O. Cleavage of cohesin rings coordinates the separation of centrioles and chromatids. Nat Cell Biol. 2011;13:966–972. doi: 10.1038/ncb2280. [DOI] [PubMed] [Google Scholar]
- 24.Oliveira RA, Nasmyth K. Cohesin cleavage is insufficient for centriole disengagement in Drosophila. Curr Biol. 2013;23:R601–603. doi: 10.1016/j.cub.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 25.Cabral G, Sans SS, Cowan CR, Dammermann A. Multiple mechanisms contribute to centriole separation in C. elegans. Curr Biol. 2013;23:1380–1387. doi: 10.1016/j.cub.2013.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawo S, Hasegan M, Gupta GD, Pelletier L. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat Cell Biol. 2012;14:1148–1158. doi: 10.1038/ncb2591. [DOI] [PubMed] [Google Scholar]
- 27.Mennella V, Agard DA, Bo H, Pelletier L. Amorphous no more: subdiffraction view of the pericentriolar material architecture. Trends Cell Biol. 2013 doi: 10.1016/j.tcb.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol Open. 2012;1:965–976. doi: 10.1242/bio.20122337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mennella V, et al. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat Cell Biol. 2012;14:1159–1168. doi: 10.1038/ncb2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu J, Glover DM. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2012;2:120104. doi: 10.1098/rsob.120104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haren L, Stearns T, Luders J. Plk1-dependent recruitment of gamma-tubulin complexes to mitotic centrosomes involves multiple PCM components. PLoS One. 2009;4:e5976. doi: 10.1371/journal.pone.0005976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim S, Rhee K. Importance of the CEP215-pericentrin interaction for centrosome maturation during mitosis. PLoS One. 2014;9:e87016. doi: 10.1371/journal.pone.0087016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee K, Rhee K. PLK1 phosphorylation of pericentrin initiates centrosome maturation at the onset of mitosis. J Cell Biol. 2011;195:1093–1101. doi: 10.1083/jcb.201106093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santamaria A, et al. The Plk1-dependent phosphoproteome of the early mitotic spindle. Mol Cell Proteomics. 2011;10:M110 004457. doi: 10.1074/mcp.M110.004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conduit PT, et al. The centrosome-specific phosphorylation of Cnn by Polo/Plk1 drives Cnn scaffold assembly and centrosome maturation. Dev Cell. 2014;28:659–669. doi: 10.1016/j.devcel.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barrera JA, et al. CDK5RAP2 regulates centriole engagement and cohesion in mice. Dev Cell. 2010;18:913–926. doi: 10.1016/j.devcel.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khodjakov A, Rieder CL. The sudden recruitment of gamma-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J Cell Biol. 1999;146:585–596. doi: 10.1083/jcb.146.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol. 2013 doi: 10.1038/nrm3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guderian G, Westendorf J, Uldschmid A, Nigg EA. Plk4 trans-autophosphorylation regulates centriole number by controlling betaTrCP-mediated degradation. J Cell Sci. 2010;123:2163–2169. doi: 10.1242/jcs.068502. [DOI] [PubMed] [Google Scholar]
- 40.Cunha-Ferreira I, et al. The SCF/Slimb ubiquitin ligase limits centrosome amplification through degradation of SAK/PLK4. Curr Biol. 2009;19:43–49. doi: 10.1016/j.cub.2008.11.037. [DOI] [PubMed] [Google Scholar]
- 41.D’Angiolella V, et al. SCF(Cyclin F) controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature. 2010;466:138–142. doi: 10.1038/nature09140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D’Angiolella V, Esencay M, Pagano M. A cyclin without cyclin-dependent kinases: cyclin F controls genome stability through ubiquitin-mediated proteolysis. Trends Cell Biol. 2013;23:135–140. doi: 10.1016/j.tcb.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J, et al. USP33 regulates centrosome biogenesis via deubiquitination of the centriolar protein CP110. Nature. 2013;495:255–259. doi: 10.1038/nature11941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pagan J, Pagano M. FBXW5 controls centrosome number. Nat Cell Biol. 2011;13:888–890. doi: 10.1038/ncb2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Puklowski A, et al. The SCF-FBXW5 E3-ubiquitin ligase is regulated by PLK4 and targets HsSAS-6 to control centrosome duplication. Nat Cell Biol. 2011;13:1004–1009. doi: 10.1038/ncb2282. [DOI] [PubMed] [Google Scholar]
- 46.Holland AJ, Lan W, Niessen S, Hoover H, Cleveland DW. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J Cell Biol. 2010;188:191–198. doi: 10.1083/jcb.200911102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koepp DM, et al. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- 48.Strohmaier H, et al. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413:316–322. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- 49.Cizmecioglu O, et al. Plk2 regulates centriole duplication through phosphorylation-mediated degradation of Fbxw7 (human Cdc4) J Cell Sci. 2012;125:981–992. doi: 10.1242/jcs.095075. [DOI] [PubMed] [Google Scholar]
- 50.Mayor T, Hacker U, Stierhof YD, Nigg EA. The mechanism regulating the dissociation of the centrosomal protein C-Nap1 from mitotic spindle poles. J Cell Sci. 2002;115:3275–3284. doi: 10.1242/jcs.115.16.3275. [DOI] [PubMed] [Google Scholar]
- 51.Soucy TA, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 52.Lydeard JR, Schulman BA, Harper JW. Building and remodelling Cullin-RING E3 ubiquitin ligases. EMBO Rep. 2013;14:1050–1061. doi: 10.1038/embor.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Z, et al. Conserved motif of CDK5RAP2 mediates its localization to centrosomes and the Golgi complex. J Biol Chem. 2010;285:22658–22665. doi: 10.1074/jbc.M110.105965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leonhardt H, et al. Dynamics of DNA replication factories in living cells. J Cell Biol. 2000;149:271–280. doi: 10.1083/jcb.149.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bond J, et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet. 2005;37:353–355. doi: 10.1038/ng1539. [DOI] [PubMed] [Google Scholar]
- 57.Rauch A, et al. Mutations in the pericentrin (PCNT) gene cause primordial dwarfism. Science. 2008;319:816–819. doi: 10.1126/science.1151174. [DOI] [PubMed] [Google Scholar]
- 58.Zimmerman WC, Sillibourne J, Rosa J, Doxsey SJ. Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol Biol Cell. 2004;15:3642–3657. doi: 10.1091/mbc.E03-11-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matsuo K, Nishimura T, Hayakawa A, Ono Y, Takahashi M. Involvement of a centrosomal protein kendrin in the maintenance of centrosome cohesion by modulating Nek2A kinase activity. Biochem Biophys Res Commun. 2010;398:217–223. doi: 10.1016/j.bbrc.2010.06.063. [DOI] [PubMed] [Google Scholar]
- 60.Buchman JJ, et al. Cdk5rap2 interacts with pericentrin to maintain the neural progenitor pool in the developing neocortex. Neuron. 2010;66:386–402. doi: 10.1016/j.neuron.2010.03.036. [DOI] [PubMed] [Google Scholar]
- 61.Jeong YT, et al. FBH1 promotes DNA double-strand breakage and apoptosis in response to DNA replication stress. J Cell Biol. 2013;200:141–149. doi: 10.1083/jcb.201209002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carrano AC, Pagano M. Role of the F-box protein Skp2 in adhesion-dependent cell cycle progression. J Cell Biol. 2001;153:1381–1390. doi: 10.1083/jcb.153.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tighe A, Staples O, Taylor S. Mps1 kinase activity restrains anaphase during an unperturbed mitosis and targets Mad2 to kinetochores. J Cell Biol. 2008;181:893–901. doi: 10.1083/jcb.200712028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.MacCoss MJ, et al. Shotgun identification of protein modifications from protein complexes and lens tissue. Proc Natl Acad Sci U S A. 2002;99:7900–7905. doi: 10.1073/pnas.122231399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Florens L, Washburn MP. Proteomic analysis by multidimensional protein identification technology. Methods Mol Biol. 2006;328:159–175. doi: 10.1385/1-59745-026-X:159. [DOI] [PubMed] [Google Scholar]
- 66.Zhang Y, Wen Z, Washburn MP, Florens L. Effect of dynamic exclusion duration on spectral count based quantitative proteomics. Anal Chem. 2009;81:6317–6326. doi: 10.1021/ac9004887. [DOI] [PubMed] [Google Scholar]
- 67.McDonald WH, et al. MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun Mass Spectrom. 2004;18:2162–2168. doi: 10.1002/rcm.1603. [DOI] [PubMed] [Google Scholar]
- 68.Zhang Y, Wen Z, Washburn MP, Florens L. Improving proteomics mass accuracy by dynamic offline lock mass. Anal Chem. 2011;83:9344–9351. doi: 10.1021/ac201867h. [DOI] [PubMed] [Google Scholar]
- 69.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. βTrCP targets Cep68 for degradation during mitosis.
a) Cep68 immunofluorescence in interphase and metaphase. U-2OS cells were transfected with control siRNA or Cep68 siRNA and analyzed 48 hours post-transfection. Cells were fixed and analyzed by immunofluorescence using Cep68 antibodies (red) and γ-tubulin antibodies (green). The areas in the white boxes are shown at higher magnification directly below the corresponding image. Cep68 is absent from mitotic centrosomes. Depletion of Cep68 by siRNA demonstrates that the Cep68 antibody specifically recognizes Cep68. Scale bars represent 1 μm.
b) Cep68 levels during a release from prometaphase. HeLa cells stably expressing FLAG-HA-Cep68 were synchronized in prometaphase with nocodazole. Round, prometaphase cells were collected by mitotic shake-off and released into freh media. Cells were collected at the indicated times and lysed for immunoblotting as indicated.
c) Cep68 degradation is mediated by βTrCP. HeLa cells stably expressing FLAG-HA-Cep68 were transfected with βTrCP siRNA. Cells were synchronized by double-thymidine arrest and released into fresh media for the indicated times, before processing as in Figure 1a.
d) Multidimensional Protein Identification Technology analysis of a FLAG-Cep68 immunopurification from HEK293T cells, listing the number of spectra, peptides, and the dNSAF (distributed normalized spectra abundance factor) for the indicated co-purifying proteins.
e) βTrCP interacts with Cep68. HEK293T cells were transfected with FLAG-tagged centrosomal proteins (CEPs). Cell lysates were immunoprecipitated with an anti-FLAG resin, and immunoprecipitates were probed with antibodies to βTrCP1. PLK1 and PLK4 are known βTrCP-interacting proteins. Asterisks denote expression of FLAG-tagged CEPs.
f) Different siRNA sequences targeting βTrCP prevent the downregulation of Cep68 in prometaphase, whereas siRNA targeting Cdc20 does not. HeLa cells were transfected with the indicated siRNA sequences. Prometaphase cells (PM) were harvested by mitotic shake-off after overnight nocodazole treatment. NS, non-synchronized. Please note that Cdc20 silencing synchronizes cells in PM, as shown by the phosphorylation state of Cdc27 (see lane 8).
Supplementary Figure 2. Mapping of the βTrCP degron in Cep68 and identification of PLK1 as the kinase phosphorylating Cep68 degron.
a) Schematic representation of Cep68 mutants used in mapping experiments. Cep68 mutants found to interact with βTrCP or Cep215 are indicated by the symbol (+). ‘−/+’ denotes reduced binding.
b) Mapping the βTrCP binding region in Cep68 with Cep68 truncation mutants. HEK293T cells were transfected with empty vector (EV), FLAG-tagged Cep68, or the indicated FLAG-tagged Cep68 constructs. Whole cell extracts were immunoprecipitated with an anti-FLAG resin, and immunoprecipitates were immunoblotted with the indicated antibodies.
c) Alignment of the βTrCP binding motif in Cep68 orthologs and previously reported βTrCP substrates. Critical amino acids required for βTrCP binding are highlighted in red.
d) Ser332 is required for binding to βTrCP1. HEK293T cells were transfected with empty vector (EV), FLAG-tagged Cep68, or the indicated FLAG-tagged Cep68 mutants. Cell lysates were immunoprecipitated with an anti-FLAG resin, and immunoprecipitates were probed with the indicated antibodies.
e) PLK1 inhibition prevents Cep68 degradation in prometaphase. HeLa cells expressing FLAG-HA-Cep68 were synchronized by double-thymidine arrest, as in Figure 1b. Where indicated, cells were treated with a PLK1 inhibitor (BI2536), an Aurora kinase inhibitor (VX680), or an Eg5 inhibitor (monastrol) for three hours prior to their collection and analysis by immunoblotting.
f) PLK1 phosphorylates Cep68 on Ser332 in vitro. Recombinant, purified GST-Cep68, GST-Cep68(S332A), or GST alone were incubated with ATP and increasing amounts of purified PLK1. Proteins were detected by immunoblotting as indicated.
g) PLK1 inhibition prevents the in vivo phosphorylation of Cep68 on Ser332. HeLa cells expressing either inducible FLAG-Cep68 or FLAG-Cep68(S332A) were released from a double-thymidine arrest. Seven hours after release, cells were treated with nocodazole and, where indicated, either BI2536 (a PLK1 inhibitor) or MLN4924 (a CRL inhibitor). Cells were then harvested at the indicated time points. Cep68 or Cep68(S332A) was immunoprecipitated from cell lysates using an anti-FLAG resin. Whole cell lysates (WCL) and immunoprecipitates were immunoblotted as indicated.
h) Analysis of Cep68 binding to PLK1 following BI2536 treatment by Multidimensional Protein Identification Technology. The table lists the total number of spectral counts and the dNSAF values for Cep68 and PLK1 in the untreated and the treated samples.
Supplementary Figure 3. Cep68 degradation does not promote centrosome separation and bipolar spindle assembly.
a) HA-Cep68 is expressed at near to physiological levels. HeLa cells or HeLa cells stably expressing pBABE-HA-tagged Cep68, Cep68(S332A), or Cep68(Δ331-337) were synchronized by a double-thymidine arrest and harvested at the G1/S transition, in G2 phase (eight hours after release), or in prometaphase (PM). Cell lysates were immunoblotted as shown.
b) Bipolar spindles assemble in the presence of Cep68(S332A). Cells expressing inducible FLAG-tagged Cep68 or Cep68(S332A) were fixed and stained with anti-FLAG and antiα–tubulin antibodies. Cells in metaphase were analyzed by immunofluorescence. This experiment was reproduced twice.
c) Expression of FLAG-Cep68(S332A) does not prevent centrosome separation in early mitosis. Cells expressing inducible FLAG-tagged Cep68 or Cep68(S332A) were fixed and analyzed by immunofluorescence. The percentage of cells with separated centrosomes in prophase or prometaphase was scored. ≥100 cells were counted for each condition from one experiment. n=cell number.
d) Time spent in mitosis for cells expressing Cep68 or Cep68(S332A). Cells expressing inducible FLAG-tagged Cep68 or Cep68(Δ331-338) were synchronized by double thymidine release. Live cell microscopy was used to calculate time in mitosis (judged from rounded cell morphology). Where indicated, monastrol (50 μm) was added to cells eight hours after release. Bars represent mean ± standard deviation (S.D.). n=30 cells per condition. ns=not significant. This experiment was performed once.
e) Centrosome disjunction is not inhibited by expression of Cep68(S332A). Cells expressing inducible FLAG-Cep68 or FLAG-Cep68(S332A) were arrested with monopolar spindles in prometaphase with 100 μm monastrol. The distance between γ-tubulin foci was measured using Softworx software. Cep68: n=28; Cep68(S332A): n=27. Bars represent mean ± standard deviation (S.D.). ns=not significant.
Supplementary Figure 4. Cep215 levels are reduced at the intercentrosomal linker in Cep68 depleted cells.
a) Cep68 depletion results in removal of Cep215 from the intercentrosomal linker in interphase. U-2OS cells were transfected for 72 hours with siRNAs targeting Cep68 or a non-targeting control sequence. Cells were fixed and stained with antibodies to γ-tubulin (green) and the specified centrosome cohesion factors (red). After Cep68 silencing, only a small amount of Cep215 is observed on interphase centrioles. The graph represents the relative intensity of Cep215 after Cep68 depletion. Bars represent mean ± standard deviation (S.D.). Control siRNA: n=10 cells; Cep68 siRNA: n=11 cells. ***P=0.0006.
b) Cep68 knockdown does not affect the total levels of Cep215. U-2OS cells were transfected with siRNAs targeting Cep68 or Cep215. Seventy-two hours post-transfection, cells were lysed for immunoblotting as indicated.
Supplementary Figure 5. The interaction between Cep215 and Cep68 is not mediated by PCNT.
a) Cep68 interacts with Cep215 and PCNT independently. Cells expressing doxycycline (DOX)-inducible FLAG-tagged Cep68 or Cep68(S332A) were transfected with the indicated siRNA sequences. Cep68 or Cep68(S332A) were immunoprecipitated from whole cell lysates (WCL) with anti-FLAG resin, and both immunoprecipitates and WCLs were immunoblotted with the indicated antibodies. Cep215 interacts with Cep68 when PCNT is depleted in interphase and mitosis. Likewise, PCNT interacts with Cep68(S332A) when Cep215 is depleted. AS, asynchronous. PM, prometaphase. The asterisk denotes a non-specific band.
b) PCNT and PCNT(R2231A) localization in cytokinesis. HeLa cells were transiently transfected with FLAG-tagged PCNT or PCNT(R2231A). Cells in cytokinesis were stained for immunofluorescence using antibodies recognizing FLAG (red) and CP110 (green). Scale bars represent 1 μm.
Supplementary Figure 6. Cep68 degradation is required for centriole separation after disengagement.
Quantification of cells containing 1 c-Nap1 dot in cells expressing Cep68(S332A) using conventional microscopy. HeLa cells expressing HA-tagged Cep68 or Cep68(S332A) were synchronized by double-thymidine arrest and allowed to progress into the next G1 phase (15 hours after release). Where indicated, cells were transfected with Cep215 siRNA during the first release from double-thymidine arrest. G1 cells were fixed and stained for c-Nap1 (green), centrin 2 (red), and Cep68 (α-HA) (blue) to determine their engagement status. The magnification bar represents 1 μm. The areas in the white boxes are shown at higher magnification directly above the corresponding image. The graph shows the quantification of disengagement. G1 cells with 1: 2 ratio of c-Nap1:Centrin 2 foci were scored as engaged. n≥100 cells for each experiment from three independent experiments were analysed. **P ≤ 0.01. Bars represent the mean ± standard deviation (S.D.) from n= three experiments.