SUMMARY
SMU.1745c, encoding a putative transcriptional regulator of the MarR family, maps to a location proximal to the fab gene cluster in Streptococcus mutans. Deletion of the SMU.1745c (fabTSm) coding region resulted in a membrane fatty acid composition comprised of longer-chained, unsaturated fatty acids (UFA), compared with the parent strain. Previous reports have indicated a role for FabT in regulation of genes in the fab gene cluster in other organisms, through binding to a palindromic DNA sequence. Consensus FabT motif sequences were identified in S. mutans in the intergenic regions preceding fabM, fabTSm and fabK in the fab gene cluster. Chloramphenicol acetyltransferase (cat) reporter fusions, using the fabM promoter, revealed elevated transcription in a ΔfabTSm background. Transcription of fabTSm was dramatically elevated in cells grown at pH values of 5 and 7 in the Δ fabTSm background. Transcription of fabTSm was also elevated in a strain carrying a deletion for the carbon catabolite repressor CcpA. Purified FabTSm and CcpA bound to the promoter regions of fabTSm and fabM. Hence, the data indicate that FabTSm acts as a repressor of fabM and fabTSm itself and the global regulator CcpA acts as a repressor for fabTSm.
Keywords: acid stress response, fatty acid synthesis, Streptococcus mutans
INTRODUCTION
Streptococcus mutans, an etiologic agent of dental caries in humans, colonizes human teeth via the production of an extracellular glucan matrix that acts to bind the organism to oral hard surfaces and initiates the accumulation of glucans with multiple species of oral bacteria, leading to the formation of the niche referred to as dental plaque (Bowen & Koo, 2011). As a result of glucan production, S. mutans remains in the oral cavity throughout human lifetimes. Following its colonization of teeth, S. mutans is confronted with a variety of environmental stressors, including the need to survive the acidic conditions in plaque that arise from the metabolism of dietary carbohydrates to organic acids. The accumulation of acids, without removal of plaque, leads to erosion of tooth enamel and the progression of dental caries (Loesche, 1986; Nyvad et al., 2013).
A key acid-adaptive strategy employed by S. mutans is the ability to effect substantial changes in membrane fatty acid composition in response to external acidification (Fozo & Quivey, 2004b; Fozo et al., 2004; Lemos & Burne, 2008). As pH values in plaque fall, the membrane fatty acid composition changes from a profile predominant in short-chained, saturated fatty acids (SFA) to a profile containing elevated levels of long-chained, unsaturated fatty acids (UFA) (Quivey et al., 2000; Fozo & Quivey, 2004b). Loss of UFAs results in extreme sensitivity to acidic pH values by S. mutans, in vitro, and to greatly diminished caries in a rat model (Fozo et al., 2007).
Clearly, membrane fatty acid composition in S. mutans is important to the pathogenesis of the organism. However, little is presently known about the regulatory mechanisms for controlling production of its membrane fatty acid repertoire in response to changes in the external environment. Reports from studies conducted with Streptococcus pneumoniae and Lactococcus lactis have shown that a transcriptional regulator, referred to as FabT, is encoded at a locus immediately upstream of the fab gene cluster (Lu & Rock, 2006; Eckhardt et al., 2013). FabTSp binds to a DNA sequence located upstream of fabT and fabK, but not upstream of fabM. Loss of FabTSp resulted in a reduced growth rate in medium held at a pH value of 5.5 (Lu & Rock, 2006; Eckhardt et al., 2013). Subsequent efforts showed that FabTSp binds acyl-acyl carrier protein (ACP) as a co-effector for regulation and that C18:1, cis-vaccenic acid, provided the strongest effects on binding of FabTSp to its target site (Jerga & Rock, 2009). Similarly, FabTLl acts to regulate fatty acid biosynthesis in L. lactis, via a consensus motif located upstream of fabTLl, fabD, accC and fabI and deletion of fabTLl led to an increase in saturated acyl chains in the membrane of the organism (Eckhardt et al., 2013). Streptococcus mutans occupies a very different ecological niche from S. pneumoniae, although all of the fab genes, including the relative position of the FabM cis-trans isomerase-encoding gene, are identically organized in their respective genomes (Tettelin et al., 2001; Ajdic et al., 2002). Gene organization in L. lactis is comparable to the streptococcal arrangement; however, L. lactis lacks fabM and possesses a fabI gene and a second fabZ elsewhere in the genome (Wegmann et al., 2007).
In the present study, we show that the gene at locus SMU.1745c (as annotated by NCBI) encodes a transcriptional regulator that is orthologous to the S. pneumoniae (Lu & Rock, 2006; Jerga & Rock, 2009) and the L. lactis FabT (Eckhardt et al., 2013). Data from biochemical and physiological experiments revealed significant differences between the S. mutans approach to the use of FabT as a regulator of fatty acid production and the regulatory scheme shown in the orthologs.
We report here that loss of fabTSm in S. mutans does not exert gross changes in membrane fatty acid composition; however, loss of FabTSm resulted in broad changes to the S. mutans transcriptome following growth at pH values of 5 and 7. Results from experiments using cat-reporter fusions indicated an approximately two-log-order dynamic range of fatty acid gene transcription that was highly dependent on external pH values. We also provide evidence for the involvement of the global regulator CcpA in fab gene transcription in response to environmental changes. We conclude that relatively subtle changes in membrane composition have dramatic consequences for cellular homeostasis in S. mutans, underscoring its central role in the ability of the organism to cope with changes in external pH values.
METHODS
Bacterial strains
Streptococcus mutans UA159 (Murchison et al., 1986), the genomic type strain (Ajdic et al., 2002), was used in these studies. MU1591, the fabTSm deletion strain, is derived from UA159. Deletion of the fabTSm locus, SMu1591 (GenBank designation: SMU.1745c) was performed essentially as previously described (Lau et al., 2002; Sheng et al., 2010; Santiago et al., 2012). Strain MU1591 was created by replacement of the SMU.1745c open reading frame with a non-polar erythromycin-resistance cassette (ErmR) flanked by approximately 400 bp upstream and downstream of the gene using primer pair MU1591P7UPF (5’-TAA GAAATAATCAATCGC-3′) and MU1591P8DNR (5’-AAGCCTTTGTGAATGTCG-3′) to facilitate homologous recombination. The intergenic region preceding the SMU.1745c coding region was left intact upstream of the erythromycin-resistance cassette. Strains were maintained on brain–heart infusion (BHI) agar medium (BD/Difco, Franklin Lakes, NJ) and supplemented with erythromycin (Erm) at 5 µg m−1, where necessary. Batch cultures were grown in tryptone–yeast extract (TY) medium supplemented with 1% glucose. Steady-state cultures were grown in a BioFlo2000 (New Brunswick, Edison, NJ) in TY medium at a dilution rate of D = 0.24 h−1 as previously described (Fozo & Quivey, 2004b; Kuhnert et al., 2004). Cells were limited by glucose (2.3 mm) and pH maintenance was achieved by addition of 2 m KOH. Steady-state cells used in these studies were harvested after a minimum of 10 generations at each pH value.
Growth rate and yield were assessed using a Bioscreen C automated growth reading system (Growth Curves USA, Piscataway, NJ) essentially as described elsewhere (Kajfasz et al., 2010; Derr et al., 2012).
Complementation of fabTSm locus
Primer pair 1591 PromBglUp2 and 1591 PromBglDwn were used for polymerase chain reaction (PCR) amplification of the fabTSm coding region and cognate promoter fragment from S. mutans UA159. The purified amplicon was cloned into pCRBlunt (Life Technologies, Carlsbad, CA) as per the manufacturer’s direction, resulting in the plasmid pCRmarRLP8. The BglII restriction sites introduced by PCR were used to excise the fabTSm plus promoter fragment for subsequent cloning into pSUGK-Bgl, an integration vector (Derr et al., 2012), and transformants were selected for KmR. Proper integration of the fabTSm promoter plus coding region into the BglII site of pSUGK-Bgl, in the opposite orientation to gtfA, was determined by colony PCR with primer pair 1591 PromBglDwn and gtfAseqKan (see Table S4). The appropriate construct, pSUGKmarRLP7, was transformed into MU1591 and colonies were selected on BHI agar medium containing kanamycin (1 mg ml−1). Transformants were screened by colony PCR with the primer pair 1591 PromBglDwn and gtfAseqKan, and also by Southern blotting with a gtfA-specific DNA probe (data not shown). One such construct, named S. mutans UR294, contained the fabTSm gene, preceded by its cognate promoter, in the gtfA locus.
Stress tolerance assays
Samples were taken from steady-state cultures of S. mutans UA159 (parent) and MU1591 (ΔfabTSm deletion strain) for comparison of sensitivity to acid and hydrogen peroxide (Belli & Marquis, 1991; Quivey et al., 1995). Briefly, strains were tested for sensitivity to acid-mediated killing by harvesting steady-state samples from the chemostat at culture pH values of 7 and 5, resuspending cell pellets in 0.1 m glycine, pH 2.5, and stirring at room temperature. Aliquots were removed at 0, 15, 30 and 60-min intervals, serially diluted, and plated on solid BHI medium. Hydrogen peroxide sensitivity assays were also performed on the cells as previously described (Belli & Marquis, 1991; Quivey et al., 1995). Briefly, steady-state samples harvested from the chemostat, at culture pH values of 7 and 5, were resuspended in BHI medium, and hydrogen peroxide was added to a final concentration of 16.3 mm. Aliquots were removed at 0, 15, 30 and 60 min, serially diluted, and plated on BHI agar medium. Viable cells from each condition were counted and used to calculate log (Nt/No), where Nt = number of colonies obtained at a specific time point, and N0 = number of colonies at time zero.
Membrane fatty acid determination
Membrane fatty acid composition was analysed in cultures of the parent strain (S. mutans UA159), the ΔfabTSm strain (MU1591), and the fabTSm-complemented strain (UR294) grown in TYG medium in batch conditions overnight in either a 5% [volume/volume (v/v)] CO2/95% air incubator (batch), in steady-state at pH values of 7 and 5 (described above), and in biofilms. Biofilm cultures of the parent strain and MU1591 (ΔfabTSm strain) were grown on glass slides in TY medium supplemented with 1% sucrose (TYS). Slides were transferred to tubes containing fresh medium every day for 5 days. At the end of the growth period, biofilms were harvested from glass slides and sonicated to break up chains.
Harvested cell pellets from each growth condition were washed twice with sterile H2O, and stored at −80°C until the samples were sent to Microbial ID, Inc. (Newark, DE) for analysis. Membrane fatty acid content was determined by GC-FAME as described elsewhere (Bligh & Dyer, 1959; Fozo & Quivey, 2004b).
Transcriptional assays
Reverse transcription (RT-) PCR were performed to determine the extent of co-transcription in the fatty acid biosynthesis cluster. Primer pairs TS12/TS8, TS21/TS10, TS14/TS9, TS22/TS7, TS20/TS11 (detailed in the Table S4) were used to examine transcription across the non-coding regions between fabM/fabTSm, acp/fabK, fabH/acp, fabG/fabF and fabF/accB, respectively. Total RNA (43 ng) isolated from a steady-state culture of S. mutans UA159 grown to a pH value of 7 was incubated with the anti-sense primer in each primer pair with or without SuperScript II Reverse Transcriptase as per the manufacturer’s instructions (Life Technologies). Each primer pair was also used in a standard PCR with UA159 genomic DNA as a positive control.
Single-copy reporter gene fusions were constructed using PCR to amplify the promoter fragments for fabTSm (SMU.1745c), fabM (SMU.1746c) and fabK (SMU.1742c). Transcriptional activity was determined from either an intact promoter fragment or from a truncated promoter fragment lacking the distal FabTSm binding site (described in Fig. 4). Primer pairs used are detailed (see Table S4). Amplicons were subcloned into pCRBlunt (Life Technologies) and appropriate constructions were verified by nucleotide sequencing. Using incorporated restriction sites (SacI/BglII), promoter fragments were subcloned into an integration vector containing a promoterless chloramphenicol acetyltransferase (cat) gene derived from Staphylococcus, pJL84 (Santiago et al., 2012). Integration of the promoter-CAT construction occurs within the intergenic regions between mtlA (SMU.1185c) – glmS (SMU.1187c) and mtlD (SMU.1182c) – phnA (SMU.1180c). Streptococcus mutans UA159 and MU1591 were transformed with the pJL84 constructs and colonies were selected on BHI agar medium containing kanamycin (1 mg ml−1). Proper integration of the specific promoter-cat constructs was verified by colony PCR using the primer pair CATJL and ICphnA (see Table S4) and nucleotide sequencing using the CATJL primer. The resulting strains UR211 (fabM-cat in UA159), UR221 (fabK-cat in UA159), UR222 (fabTSm-cat in UA159), UR237 (fabK-cat in MU1591), UR242 (fabTSm-cat in MU1591), UR243 (fabM-cat in MU1591), UR266 (fabK truncated promoter-cat in UA159), UR272 (fabM truncated promoter-cat in UA159), UR273 (fabK truncated promoter-cat in MU1591) and UR274 (fabM truncated promoter-cat in MU1591) are detailed in Table 1.
Figure 4.
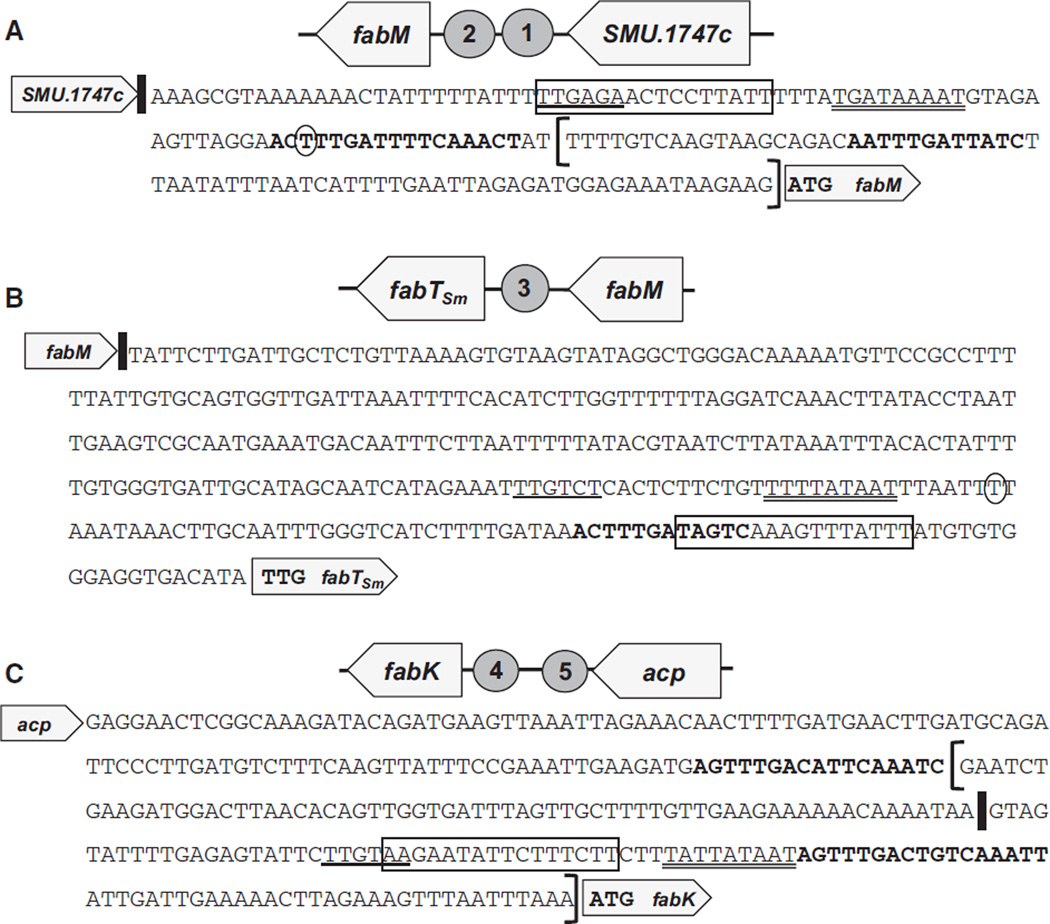
Intergenic regions (IGRs) containing consensus FabT binding sites. In all panels: the putative −35 sequence (single underline), −10 sequence (double underline), and predicted transcriptional start site (circled nucleotide in A and B) (as predicted by BPROM, Softberry, promoter prediction); vertical bars indicate translational stop sites; sequence in boldface is the predicted FabTSm binding motif and boxed sequence is the predicted cre site (predicted by RegPrecise: Novichkov et al., 2010). Brackets in (A) and (C) indicate the shorter, truncated promoters lacking one FabTSm motif used in transcriptional fusion assays. (A) IGR preceding fabM. (B) IGR preceding fabTSm. (C) IGR preceding fabK.
Table 1.
Strains and plasmids
| Strains | Genotype description | References |
|---|---|---|
| Streptococcus mutans | ||
| UA159 | Genomic type strain | Murchison et al. (1986), Ajdic et al. (2002) |
| MU1446 | ccpA deletion strain | Santiago et al. (2013) |
| MU1591 | fabTSm deletion strain | This study |
| UR211 | fabM-cat in UA159 | This study |
| UR221 | fabK-cat in UA159 | This study |
| UR222 | fabTSm-cat in UA159 | This study |
| UR237 | fabK-cat in MU1591 | This study |
| UR242 | fabTSm-cat in MU1591 | This study |
| UR243 | fabM-cat in MU1591 | This study |
| UR266 | fabK truncated promoter-cat in UA159 | This study |
| UR272 | fabM truncated promoter-cat in UA159 | This study |
| UR273 | fabK truncated promoter-cat in MU1591 | This study |
| UR274 | fabM truncated promoter-cat in MU1591 | This study |
| UR294 | fabTSm complement strain | This study |
| UR310 | fabTSm-cat in MU1446 | This study |
| UR313 | fabM-cat in MU1446 | This study |
| Escherichia coli | ||
| Rosetta(λDE3) | F- ompT hsdSB(rB−mB−) gal dcm (DE3) pRARE (CmR) | Novagen |
| TOP10 | F- mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu) 7697 galU galK rpsL (StrR) endA1 nupG |
Invitrogen |
| Plasmids | ||
| pCRBlunt | Cloning vector for blunt end PCR products | Invitrogen |
| pET19b | N-terminal His-tag expression vector | Novagen |
| pGEM7Zf+ | Cloning vector | Promega |
| pJL84 | Promoterless cat integration vector | Santiago et al. (2012) |
| pJLmarR | Promoter of SMU.1745c (fabTSm) in pJL84 | This study |
| pJLfabK | Promoter of SMU.1742c (fabK) in pJL84 | This study |
| pJLfabKtrunc | fabK promoter missing distal FabTSm binding site in pJL84 | This study |
| pJLfabM | Promoter of SMU.1746c (fabM) in pJL84 | This study |
| pJLfabMtrunc | fabM promoter missing distal FabTSm binding site in pJL84 | This study |
| pSUGK-Bgl | Low-copy cloning vector derived from pBGK & pSU20 with BglII cloning site | Derr et al. (2012) |
| pSUGmarRLP7 | pSUGK with fabTSm coding region and native promoter | This study |
| pCRmarRLP8 | pCRBlunt with fabTSm coding region and native promoter | This study |
Reporter constructs for examining the global regulator CcpA were constructed in a similar manner as detailed above. The fabTSm-cat construct, pJLmarR, and the fabM-cat construct, pJLfabM, were transformed into S. mutans MU1446, carrying a deletion in ccpA (Santiago et al., 2013) (Table 1). Colonies were selected on BHI agar medium containing kanamycin (1 mg m−1). Proper integration of the specific promoter-cat constructs was verified by colony PCR using the primer pair CATJL and ICphnA (see Table S4) and the strains were named S. mutans UR310 and UR313, respectively.
The CAT assays, as modified for oral streptococci, were performed as previously described (Shaw et al., 1979; Chen et al., 1998; Kuhnert et al., 2004). Briefly, all reporter strains were grown in batch culture to a final optical density at 600 nm of approximately 0.5 in a 5% (v/v) CO2/95% air incubator in BHI medium titrated to pH 5 with HCl or buffered to pH 7 with 50 mm KPO4 buffer; or in unbuffered BHI medium or in FMC minimal medium +1% glucose (Terleckyj & Shockman, 1975; Terleckyj et al., 1975). Cells were harvested and stored at −80°C until whole cell lysates were prepared. Reactions were initiated by the addition of 20 µl 5 mm chloramphenicol. Time–course of the reactions was followed by absorbance at 412 nm. Rates were calculated as nmol min−1 of product formed. Protein concentration was estimated using the BioRad protein assay reagent (BioRad, Hercules, CA) (Bradford, 1976). CAT activity units were calculated as nmol min−1 mg protein−1.
RNA purification
RNA used in the experiments outlined here was isolated from four, independent, steady-state cultures of S. mutans UA159 and MU1591 grown to pH values of 7 and 5 and purified as previously described (Abranches et al., 2006; Baker et al., 2014). Briefly, S. mutans cells were homogenized in a MiniBead Beater 8 (BioSpec Products, Bartlesville, OK). Cell lysates were subjected to three hot acid–phenol–chloroform extractions, and the nucleic acid was precipitated overnight at −20°C. RNA pellets were resuspended in nuclease-free H2O and treated with DNase I (Ambion, Austin, TX) at 37°C for 30 min. The RNA was purified again using an RNeasy minikit (Qiagen, Valencia, CA), including a second on-column DNase treatment, performed as recommended by the supplier. RNA concentrations were determined using a NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA), and samples were run on a 1.2% FlashGel (Lonza Inc., Allendale, NJ) to verify RNA integrity.
Microarray analysis
Streptococcus mutans UA159 microarray chips were provided by The Institute for Genomic Research (TIGR; JCVI). A reference RNA, isolated from S. mutans UA159 cells grown in BHI medium to an optical density at 600 nm of 0.5, was used in every experiment. The reference RNA was purified as above, aliquoted, and stored at −80°C.
All sample RNAs were purified as described above and used to generate cDNA according to the protocol provided by the Pathogen Functional Genomics Resource Center (PFGRC) at the J. Craig Venter Institute (ftp://ftp.jcvi.org/pub/data/PFGRC/MAIN/pdf_files/protocols/M007.pdf) with minor modifications as detailed previously (Abranches et al., 2006; Baker et al., 2014). Labeled cDNA from four independent cultures of S. mutans UA159 and MU1591 grown to steady-state pH values of 7 and 5 were hybridized overnight at 42°C with labeled reference cDNA, generating a total of 16 slides. Hybridization was carried out in a MAUI 4-Bay Hybridization system (BioMicro Systems, Inc., Salt Lake City, UT), washed according to PFGRC protocols, and scanned using a GenePix 4000b Microarray Scanner (Molecular Devices, Inc., Sunnyvale, CA). After scanning, single channel images were gridded using tigr Spotfinder, normalized and analysed using the method previously outlined (Abranches et al., 2006) and available from the tm4 Microarray Software Suite (http://www.tm4.org/). Statistical analysis was carried out using BRB array tools (http://linus.nci.nih.gov/BRB-ArrayTools.html) with a cutoff P-value of 0.01 and at least two-fold change in expression. The microarray data have been deposited in the NCBI Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) under GEO accession number GSE36960.
Microarray results were validated by performing real-time PCR on a subset of genes. Primers for the genes tested are listed in Table S4.
Real-time PCR analysis
Real-time quantitative RT-PCR was used to validate microarray experiments. For this, 0.5 µg RNAs isolated (as described above) from four, independent steady-state cultures of S. mutans UA159 and MU1591, grown to pH values of 7 and 5, were used to synthesize cDNA using the High Capacity cDNA Reverse Transcription Kit (Life Technologies) containing random primers. Control reactions were also conducted to determine the contribution of any DNA signal. Gene-specific primers (Table S4) used in all real-time PCR experiments were designed using Beacon Designer 2.0 software (Premier Biosoft International, Palo Alto, CA). Standard curves for each gene were prepared as described elsewhere (Abranches et al., 2006; Buckley et al., 2014). Briefly, the concentration of the purified amplicon derived from gene-specific primers was determined using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). The template concentration was used to calculate the template copy number, and serial dilutions were performed to create a standard curve. Reactions were carried out in a StepOne Plus Real Time PCR System (Life Technologies) using Power SYBR Green Master Mix (Life Technologies) with three replicates per sample and wells containing dH2O were used to determine background levels of fluorescence. Melt curves of the amplification reactions were verified to contain a single peak and reaction mixtures from random wells were examined by gel electrophoresis to ascertain that a single band was obtained. The relative quantities for each replicate of a sample were averaged and a standard deviation was calculated.
Expression of FabTSm
The S. mutans fabTSm gene was PCR amplified using primer pair MarR Fwd XhoI and MarR Rev BamHI (Table S4). The resulting amplicon was digested and cloned into the XhoI and BamHI sites of pET19b (Novagen, Rockland, MA). Proper construction was verified by nucleotide sequencing and a correct clone was transformed into E. coli Rosetta competent cells (Novagen) for protein expression. One such transformant was grown at 37°C, with shaking, in LB medium containing ampicillin (100 µg m−1) to an OD600 of 0.6, then induced with 1 mm IPTG (isopropyl β-d-galactopyranoside) and allowed to grow overnight, with shaking, at 20°C. Cells were harvested and lysed with addition of lysozyme to 1 mg m−1 and mechanical disruption in a Mini BeadBeater (BioSpec Products). Soluble proteins were applied to a Ni-nitrilotriacetic acid (NTA) agarose column (Qiagen) and washed with 50 mm NaH2PO4, 300 mm NaCl, 20 mm imidazole, pH 8 as per the manufacturer’s instructions for purification of native protein. His-tagged proteins were eluted with 50 mm NaH2PO4, 300 mm NaCl, 250 mm imidazole, pH 8. Eluant fractions containing recombinant FabTSm protein, as determined by sodium dodecyl sulfate– polyacrylamide gel electrophoresis, were dialysed against 10 mm Tris–HCl, pH 8; 50 mm NaCl; 10% glycerol overnight in preparation for the electrophoretic mobility shift assay (EMSA).
Protein concentration was determined using the method of Bradford (Bradford, 1976) with the BioRad Protein Assay Reagent. Presence of the 6X His tag was verified by Western blot analysis using α-His IgG primary antibody (GE Healthcare, Piscataway, NJ) (data not shown).
Electrophoretic mobility shift assay
Target DNAs containing the full-length intergenic regions preceding fabTSm (323 bp), fabM (163 bp) and fabK (300 bp) were PCR amplified from plasmids pJLmarR, pJLfabM, pJLfabK, respectively (Table 1). The truncated fabM promoter (79 bp) was PCR amplified from the vector pJLfabMtrunc (Table 1). The gel-purified PCR amplicons were end-labeled with [γ-32P]ATP (Perkin Elmer, Waltham, MA) using T4 Polynucleotide Kinase as per the manufacturer’s instructions (Life Technologies).
The EMSAs involving FabTSm were performed essentially as described (Lu & Rock, 2006), with modifications. Briefly, the DNA binding reactions containing 32P-labeled probe (approximately 5000 counts min−1) and purified, His-tagged FabTSm (1000 nm) were incubated for 45 min at room temperature (25°C) in a buffer consisting of 10 mm Tris–HCl, pH 8; 50 mm NaCl; 10 mm EDTA; 10% (v/v) glycerol; 1 mm dithiothreitol in a total volume of 20 µl. For competition experiments, 200-fold excess, cold promoter DNA or 1 µg poly(dG-dC) (Roche Applied Science, Indianapolis, IN) was added to the binding reaction components, including FabTSm, and allowed to preincubate 15 min at room temperature before addition of the labeled probe. Bovine serum albumin (1000 nm) was used as a control for specificity of DNA binding. When used, long-chain acyl-acyl carrier protein, C18:1 acyl-ACP (gift of C. Rock), was added at a concentration of 1500 nm. Samples were separated on 6% non-denaturing gels in 1 × Tris–borate EDTA.
EMSAs involving CcpA [gift of R. Burne; (Abranches et al., 2008)] were performed as previously described. Briefly, DNA fragments containing the full-length intergenic regions preceding fabM, fabTSm and fabK were created as described above. A representative binding reaction (25 µl) consisted of the following mixture: binding buffer [20 mm Tris–HCl, pH 8; 8.7% (v/v) glycerol, 1 mm EDTA; 5 mm MgCl2; 250 mm KCl; 0.5 mm dithiothreitol; 2 µg bovine serum albumin], radiolabeled DNA probe (3000 counts min−1) and purified recombinant CcpA (3000 nm). Binding reactions were incubated at 37°C for 45 min. CcpA-DNA binding was assessed using an 8% non-denaturing gel in 1 × Tris–glycine buffer.
The gels were exposed to a phosphorimager screen and binding was detected with a Molecular FX phosphorimager and BioRad Quantity One software (BioRad).
RESULTS
Identification of the FabTSm ortholog in S. mutans
The SMU.1745c open reading frame in S. mutans UA159, annotated as a transcriptional regulator of the MarR family (Sulavik et al., 1995; Ajdic et al., 2002) (http://www.ncbi.nlm.nih.gov/gene/1028966), shares homology with the previously characterized regulator of membrane fatty acid biosynthesis in S. pneumoniae and L. lactis, both denoted as FabT (Lu & Rock, 2006; Jerga & Rock, 2009; Eckhardt et al., 2013). The deduced amino acid sequence encoded by SMU.1745c is approximately 60% identical and 79% similar to FabT from S. pneumoniae and 36% identical and 57% similar to the L. lactis ortholog. Hereafter, we will refer to the S. mutans ortholog as FabTSm.
Using a PCR-based approach, the fabTSm coding region was deleted, leaving in its place, a non-polar, in-frame erythromycin-resistance marker (ErmR), transcriptionally driven by the native fabTSm promoter. The method of deleting fabTSm was accomplished such that its removal did not eliminate transcription of the immediate downstream gene, fabH (SMU.1744c), as determined from real-time PCR measurements of fabH transcripts in the ΔfabTSm strain (see Fig. S2). Deletion of fabTSm was verified by colony blotting and real-time PCR experiments using RNA isolated from the deletion strain, referred to as MU1591 (see Fig. S1).
Stress tolerance of the fabTSm mutant strain
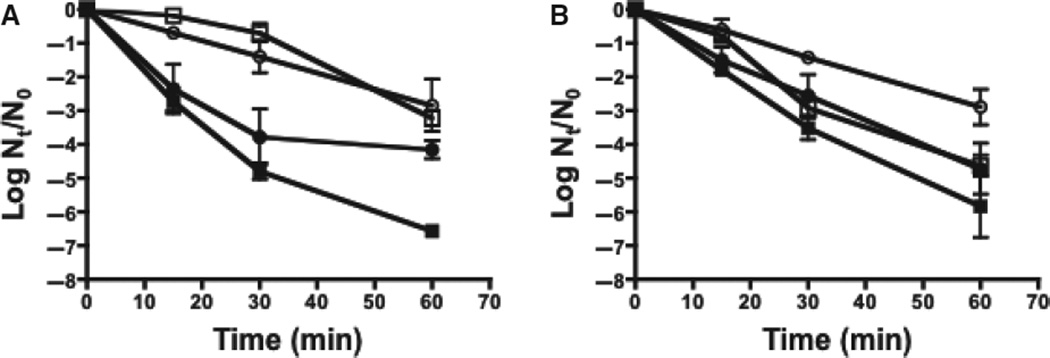
In our previous studies, loss of the fabM gene resulted in a greatly diminished growth rate and a high degree of acid-sensitivity in S. mutans (Fozo & Quivey, 2004b). In contrast, deletion of fabTSm did not affect the final yield, compared with growth of the parent strain, under various growth conditions, including rich medium (BHI) or in medium adjusted to an acidic pH value (BHI pH 5.4); or, in the presence of H2O2 or 8-hydroxyquinoline (see Table S1). In fact, the ΔfabTSm strain exhibited a shorter relative doubling time under acidic (pH 5.4) conditions (Table S1). Previous studies have shown that S. mutans mounts a substantial, and effective, response to acid and oxidative stress following growth at low pH (Belli & Marquis, 1991). Accordingly, the ΔfabTSm strain was further tested for its ability to withstand severe acid and oxidative challenges following growth in steady-state cultures held at pH values of 7 or 5 (Fig. 1). The fabTSm deletion strain was more acid-resistant when grown at pH 7 than the parent strain, and slightly more acid-sensitive when grown at pH 5. The fabTSm mutant strain also exhibited a tolerance to peroxide when grown at both pH 7 and pH 5, compared with the parent strain. Hence, overall, loss of FabTSm resulted in relatively modest physiological effects, under the conditions tested.
Figure 1.
Stress sensitivity assays. Streptococcus mutans UA159 (□) and MU1591 (ΔfabTSm) (○) were grown to steady-state in chemostat cultures to pH values of 5 (open symbol) and 7 (filled symbol). Data are from three independent cultures and represented as Log N/N0. (A) Survival of strains following acid challenge (pH 2.5). (B) Survival of strains following peroxide challenge (16.3 mm).
Loss of FabTSm affects membrane fatty acid composition
Under all conditions tested, deletion of fabTSm resulted in a membrane fatty acid composition with an elevated UFA : SFA ratio compared with the parent strain, UA159 (Table 2A; and see Table S3). With the exception of cells grown to steady-state in cultures at pH 5, which nevertheless exhibited the same trend, deletion of fabTSm resulted in a membrane fatty acid composition consisting of a significantly elevated ratio of long-chained (C18 and C20) to short-chained (C10, C12, C14, and C16) fatty acids, compared with UA159 (Table 2B). The increase in UFAs observed in the S. mutans ΔfabT strain is dissimilar to the effect seen in fabT mutant strains of S. pneumoniae and L. lactis, and suggested a role for FabT regulation of fabM expression in S. mutans.
Table 2.
The unsaturated fatty acid : saturated fatty acid (UFA : SFA) ratio is increased in cultures of MU1591
| Strain | pH | UFA : SFA | ||
|---|---|---|---|---|
| (A) | ||||
| UA159 | 7 | 0.60 ± 0.081 | ||
| UA159 | 5 | 2.00 ± 0.042 | ||
| ΔfabTSm | 7 | 0.78 ± 0.081 | ||
| ΔfabTSm | 5 | 2.19 ± 0.442 | ||
| Strain | pH | Long-chained : Short-chained | ||
| (B) | ||||
| UA159 | 7 | 0.58 ± 0.101 | ||
| UA159 | 5 | 3.19 ± 0.72 | ||
| ΔfabTSm | 7 | 0.82 ± 0.081 | ||
| ΔfabTSm | 5 | 4.33 ± 0.29 | ||
Streptococcus mutans UA159 and MU1591 (ΔfabTSm) were grown to steady-state in chemostat culture to pH values of 7 and 5 in TY medium + 1% glucose.
Panel A: Values represent the UFA : SFA ratios for each culture condition ± standard deviation; n = 3.
Panel B: Values represent the ratio of long-chained membrane fatty acids (C18 or C20) vs. short-chained membrane fatty acids (C10 through C16) ± standard deviation; n = 3.
In both panels:
P< 0.05;
P< 0.005 for the pairwise comparisons between parent strain and MU1591, as determined by Student’s t-test.
The fabTSm complement strain, UR294, behaved similarly to the parent strain, with respect to the ratio of long-chained : short-chained fatty acids; yet, the UFA : SFA ratio was more similar to MU1591 (see Table S2). We attribute the intermediate phenotype of the complement strain (between the parent and deletion strain) to the fact that the complementing copy of fabTSm was expressed from an ectopic locus in the genome (gtfA, SMU.881) and, therefore, may have been removed from potential native regulatory elements. However, results from quantitative real-time PCR showed that fabTSm expression levels in the complement strain, UR294, were similar to the parent strain, indicating no effect on transcription of the gene (Fig. S1).
Identification of FabT binding motifs in the fab gene cluster
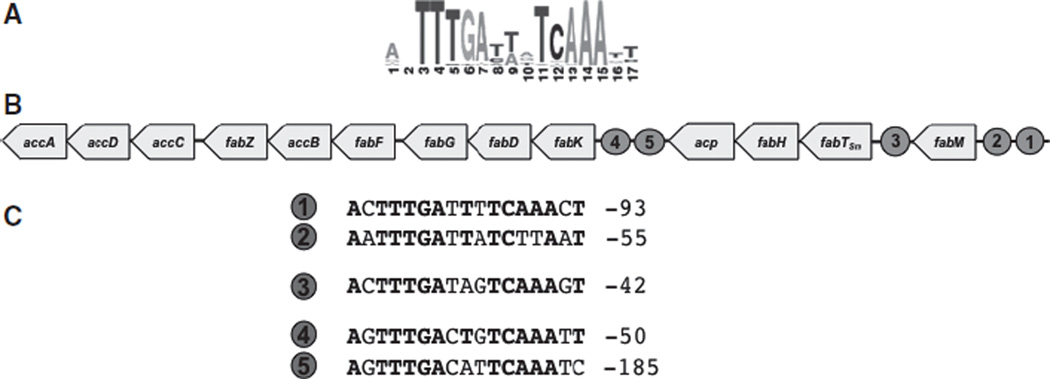
A consensus FabT binding site in Streptococcaceae has been described in the RegPrecise collection of manually curated regulons (http://regprecise.lbl.gov/RegPrecise/regulog.jsp?regulog_id=3571) (Novichkov et al., 2010) (Fig. 2A). The putative motif is shared among various streptococcal species, including S. mutans. The predictions included the possibility of FabT binding sites in promoter regions of the fabM, fabK and fabTSm genes of S. mutans, similar to the predicted, and experimentally verified, locations in the S. pneumoniae genome (Lu & Rock, 2006) and the L. lactis genome (Eckhardt et al., 2013). In S. mutans, there are two sites predicted upstream of both fabK and fabM, and one site upstream of fabTSm (Fig. 2B,C), suggesting that FabTSm may have a role in the expression of these genes.
Figure 2.
FabTSm conserved motifs. (A) Conserved motif for FabTSm binding depicting probability of nucleotide at each position (from http://regprecise.lbl.gov/RegPrecise/gmregulon.jsp?gmproject_id=6926) (Novichkov et al., 2010). (B) Genetic organization of fatty acid biosynthetic genes in Streptococcus mutans and locations of putative FabTSm binding sites adjacent to genes regulated (designated by the numbered circles). (C) Nucleotide sequence of proposed binding site(s) where number on left corresponds to numbered circle in (B) and number on right represents location relative to translational start site. Text in bold indicates conserved sequence.
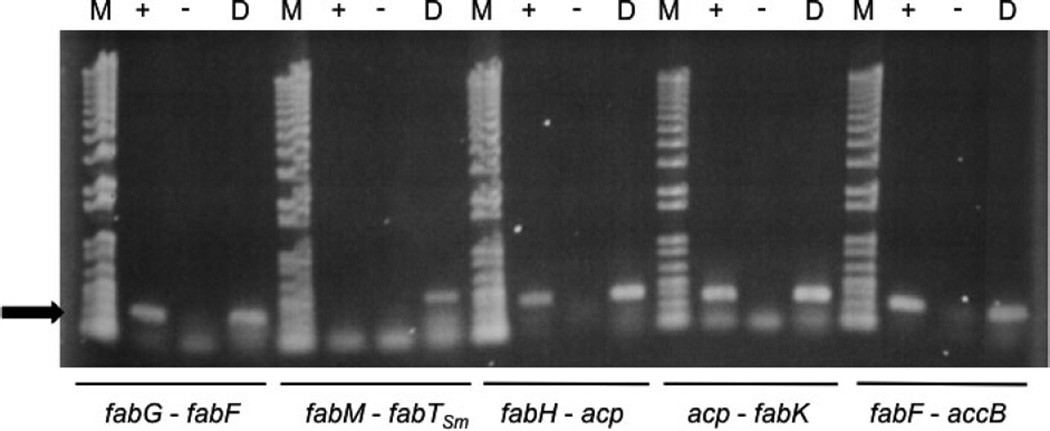
The presence of small intergenic regions (IGRs), between reading frames in the S. mutans fab gene cluster, suggested that the clusters of open reading frames might be co-transcribed. To test this hypothesis, we performed reverse-transcription reactions, using RNA prepared from cells grown at pH 7, between the following loci: fabM and fabTSm, fabH and acp, acp and fabK, fabG and fabF, and fabF and accB (Fig. 3). The results revealed co-transcription across all of the junctions tested, except the IGR between fabM and fabTSm. In addition, the results of primer extension analysis identified a potential start site of transcription between acp and fabK (data not shown). These data suggested the possibility that multiple transcripts could be involved in fab transcription and that, probably, the genes downstream of fabTSm are part of the same transcript.
Figure 3.
Co-transcription of genes in fab gene cluster. Total RNA (43 ng) isolated from a steady-state culture of Streptococcus mutans UA159 grown to a pH value of 7 was incubated with the anti-sense primer in each primer pair with (+) or without (−) SuperScript II Reverse Transcriptase. Each primer pair was also used in a standard PCR with UA159 genomic DNA as a positive control (D). Primers spanned the intergenic spaces between fabG/fabF, fabM/fabTSm, fabH/acp, acp/fabK and fabF/accB as indicated in the figure. M = 1 kb+ DNA Ladder. RT-PCR or PCR products are indicated by arrow at left of figure.
Loss of FabTSm affects transcriptional regulation of fatty acid biosynthetic genes
The data so far suggested that FabTSm might influence expression of the fatty acid biosynthesis genes via consensus motifs, specifically fabM and fabK, and that transcription of the downstream fab genes may be driven from the promoter preceding fabTSm. To investigate the level of transcriptional regulation via the putative FabTSm motifs, we created cat-promoter fusion constructs that included potential binding sites for FabTSm in the promoter regions of fabTSm, fabK and fabM (Figs 2B and 4). Loss of fabTSm led to elevated fabTSm promoter expression in cells grown at pH values of 5 or 7 (Table 3A). Loss of fabTSm also correlated with significantly elevated fabM expression in cells grown at pH values of 5 or 7 (Table 3C). These findings support, and provide at least one mechanism for, the increased UFA : SFA ratio seen in the S. mutans fabT deletion strain. In contrast, in the S. pneumoniae FabT regulon, FabTSp did not exhibit transcriptional control over fabM expression, but did exhibit a reduction in the UFA : SFA ratio in the membrane of the fabTSp mutant strain (Lu & Rock, 2006). The L. lactis ΔfabT strain also exhibited a higher percentage of SFAs in the membrane; however, the organism does not encode a fabK ortholog (Wegmann et al., 2007; Eckhardt et al., 2013). The authors suggest that fabI, a trans-2-enoyl-ACP reductase, may act as a fabK substitute, and FabTLl has been shown to control fabI expression. Interestingly, fabK transcription in S. mutans did not appear to originate from a promoter in the IGR directly upstream of the start site of translation. In fact, promoter-cat reporter fusions from this IGR did not yield chloramphenicol acetyl-transferase activity above background in either the parent strain or the fabTSm deletion strain under the conditions tested (data not shown).
Table 3.
FabTSm and CcpA regulate transcription of fabTSm and fabM
| (A) fabTSm transcription: pH effect | ||
| Background | pH 5 | pH 7 |
| UA159 | 134.83 ± 7.05*,†,ø | 93.79 ± 7.68*,^,¥ |
| ΔfabTSm | 224.58 ± 43.47† | 203.86 ± 9.14^ |
| ΔccpA | 140.87 ± 11.17ø,# | 157.51 ± 8.09¥,# |
| (B) fabTSm transcription: minimal media | ||
| Background | BHI | FMC |
| UA159 | 143.58 ± 12.78*,† | 133.25 ± 15.26*,¥ |
| ΔccpA | 170.55 ± 5.97† | 172.39 ± 29.18¥ |
| (C) fabM transcription | ||
| Background | pH 5 | pH 7 |
| UA159 | 10.62 ± 1.59*,†,# | 2.45 ± 0.68*,^,ø |
| ΔfabTSm | 18.25 ± 0.99¥,† | 5.19 ± 0.65¥,^ |
| ΔccpA | 4.64 ± 1.39# | 4.24 ± 1.01ø |
Transcriptional activity from promoters of fabTSm and fabM was measured using a chloramphenicol acetyltransferase reporter system. Cultures of Streptococcus mutans UA159, MU1591 (ΔfabTSm), or MU1446 (ΔccpA) carrying promoter-cat fusion of interest, were grown in brain–heart infusion (BHI) medium titrated to a pH value of 5 with HCl or buffered to a pH value of 7 with 50 mm KPO4 buffer (Panel A and C), or in BHI medium or FMC minimal medium + 1% (vol/vol) glucose in a 5% (vol/vol) CO2/95% air atmosphere (Panel B). Panel A: fabTSm promoter-cat; Panel B: fabTSm promoter-cat; Panel C: fabM promoter-cat. Activity from the promoters is represented as nmol chloramphenicol acetylated min−1 mg protein−1± standard deviation, n = 3. Statistical significance, between pairs, was determined by Student’s t-test, where pairs of symbols indicate P < 0.005.
The promoter regions of fabM and fabK revealed two potential binding sites for FabTSm (Fig. 4A,C, respectively). We wished to determine whether both sites were necessary for binding of the potential regulator. A truncated version of the full-length fabK promoter (indicated by brackets in Fig. 4C) was tested that did not include the distal FabTSm binding site. This smaller promoter fragment showed no difference from full-length fabK-promoter activity; that is, there was still no activity above background levels even in the absence of one of the motifs (data not shown). However, removal of the distal FabTSm binding site upstream of the fabM coding region, that is, using only the region bracketed in Fig. 4A, abolished fabM transcriptional activity, probably as the result of the lack of −10/−35 promoter sequences and the start site of transcription (data not shown).
Local effects of fabTSm deletion
The transcriptional data indicated that the presence of the FabTSm motifs correlated with expression of fabTSm and fabM. We next wanted to examine if the absence of fabTSm would affect transcription of genes flanking fabTSm, indicating that the promoter in front of fabTSm may be important for regulation of their expression. In the case of fabH, acp and fabK, the loss of fabTSm resulted in a statistically significant reduction in the copy number from cultures grown to steady-state at pH 7, when compared with copy number from cultures of the parent strain grown under the same conditions (Fig. S2A–C). Expression of acp and fabK is not influenced by pH value in the parent strain, whereas in the ΔfabTSm strain, the expression of these genes is elevated at low pH (Fig. S2). We infer from the transcriptional data (Fig. S2) and the reverse transcription reactions (Fig. 2) that the promoter in front of fabTSm may play a role in the expression of the fab genes from fabTSm through fabF. Further, we show an effect on transcription of fabM, in the absence of fabTSm, that mimics the effects on the downstream genes in the cluster (Fig. S2D).
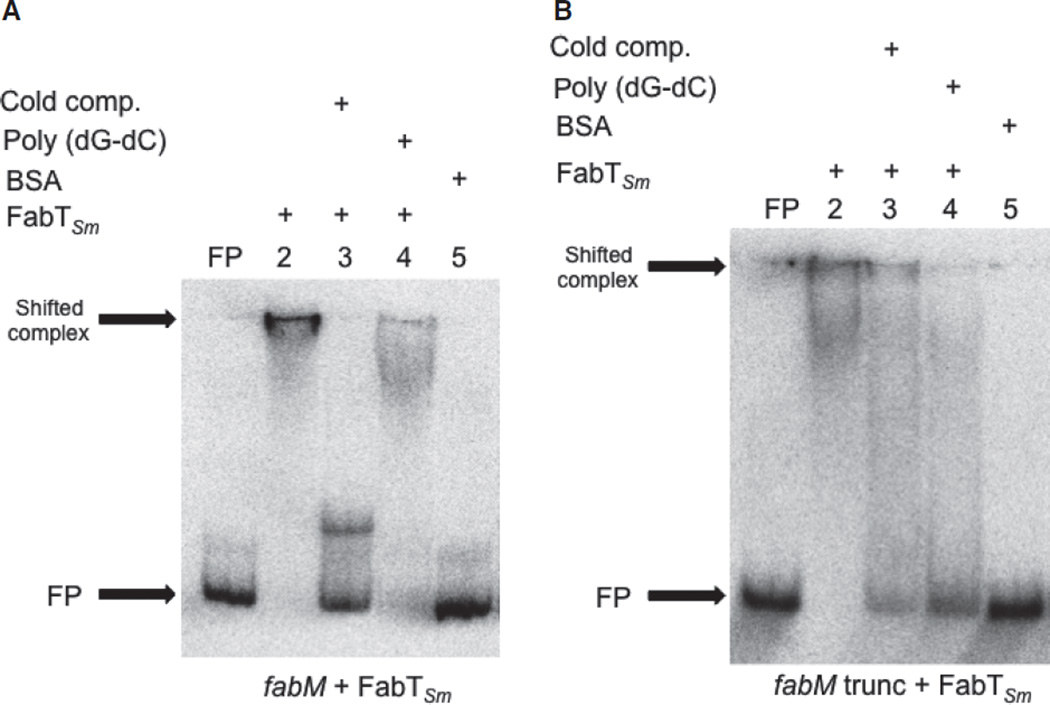
Differential binding of FabTSm to fabTSm and fabM promoters
To verify that the promoter elements affected by the presence of FabTSm were the reason for the observed changes in transcription, we performed EMSAs to investigate the ability of FabTSm to bind directly to DNA fragments derived from the full-length promoter–fusion constructs (illustrated in Fig. 4). The results revealed that the promoter region of fabM bound specifically to FabTSm (Fig. 5A; lane 2), compared with the absence of a shift in the presence of BSA (Fig. 5A; lane 5). The presence of a non-specific competitor, poly(dG-dC), did not compete for FabTSm binding of the target DNA sequence (Fig. 5A; lane 4). The specificity of FabTSm binding to the fabM promoter was also supported by the observation that non-radiolabelled, fabM promoter DNA was an effective competitor for protein binding to labeled DNA (Fig. 5A; lane 3). Similar results were observed when we examined the ability of FabTSm to bind its cognate promoter (see Fig. S3, lane 2 compared with lane 1), suggesting a role for FabTSm in auto-regulation supported by transcription assay data (Table 3A).
Figure 5.
FabTSm binds to the fabM promoter. Electrophoretic mobility shift assays demonstrating binding of FabTSm to the fabM promoter regions detailed in Fig. 4: the full-length fabM promoter region (A), or the truncated fabM promoter region lacking the distal FabTSm motif (B). For each panel, FP = free probe (5000 counts min−1) (no protein added); lane 2 = promoter + FabTSm (1000 nm) (binding reaction); lane 3 = promoter + FabTSm (1000 nm) + cold competitor (200-fold excess unlabeled, cognate promoter DNA); lane 4 = promoter + FabTSm (1000 nm) + unlabeled, non-specific competitor (1 µg poly (dG-dC)); lane 5 = promoter + BSA (1000 nm).
A previous study in S. pneumoniae demonstrated the ability of various length acyl-ACPs to enhance the affinity of FabTSp for target DNA sequences (Lu & Rock, 2006; Jerga & Rock, 2009). We examined the contribution of C18:1 acyl-ACP, from S. pneumoniae, to the ability of FabTSm to bind to the promoters of interest, fabTSm, fabM and fabK. The results indicate that the addition of the long chain-length ACP did not act as a co-effector for FabTSm and did not contribute to greater specificity of the repressor for its cognate motif when concentrations of FabTSm are limiting (data not shown).
We observed that fabM expression was affected by the presence of the FabTSm motifs within the promoter. In an effort to determine whether the FabTSm repressor was able to bind to the proximal FabTSm motif in the fabM promoter (Fig. 4A, bracketed sequence), we performed an EMSA using this fabM-truncated promoter as a probe and, indeed, the presence of only one FabTSm motif was sufficient to allow binding of the probe to the protein (Fig. 5B, lane 2). This DNA–protein affinity was also evident at lower concentrations of FabTSm, but again, the presence of the acyl-ACP did not enhance binding (data not shown). FabTSm also bound to the fabT-motif in the IGR preceding fabK, suggesting a potential role for FabTSm in regulating fabK (data not shown).
Global effects of fabTSm deletion
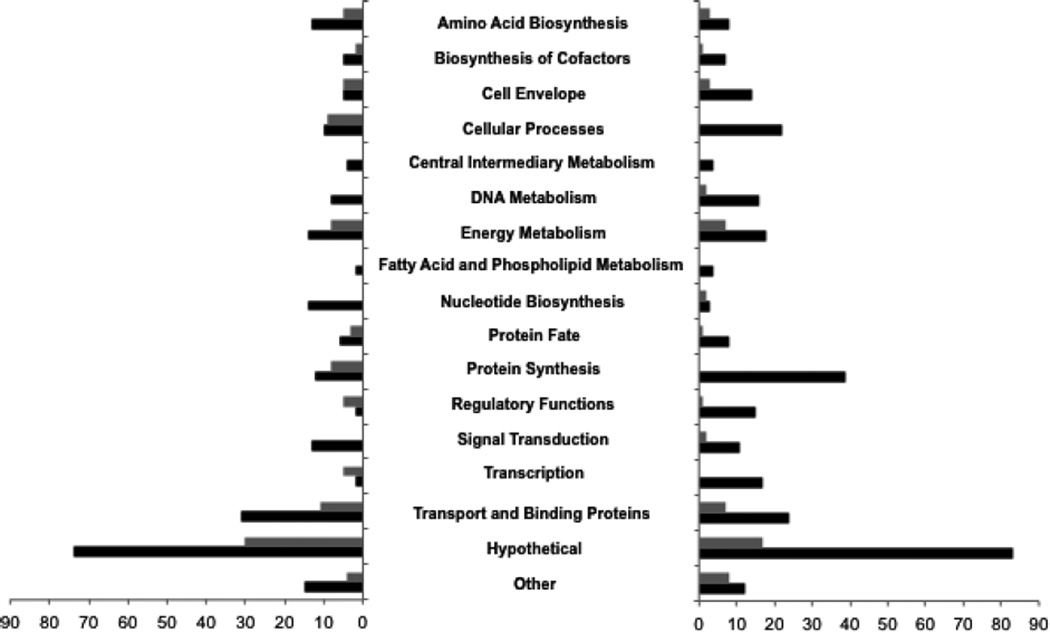
Given the evidence so far that fabM expression is governed by FabTSm, and that our previous work has shown that the loss of fabM resulted in dramatic sensitivity to acidic conditions (Fozo & Quivey, 2004b), we were interested in potential global effects of the loss of the FabTSm regulator. FabTSm is also a member of the larger MarR transcriptional regulator family and we hypothesized that deletion of the fabTSm locus may impact the transcriptome of the organism. Microarray analysis was performed to ascertain potential global effects of the loss of FabTSm. RNA isolated from steady-state cultures of the ΔfabTSm and parent strain, UA159, grown at pH values of 7 and 5, was used examine changes due to environmental pH conditions in the transcriptome of the deletion strain, compared with the parent strain. Using a two-fold cutoff for expression with a P-value ≤ 0.01, there were 647 genes, over 30% of the genome, whose transcription was affected. Our observations may reflect the broad ramifications of altering the UFA : SFA ratio in an organism whose membrane is finely tuned to changes in external pH, indicating the ability of S. mutans to adjust to stress. We found 230 genes that were upregulated and 305 genes down-regulated at pH 7 in ΔfabTSm, and 94 genes upregulated and 55 genes downregulated at pH 5 in ΔfabTSm, compared with the parent strain. Furthermore, relative to expression levels in the parent strain, the genes annotated as encoding hypothetical proteins represented the largest functional class of differentially-regulated genes, whether up or down, for both pH 7-grown and pH 5-grown cells. Other highly impacted functional classes, those for which the most genes were either upregulated or downregulated, under each condition tested, included transport and binding proteins, protein synthesis, cellular processes and energy metabolism (Fig. 6). Using the FabTSm consensus motif (Fig. 2A) to search the genome for potential binding sites, with zero mismatches, via Virtual Footprint (Münch et al., 2005), 34 candidate genes were identified, and 17 of these genes appeared in the microarray data set within the parameters that we established.
Figure 6.
Loss of FabTSm results in changes in global transcription. RNA isolated from cultures of Streptococcus mutans UA159 and MU1591 (ΔfabTSm) grown to steady-state pH values of 5 or 7 were used to synthesize cDNA for microarray analysis. Functional classes down-regulated in the MU1591 strain compared with parent strain are on the left graph, functional classes up-regulated in the deletion strain are on the right graph. Gray bars represent number of genes differentially expressed in steady-state cultures grown to a pH value of 5, black bars represent number of genes differentially expressed in steady-state cultures grown to a pH value of 7. A two-fold cutoff was used for change in expression with a P-value ≤ 0.01, as determined by BRB-Array tools; n = 4.
Coordinated regulation of fatty acid biosynthesis
The number of genes affected by the loss of the FabTSm regulator, approximately 30% of the reading frames in S. mutans, clearly suggested the important role of UFA : SFA content in the physiology of S. mutans, as well as the magnitude of the response by the organism to maintain homeostasis in the absence of FabTSm. In an effort to classify the genes included in our array data sets, we used MEME analysis (Bailey & Elkan, 1994; Bailey et al., 2009) to predict shared motifs among the intergenic regions preceding open reading frames that were differentially regulated in the absence of fabTSm, at pH 7 and pH 5, looking for commonality of the genes affected.
Allowing for the discovery of a maximum of six motifs, with any number of repetitions, three motifs were predicted with an e-value of ≤10−9. Further analysis of these predicted motifs with TOMTOM (Tool for Motif to Motif comparison; Gupta et al., 2007; Bailey et al., 2009; Tanaka et al., 2011) using the RegTransBase database of manually curated prokaryotic motifs yielded matches to three subfamilies of regulatory proteins: LacI, GntR and AraC/XylS/XylR. All three regulators are part of the helix-turn-helix (H-T-H) family of DNA-binding proteins that differ in their cognate binding sequences and location of the H-T-H domain.
We then sought to determine which of the families of regulators, encoded by streptococci, appeared in the microarray data and were differentially-regulated in response to the loss of fabTSm. A single transcript encoding a LacI regulator, two genes encoding GntR regulators, and no AraC/XylS/XylR regulators appeared in the microarray data.
LacI-family regulation
The single LacI regulator that fitted the criteria we set was SMU.1591c, encoding the global regulator CcpA (Simpson & Russell, 1998). SMU.1591c was down-regulated in the ΔfabTSm strain compared with the parent strain during growth at pH 7. CcpA is well established as a negative regulator of metabolism, that acts by binding to catabolite responsive elements (cre) and represses transcription of the downstream gene (Hueck & Hillen, 1995; Titgemeyer & Hillen, 2002; Sonenshein, 2007). CcpA has also been demonstrated to play a role in the regulation of branched-chain amino acid production by positively regulating ilvE (Santiago et al., 2013). Previous work has indicated that fabTSm is differentially transcribed in the absence of ccpA (Abranches et al., 2008) and analysis of the S. mutans UA159 genome for potential cre sites via xBASE (Chaudhuri et al., 2008) predicts the motif to be located upstream of the fabTSm, fabM and fabK genes. To test whether CcpA may be involved in fatty acid biosynthesis, we performed transcriptional reporter assays to determine the effect of the loss of ccpA on expression of fabTSm and fabM. As shown in Table 3A, in a strain carrying a ccpA deletion, the amount of CAT activity, as a measure of fabTSm transcription, was significantly elevated, when compared with the levels of activity in the parent strain background, at both a pH value of 5 and 7. The elevated CAT activity was indicative of de-repression of fabTSm by CcpA. As CcpA has been implicated in the regulation of genes involved in growth and homeostasis during low carbohydrate availability, we also tested the fabTSm promoter-cat fusion following growth of the reporter strain in FMC minimal media (Terleckyj & Shockman, 1975; Terleckyj et al., 1975). As shown in Table 3B, transcription of fabTSm was elevated in the ΔccpA strain grown in either rich or minimal medium, compared with expression levels in the UA159 strain, indicating that low nutrient availability itself does not appear to play a role in fabTSm transcription. Rather, these results are similar to those observed in the ΔccpA strain grown in buffered rich medium to pH values of 5 and 7, where the fabTSm expression levels are increased due to the loss of ccpA.
Transcription from the fabM promoter was also tested in a ΔccpA background. Here, the loss of CcpA resulted in an intermediate level of fabM expression, compared with expression in the parent strain or the ΔfabTSm strain, at both pH values of 5 and 7. In fact, culture pH had no effect on the expression of fabM in the absence of ccpA (Table 3C).
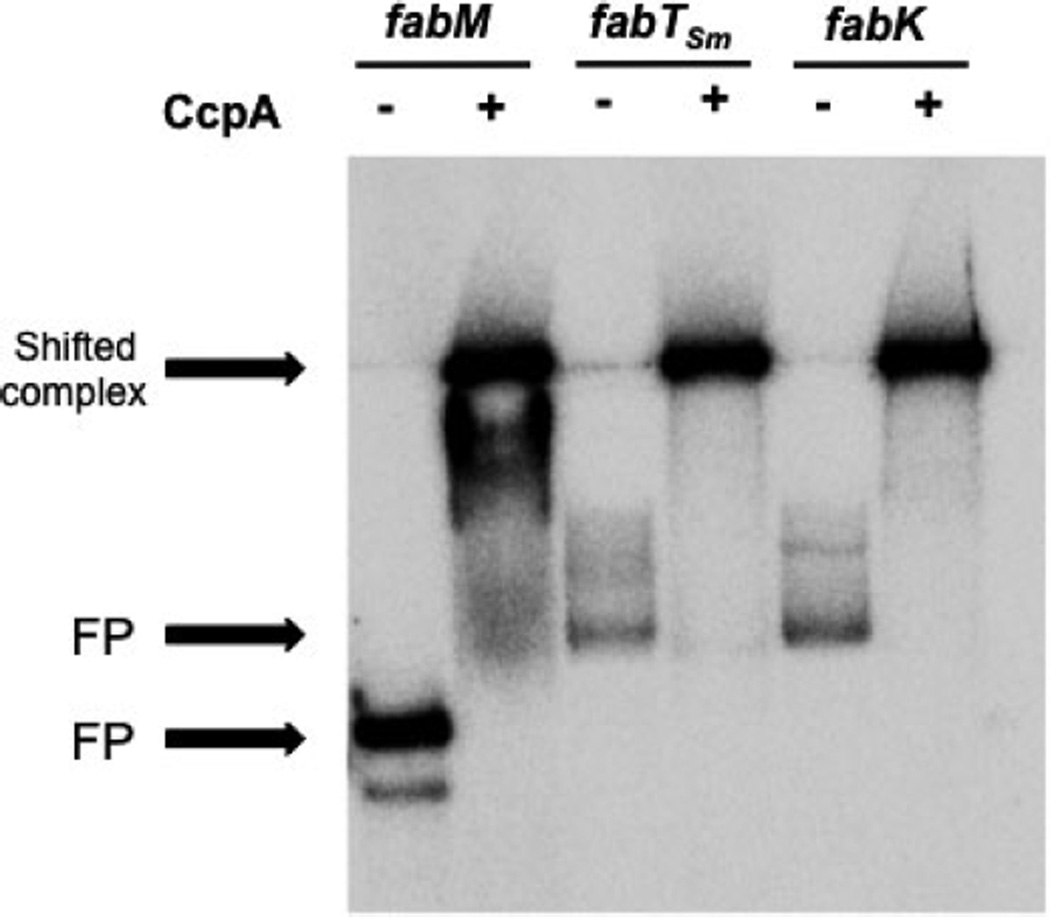
Following the transcriptional fusion experiments, we tested the ability of purified, recombinant CcpA to directly bind to the same full-length intergenic regions (shown in Fig. 4) used in the binding assays with FabTSm and in the cat-reporter assays, containing the predicted cre sites located upstream of the fabM, fabTSm and fabK coding regions. Results from EMSAs show that CcpA bound specifically to all three fragments (Fig. 7), suggesting a direct role for the carbon-catabolite repressor, CcpA, as a participant in the regulation of fatty acid biosynthesis in S. mutans.
Figure 7.
CcpA binds to intergenic regions preceding fabM, fabTSmand fabK. Electrophoretic mobility shift assay demonstrating binding of CcpA to the intergenic regions preceding fabM, fabTSm and fabK“—” = free probe (3000 counts min−1) (no protein added); “+” = probe + CcpA (3000 nm).
GntR regulation
The two GntR-encoding regulators identified in the microarrays were SMU.1065c, a hypothetical GntR regulator, and SMU.2040, orthologous to the Bacillus subtilis TreR (Helfert et al., 1995). Both S. mutans genes are annotated as regulatory proteins, based on conserved H-T-H and UTRA (UbiC transcriptional regulator-associated) domains at their N-terminus and C-terminus, respectively; but there are no previous reports on their specific function. It is of note, however, that these predicted proteins are the sole UTRA domain-containing regulators, responsible for modulating activity of bacterial transcription factors in response to a variety of ligands, in the S. mutans genome. The association of these conserved domains within the same peptide, mosaic modular association, has been described in B. subtilis for the hybrid protein AraR (Mota et al., 1999; Rigali et al., 2002). Further, these loci appear to be lethal mutations for S. mutans (Faustoferri, R.C., Hubbard, C.J. and Quivey, Jr., R.G., unpublished observation), suggesting a vital role in regulation.
DISCUSSION
In the present study, we focused on the regulatory aspects of FabTSm with respect to production of UFA in S. mutans, a well-established member of the complex microbial community found on tooth surfaces (Takahashi & Nyvad, 2011; Nyvad et al., 2013). Examination of fatty acid biosynthesis in the gram-positive organisms S. pneumoniae and L. lactis demonstrated a role for the MarR-family regulator FabT in modulating the proportion of saturated membrane fatty acids. We observed that while there are similarities with the S. pneumoniae and L. lactis regulatory systems (Lu & Rock, 2006; Jerga & Rock, 2009), there are substantial differences, and additional regulators of production of UFAs in S. mutans that indicate responsiveness to a range of environmental stresses confronting the organism in the oral cavity. These studies show that the regulators include, at least the local regulator, FabTSm, and the global regulator, CcpA.
We observed elevated transcription from the fabTSm promoter in cells grown at low pH, and transcription from the fabM promoter that was dependent on FabTSm. No detectable transcription was noted from the putative fabK promoter, despite the presence of predicted FabTSm sites. These results contrast with patterns reported in S. pneumoniae and L. lactis. We interpret the differences between the organisms, particularly with regard to pH-responsive transcription of fabM, as being an evolutionarily beneficial arrangement for S. mutans. Previous work from our group has demonstrated that the membrane composition of the organism changes rapidly, within 20 min, in response to acidification, with the purpose of protecting acid-sensitive glycolytic enzymes (Fozo & Quivey, 2004b). The acid-protective effect maintains ΔpH values (Fozo & Quivey, 2004a), shields glycolytic enzymes from the inimical effects of low pH (Bender et al., 1986; Sutton & Marquis, 1987), and elevates proton-removal from cells by concomitant up-regulation of the atp operon, producing more membrane-bound F-ATPase (Kuhnert et al., 2004). In support of this, the microarray data from the present study indicate that the organism responds aggressively to loss of FabT, with approximately one-third of its genome involved in a survival response, including the genes encoding the membrane subunits of the F-ATPase operon, atpG, atpF and atpE.
Interestingly, the occurrence of FabT binding sites appears to be universal in streptococci, as is the appearance of a fabM gene. However, the distribution of FabT binding sites within organisms varies. Existing data indicate that the sites present in Streptococcus sanguinis and Streptococcus gordonii, oral streptococci generally considered to be non-aciduric, correlate with several observable traits: reduced acid-responsiveness, relatively lower production of UFAs compared with S. mutans, and membrane F1F0-ATPase activity that is both reduced in specific activity and in abundance of enzyme itself (Sutton & Marquis, 1987; Fozo et al., 2004; Sheng & Marquis, 2007). For example, in S. sanguinis, two FabTSm binding sites are predicted upstream of the fabTSs ortholog, but only one site upstream of fabK and fabM; whereas, there is a single, predicted, binding site upstream of each of these genes in S. gordonii and S. pneumoniae. Hence, we hypothesize that the distribution of FabT binding sites in streptococci is part of an overall system contributing to the aciduricity of a given member of the genus. Indeed, our previous studies have shown that loss of UFA-producing capacity results in diminished ΔpH values in S. mutans and elevated transcription of the F-ATPase operon, apparently in an attempt to maintain intracellular homeostasis. Non-aciduric microorganisms such as S. sanguinis, S. gordonii, S. pneumoniae and L. lactis also have pH optima for their respective F1F0-AT-Pase enzymes that are between 1 and 1.5 pH units higher than the optimum pH for the enzyme in S. mutans (Sutton & Marquis, 1987; O’Sullivan & Condon, 1999; Martin-Galiano et al., 2001).
It is also important to consider that in S. pneumoniae and in L. lactis, loss of FabT results in a decrease in the UFA : SFA ratio (Lu & Rock, 2006; Eckhardt et al., 2013). Here, we demonstrate that FabTSm appears to interact with, and affect transcription from the fabM and fabTSm promoters, thereby increasing the UFA : SFA ratio. It is well established that loss of UFAs in S. mutans results in acid-sensitivity, a reduction in ΔpH across the membrane, and reduction in virulence potential (Fozo & Quivey, 2004a, b; Fozo et al., 2007); though the role of membrane unsaturated fatty acids in the acid tolerance response of S. mutans is still to be elucidated at the biophysical level.
In addition to the local regulator FabTSm, the presence of cre sites upstream of the fabM, fabTSm and fabK genes suggested that CcpA, the global regulator of carbon metabolism genes in bacteria (Bruckner & Titgemeyer, 2002), including S. mutans (Simpson & Russell, 1998; Abranches et al., 2008), may play a role in regulation of these fab genes. This hypothesis is borne out by the data from reporter fusions and EMSAs reported here: loss of CcpA leads to elevated fabTSm transcription, and as FabTSm acts to repress fabM, transcription of fabM, in a ΔccpA background, is also repressed. We interpret the data to indicate that regulation of membrane fatty acids is consistent with global regulation of carbon metabolism in cells. The production of membrane fatty acids, necessarily coordinated with central carbon metabolism, is now firmly established, and has been shown to be a part of the regulons for the global pathways controlled by CcpA and CodY (Santiago et al., 2012, 2013).
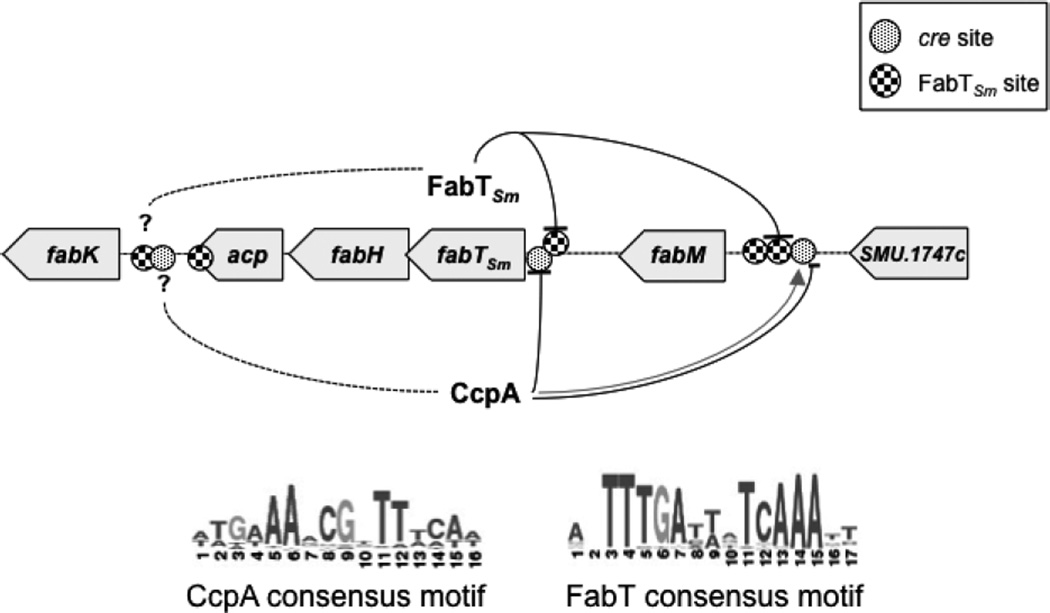
In summary, we have shown that the regulation of unsaturated fatty acid production in S. mutans exhibits a use of the FabT regulator that is different from the S. pneumoniae and L. lactis systems, which we attribute to the organism’s distinct microbiological niche. Wide changes occur in carbohydrate availability and pH values in the oral cavity, in conjunction with the presence of abundant numbers of competing organisms. An increased proportion of membrane UFAs gives S. mutans a survival advantage in this niche, potentially providing an altered milieu for membrane proteins involved in the acid-adaptive response (Abranches, J., Faustoferri, R.C., Hubbard, C.J., Courtney, M., and Quivey, Jr., R.G., in preparation). Hence, it is reasonable to propose a model that accounts for the involvement of FabTSm, a local regulator of fatty acid biosynthesis, with specific binding motifs that function as part of a rapid response to decreasing pH values (Fig. 8). The model also incorporates CcpA, a regulator that responds to carbon availability and a key regulator of overall cell growth and metabolism. These two transcriptional regulators act upon the genes responsible for fatty acid biosynthesis, enabling S. mutans to endure the decrease in pH resulting from fermentation end products and concurrent depletion of available carbon sources. Future studies will explore other global regulators, such as the GntR orthologs, that may also participate in the modulation of UFA production in S. mutans in response to environmental stress.
Figure 8.
Proposed model for regulation of fatty acid biosynthesis gene cluster by FabTSm and CcpA. Location of binding sites for CcpA (cre sites) and FabTSm are shown with respect to their relative position to the coding regions in the fatty acid biosynthetic gene cluster. FabTSm binds to promoter elements in front of fabTSm fabM and fabK. FabTSm acts as a repressor of fabTSm and fabM transcription; however, the magnitude of repression is dependent on environmental pH. Under acidic growth conditions (pH 5), CcpA binds to the intergenic region preceding fabTSmacting as a repressor. In acidic growth conditions, CcpA also binds to the intergenic region preceding fabM thereby activating transcription (gray line with arrow). However, at neutral pH (pH 7), CcpA represses fabM expression (black line with bar). CcpA also plays a role in repressing fabTSm transcription independent of environmental pH. The role for CcpA and FabTSm in regulation of fabK, if any, is currently unknown.
ACKNOWLEDGEMENTS
This study was supported by NIH/NIDCR DE-13683, DE017425 and DE017157 (RGQ) and by NIH/NIDCR T32 DE-07165 (AB, TS) and T90-DE021985 (AB, BS). The authors would like to thank Matthew Mac-Gilvray for construction of the fabM promoter construct, Dr. Alan Smith for assistance with protein purification and cloning, and Dr. Jacqueline Abranches for assistance and expertise with microarray analysis and real-time PCR. We also thank Dr. Charles Rock (St. Jude’s Children’s Hospital, Memphis, TN) for the gift of the C18:1-acyl-ACP, Dr. Robert Burne (University of Florida, Gainesville, FL) for the gift of CcpA used in the binding assays and Dr. Jose Lemos for helpful discussions.
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
REFERENCES
- Abranches J, Candella MM, Wen ZT, Baker HV, Burne RA. Different roles of EIIABMan and EIIGlc in regulation of energy metabolism, biofilm development, and competence in Streptococcus mutans. J Bacteriol. 2006;188:3748–3756. doi: 10.1128/JB.00169-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abranches J, Nascimento MM, Zeng L, et al. CcpA regulates central metabolism and virulence gene expression in Streptococcus mutans . J Bacteriol. 2008;190:2340–2349. doi: 10.1128/JB.01237-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajdic D, McShan WM, McLaughlin RE, et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci U S A. 2002;99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in bio-polymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JL, Derr AM, Karuppaiah K, et al. Streptococcus mutans NADH oxidase lies at the intersection of overlapping regulons controlled by oxygen and NAD+ levels. J Bacteriol. 2014;196:2166–2177. doi: 10.1128/JB.01542-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belli WA, Marquis RE. Adaptation of Streptococcus mutans and Enterococcus hirae to acid stress in continuous culture. Appl Environ Microbiol. 1991;57:1134–1138. doi: 10.1128/aem.57.4.1134-1138.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender GR, Sutton SV, Marquis RE. Acid tolerance, proton permeabilities, and membrane ATPases of oral streptococci. Infect Immun. 1986;53:331–338. doi: 10.1128/iai.53.2.331-338.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Bowen WH, Koo H. Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 2011;45:69–86. doi: 10.1159/000324598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bruckner R, Titgemeyer F. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol Lett. 2002;209:141–148. doi: 10.1111/j.1574-6968.2002.tb11123.x. [DOI] [PubMed] [Google Scholar]
- Buckley AA, Faustoferri RC, Quivey RG., Jr. beta-Phosphoglucomutase contributes to aciduricity in Streptococcus mutans . Microbiology. 2014;160:818–827. doi: 10.1099/mic.0.075754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri RR, Loman NJ, Snyder LA, Bailey CM, Stekel DJ, Pallen MJ. xBASE2: a comprehensive resource for comparative bacterial genomics. Nucleic Acids Res. 2008;36:D543–D546. doi: 10.1093/nar/gkm928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YY, Weaver CA, Mendelsohn DR, Burne RA. Transcriptional regulation of the Streptococcus salivarius 57.I urease operon. J Bacteriol. 1998;180:5769–5775. doi: 10.1128/jb.180.21.5769-5775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derr AM, Faustoferri RC, Betzenhauser MJ, Gonzalez K, Marquis RE, Quivey RG., Jr. Mutation of the NADH oxidase gene (nox) reveals an overlap of the oxygen- and acid-mediated stress responses in Streptococcus mutans . Appl Environ Microbiol. 2012;78:1215–1227. doi: 10.1128/AEM.06890-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt TH, Skotnicka D, Kok J, Kuipers OP. Transcriptional regulation of fatty acid biosynthesis in Lactococcus lactis . J Bacteriol. 2013;195:1081–1089. doi: 10.1128/JB.02043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozo EM, Quivey RG., Jr. The fabM gene product of Streptococcus mutans is responsible for the synthesis of monounsaturated fatty acids and is necessary for survival at low pH. J Bacteriol. 2004a;186:4152–4158. doi: 10.1128/JB.186.13.4152-4158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozo EM, Quivey RG., Jr. Shifts in the membrane fatty acid profile of Streptococcus mutans enhance survival in acidic environments. Appl Environ Microbiol. 2004b;70:929–936. doi: 10.1128/AEM.70.2.929-936.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozo EM, Kajfasz JK, Quivey RG., Jr. Low pH-induced membrane fatty acid alterations in oral bacteria. FEMS Microbiol Lett. 2004;238:291–295. doi: 10.1016/j.femsle.2004.07.047. [DOI] [PubMed] [Google Scholar]
- Fozo EM, Scott-Anne K, Koo H, Quivey RG., Jr. Role of unsaturated fatty acid biosynthesis in virulence of Streptococcus mutans . Infect Immun. 2007;75:1537–1539. doi: 10.1128/IAI.01938-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfert C, Gotsche S, Dahl MK. Cleavage of trehalose-phosphate in Bacillus subtilis is catalysed by a phospho-alpha-(1-1)-glucosidase encoded by the treA gene. Mol Microbiol. 1995;16:111–120. doi: 10.1111/j.1365-2958.1995.tb02396.x. [DOI] [PubMed] [Google Scholar]
- Hueck CJ, Hillen W. Catabolite repression in Bacillus subtilis: a global regulatory mechanism for the gram-positive bacteria? Mol Microbiol. 1995;15:395–401. doi: 10.1111/j.1365-2958.1995.tb02252.x. [DOI] [PubMed] [Google Scholar]
- Jerga A, Rock CO. Acyl-Acyl carrier protein regulates transcription of fatty acid biosynthetic genes via the FabT repressor in Streptococcus pneumoniae . J Biol Chem. 2009;284:15364–15368. doi: 10.1074/jbc.C109.002410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajfasz JK, Rivera-Ramos I, Abranches J, et al. Two Spx proteins modulate stress tolerance, survival, and virulence in Streptococcus mutans. J Bacteriol. 2010;192:2546–2556. doi: 10.1128/JB.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhnert WL, Zheng G, Faustoferri RC, Quivey RG., Jr. The F-ATPase operon promoter of Streptococcus mutans is transcriptionally regulated in response to external pH. J Bacteriol. 2004;186:8524–8528. doi: 10.1128/JB.186.24.8524-8528.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau PC, Sung CK, Lee JH, Morrison DA, Cvitkovitch DG. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J Microbiol Methods. 2002;49:193–205. doi: 10.1016/s0167-7012(01)00369-4. [DOI] [PubMed] [Google Scholar]
- Lemos JA, Burne RA. A model of efficiency: stress tolerance by Streptococcus mutans . Microbiology. 2008;154:3247–3255. doi: 10.1099/mic.0.2008/023770-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loesche WJ. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986;50:353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YJ, Rock CO. Transcriptional regulation of fatty acid biosynthesis in Streptococcus pneumoniae . Mol Microbiol. 2006;59:551–566. doi: 10.1111/j.1365-2958.2005.04951.x. [DOI] [PubMed] [Google Scholar]
- Martin-Galiano AJ, Ferrandiz MJ, de la Campa AG. The promoter of the operon encoding the F0F1 ATPase of Streptococcus pneumoniae is inducible by pH. Mol Microbiol. 2001;41:1327–1338. doi: 10.1046/j.1365-2958.2001.02597.x. [DOI] [PubMed] [Google Scholar]
- Mota LJ, Tavares P, Sa-Nogueira I. Mode of action of AraR, the key regulator of L-arabinose metabolism in Bacillus subtilis . Mol Microbiol. 1999;33:476–489. doi: 10.1046/j.1365-2958.1999.01484.x. [DOI] [PubMed] [Google Scholar]
- Münch R, Hiller K, Grote A, et al. Virtual footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics. 2005;21:4187–4189. doi: 10.1093/bioinformatics/bti635. [DOI] [PubMed] [Google Scholar]
- Murchison HH, Barrett JF, Cardineau GA, Curtiss R., 3rd Transformation of Streptococcus mutans with chromosomal and shuttle plasmid (pYA629) DNAs. Infect Immun. 1986;54:273–282. doi: 10.1128/iai.54.2.273-282.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novichkov PS, Laikova ON, Novichkova ES, et al. RegPrecise: a database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res. 2010;38:D111–D118. doi: 10.1093/nar/gkp894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyvad B, Crielaard W, Mira A, Takahashi N, Beighton D. Dental caries from a molecular microbiological perspective. Caries Res. 2013;47:89–102. doi: 10.1159/000345367. [DOI] [PubMed] [Google Scholar]
- O’Sullivan E, Condon S. Relationship between acid tolerance, cytoplasmic pH, and ATP and H+-ATPase levels in chemostat cultures of Lactococcus lactis . Appl Environ Microbiol. 1999;65:2287–2293. doi: 10.1128/aem.65.6.2287-2293.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivey RG, Jr, Faustoferri RC, Clancy KA, Marquis RE. Acid adaptation in Streptococcus mutans UA159 alleviates sensitization to environmental stress due to RecA deficiency. FEMS Microbiol Lett. 1995;126:257–261. doi: 10.1111/j.1574-6968.1995.tb07427.x. [DOI] [PubMed] [Google Scholar]
- Quivey RG, Jr, Faustoferri R, Monahan K, Marquis R. Shifts in membrane fatty acid profiles associated with acid adaptation of Streptococcus mutans . FEMS Microbiology Lett. 2000;189:89–92. doi: 10.1111/j.1574-6968.2000.tb09211.x. [DOI] [PubMed] [Google Scholar]
- Rigali S, Derouaux A, Giannotta F, Dusart J. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J Biol Chem. 2002;277:12507–12515. doi: 10.1074/jbc.M110968200. [DOI] [PubMed] [Google Scholar]
- Santiago B, MacGilvray M, Faustoferri RC, Quivey RG., Jr. The branched-chain amino acid aminotransferase encoded by ilvE is involved in acid tolerance in Streptococcus mutans . J Bacteriol. 2012;194:2010–2019. doi: 10.1128/JB.06737-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago B, Marek M, Faustoferri RC, Quivey RG., Jr. The Streptococcus mutans aminotransferase encoded by ilvE is regulated by CodY and CcpA. J Bacteriol. 2013;195:3552–3562. doi: 10.1128/JB.00394-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw WV, Packman LC, Burleigh BD, Dell A, Morris HR, Hartley BS. Primary structure of a chloramphenicol acetyltransferase specified by R plasmids. Nature. 1979;282:870–872. doi: 10.1038/282870a0. [DOI] [PubMed] [Google Scholar]
- Sheng J, Marquis RE. Malolactic fermentation by Streptococcus mutans . FEMS Microbiol Lett. 2007;272:196–201. doi: 10.1111/j.1574-6968.2007.00744.x. [DOI] [PubMed] [Google Scholar]
- Sheng J, Baldeck JD, Nguyen PT, Quivey RG, Jr, Marquis RE. Alkali production associated with malolactic fermentation by oral streptococci and protection against acid, oxidative, or starvation damage. Can J Microbiol. 2010;56:539–547. doi: 10.1139/w10-039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson CL, Russell RR. Identification of a homolog of CcpA catabolite repressor protein in Streptococcus mutans . Infect Immun. 1998;66:2085–2092. doi: 10.1128/iai.66.5.2085-2092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenshein AL. Control of key metabolic intersections in Bacillus subtilis . Nat Rev Microbiol. 2007;5:917–927. doi: 10.1038/nrmicro1772. [DOI] [PubMed] [Google Scholar]
- Sulavik MC, Gambino LF, Miller PF. The MarR repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli: prototypic member of a family of bacterial regulatory proteins involved in sensing phenolic compounds. Mol Med. 1995;1:436–446. [PMC free article] [PubMed] [Google Scholar]
- Sutton SV, Marquis RE. Membrane-associated and solubilized ATPases of Streptococcus mutans and Streptococcus sanguis . J Dent Res. 1987;66:1095–1098. doi: 10.1177/00220345870660060201. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Nyvad B. The role of bacteria in the caries process: ecological perspectives. J Dent Res. 2011;90:294–303. doi: 10.1177/0022034510379602. [DOI] [PubMed] [Google Scholar]
- Tanaka E, Bailey T, Grant CE, Noble WS, Keich U. Improved similarity scores for comparing motifs. Bioinformatics. 2011;27:1603–1609. doi: 10.1093/bioinformatics/btr257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terleckyj B, Shockman GD. Amino acid requirements of Streptococcus mutans and other oral streptococci. Infect Immun. 1975;11:656–664. doi: 10.1128/iai.11.4.656-664.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terleckyj B, Willett NP, Shockman GD. Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect Immun. 1975;11:649–655. doi: 10.1128/iai.11.4.649-655.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, Nelson KE, Paulsen IT, et al. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae . Science. 2001;293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- Titgemeyer F, Hillen W. Global control of sugar metabolism: a gram-positive solution. Antonie Van Leeuwenhoek. 2002;82:59–71. [PubMed] [Google Scholar]
- Wegmann U, O’Connell-Motherway M, Zomer A, et al. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J Bacteriol. 2007;189:3256–3270. doi: 10.1128/JB.01768-06. [DOI] [PMC free article] [PubMed] [Google Scholar]