Abstract
Protein structure comparison, an important problem in structural biology, has two main applications: (i) comparing two protein structures in order to identify the similarities and differences between them, and (ii) searching for structures similar to a query structure. Many web-based resources for both applications are available, but all are based on rigid structural alignment algorithms. FATCAT server implements the recently developed flexible protein structure comparison algorithm FATCAT, which automatically identifies hinges and internal rearrangements in two protein structures. The server provides access to two algorithms: FATCAT-pairwise for pairwise flexible structure comparison and FATCAT-search for database searching for structurally similar proteins. Given two protein structures [in the Protein Data Bank (PDB) format], FATCAT-pairwise reports their structural alignment and the corresponding statistical significance of the similarity measured as a P-value. Users can view the superposition of the structures online in web browsers that support the Chime plug-in, or download the superimposed structures in PDB format. In FATCAT-search, users provide one query structure and the server returns a list of protein structures that are similar to the query, ordered by the P-values. In addition, FATCAT server can report the conformational changes of the query structure as compared to other proteins in the structure database. FATCAT server is available at http://fatcat.burnham.org.
INTRODUCTION
Protein structure comparison has been a classic challenge in computational molecular biology for more than two decades. Thanks to the rapidly improving techniques for protein structure determination [nuclear magnetic resonance (NMR) and X-ray], the number of known protein structures is increasing quickly and we can foresee even faster growth resulting from the recent Structural Genomics Initiative and related development of high-throughput structure determination techniques (1,2). About 25 000 structures were available from the Protein Data Bank (PDB) (3) in March 2004 (see http://www.rcsb.org for the latest statistics), and the number of new weekly depositions has grown from <100 a few years ago to >300 in 2003. Typically, the first thing we want to know once a new structure is determined is whether it is similar to any known protein structures. If it is, we want to know the differences between this structure and its homologs: differences between structures are often related to functional specificity of homologs. If it is not, the importance of the new fold and (presumably) novel mechanism and function would add significance to the new structure.
Many programs addressing this challenge have been developed (4–8). Most of them, however, treat protein structures as rigid bodies even though it is well known that proteins are flexible and undergo significant structural changes as part of their normal function (9–11). To simplify the algorithms and speed up the search time, and because this is how historically the structure comparison problem has always been formulated, most of the existing programs aim at identifying the largest rigid substructure shared by two proteins. Recently several flexible protein structure alignment algorithms (12,13) have been developed, allowing for internal rearrangements in the structures during alignment. These programs have changed the paradigm of protein structure comparison from identifying common parts between protein structures to identifying and understanding rules of change in protein structures.
A server based on one such algorithm, FlexProt (13), is available at http://pc-gamba.math.tau.ac.il/FlexProt/. It implements pairwise flexible structure alignment, but until now it has had no database searching capability. Here we present a server based on FATCAT, the flexible protein structure alignment program developed in our group (12), providing both pairwise comparison (FATCAT-pairwise) and database searching for similar structures (FATCAT-search). FATCAT server is different from the popular structure comparison servers that are based on rigid-body structure comparisons, such as DALI (http://www.ebi.ac.uk/dali/) (5), VAST (http://www.ncbi.nih.gov/Structure/MMDB/mmdb.shtml) (14) and CE (http://cl.sdsc.edu/ce.html) (8). FATCAT-pairwise reports the optimal alignment, the corresponding statistical significance of the structure similarity between two structures and positions of automatically identified pivot points in the structures. FATCAT-search reports a list of structures that are statistically similar to a query from the representative set of protein structures along with pairwise alignments between each structure on this list and the query.
ALGORITHM AND IMPLEMENTATION
FATCAT starts by identifying a list of AFPs (aligned fragment pairs)—a superposition of two continuous fragments—in the two proteins to be compared. The FATCAT structure alignment is formulated as an AFP chaining process, allowing flexibility in connecting them. A rotation/translation (twist) can be introduced between two consecutive AFPs if it results in a substantially better superposition of the structures. FATCAT integrates simple extensions, gaps and twists into a unified scoring function and performs the alignment and hinge detection simultaneously using dynamic programming. Several post-processing steps are applied to refine the alignments. The significance of the similarity detected by FATCAT is evaluated by a P-value that measures the chance of getting the same similarity in two random structures. This P-value is calculated based on the empirical fitting of the extreme value distribution (EVD) to the FATCAT similarity score (15). The smaller the P-value, the more statistically significant the similarity between corresponding structures.
The FATCAT algorithm has been implemented in a fast and efficient computer program written in C++ and systematically tested on large alignment benchmarks (12). In an extensive comparison with other structure alignment programs, FATCAT has been shown to be unbiased toward introducing twists into the structure and to achieve performance that matches the rigid-structure alignment programs for all the test cases. Meanwhile, in most testing cases of pairwise alignments FATCAT outperforms the pioneering flexible alignment program FlexProt (13) by producing longer alignments with a smaller number of twists and lower Root Mean Square Deviation (RMSD) values. The FATCAT web server is implemented on a Linux redhat 9.0 platform with the Common Gateway Interface (CGI) scripts written in perl.
FATCAT-pairwise
FATCAT-pairwise requires two protein structures (in PDB format) as inputs (either by uploading local PDB files or by providing PDB codes). The server reports the structural alignment between them and the significance of their similarity as a P-value. Users can view the superposition of the structures online in web browsers that support the Chime plug-in; otherwise, a PDB file with two chains (chain A for the first protein and chain B for the second protein) and a Rasmol script for easy viewing are generated by the server for downloading. If FATCAT detects twists in the alignment, the second structure is modified by rotating/translating the rigid blocks along the pivot points. The most important measurements of the flexible similarity between two structures are the statistical significance of the similarity (P-value), the number of twists (twists), the number of equivalent positions (opt-len) and the overall RMSD (opt-rmsd). Some other values that may be useful for reference are also shown in the alignment output, such as the RMSD between unmodified structures (chain-rmsd) and the length of the alignment including gaps (align-len).
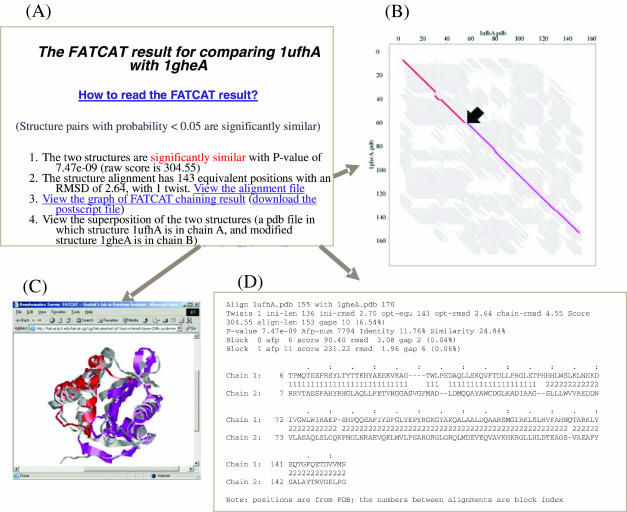
An example showing the comparison between putative acetyltransferase Yycn from Bacillus subtilis (PDB code 1ufh, chain A, with 155 residues) and tabtoxin resistance protein from Pseudomonas syringae pv. tabaci (PDB code 1ghe, chain A, with 170 residues) is shown in Figure 1. This comparison took ∼4 s. A single twist is introduced in the alignment between these two structures, which covers almost the entire proteins; otherwise, only parts of the structures, either the pink region or the red region, can be well aligned (Figure 1C).
Figure 1.
A sample FATCAT-pairwise report for structure comparison between 1ufh and 1ghe: (A) the main report page, which shows that these two structures are significantly similar (with P-value of 7.47e-09) with one twist; (B) alignment graph with the AFPs in the optimal alignment shown as colored lines and all the AFPs between the two structures shown as short gray lines in the background; (C) the online visualization of superposition between structure 1ufh and the modified structure of 1ghe using the Chime plug-in; and (D) the detailed alignment in text format, in which different measurements of the alignment are shown and alignment blocks are labeled incrementally from 1.
FATCAT-search
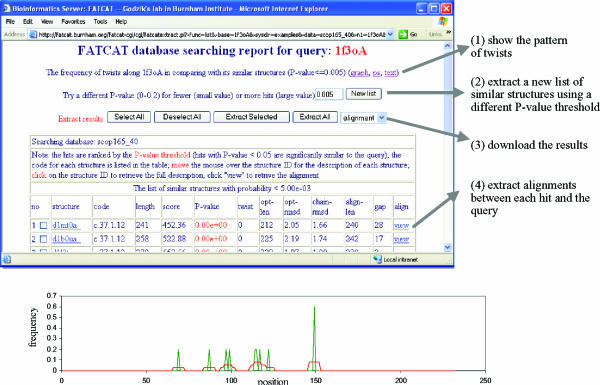
FATCAT-search can be used to search in a set of proteins for protein structures similar to an input structure. Currently, non-redundant sets based on SCOP (1.65 release) (16) and PDB (as of March 16, 2003), both at two clustering levels, are available at the server. SCOP sets contain protein domains that are classified in a four-level hierarchy so that interpreting the search results against them is easier than interpreting those against the PDB database. Therefore, SCOP is used as the default database for FATCAT-search, but users are able to choose a different database for their searches. FATCAT-search accepts a PDB file (or a PDB code) as an input and returns a list of protein structures that are similar to the query structure, ordered by the P-values. The alignment and other information for each protein on the list can be extracted by following the links on the output page. In addition to the alignment, FATCAT-search server reports the overall view of the flexibility of the query structure, showing the distribution of twists along the query protein. A sample distribution is shown in Figure 2, describing the FATCAT-search results for hypothetical Abc transporter ATP-binding protein Mj0796 (PDB code 1f3o, chain A, with 232 residues). This comparison (against 5674 structures) took ∼11 h. Strong regularities in the position of the twists (hot spots) were observed in this case.
Figure 2.
A sample FATCAT-search report for 1f3o against the SCOP (1.65 release) 40% non-redundant database. A snap-shot of the FATCAT-search main report page is shown at the top, and four functions that we implemented on the server for managing the result are listed on the right. The bottom graph shows the frequency of twists along the query protein when it is compared to its similar structures of P-value ≤ 0.005. The x-axis is the position along the query protein and the y-axis is the frequency of the twists detected in each position in all the comparisons between the query and its similar structures (raw frequency shown as the green curve and smoothed frequency using window length of seven shown as the red curve). This graph shows that there are several regions with a high frequency of twists (hot spots) in the query protein.
FUTURE PLANS
We will update the structure database regularly for FATCAT-search. Considering the CPU cost of the database searching, we are developing a FATCAT-search database so that users can extract pre-calculated results. In addition, we are planning to classify structures based on their similarity defined by FATCAT, and to extract the patterns of structural changes among structural analogs systematically. Both results will be available on the server for users' reference.
Acknowledgments
ACKNOWLEDGEMENTS
Thanks to Erik Kleist for his assistance in setting up the FATCAT server. This research was supported by NIH grant GM63208.
REFERENCES
- 1.Vitkup D., Melamud,E., Moult,J. and Sander,C. (2001) Completeness in structural genomics. Nature Struct. Biol., 8, 559–566. [DOI] [PubMed] [Google Scholar]
- 2.Lesley S.A., Kuhn,P., Godzik,A., Deacon,A.M., Mathews,I., Kreusch,A., Spraggon,G., Klock,H.E., McMullan,D., Shin,T. et al. (2002) Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline. Proc. Natl Acad. Sci., USA, 99, 11664–11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eidhammer I., Jonassen,I. and Taylor,W.R. (2001) Structure comparison and structure patterns. J. Comput. Biol., 7, 685–716. [DOI] [PubMed] [Google Scholar]
- 5.Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- 6.Boutonnet N.S., Rooman,M.J., Ochagavia,M.E., Richelle,J. and Wodak,S.J. (1995) Optimal protein structure alignments by multiple linkage clustering: application to distantly related proteins. Protein Eng., 8, 647–662. [DOI] [PubMed] [Google Scholar]
- 7.Madej T., Gibrat,J.F. and Bryant,S.H. (1995) Threading a database of protein cores. Proteins, 23, 356–369. [DOI] [PubMed] [Google Scholar]
- 8.Shindyalov I.N. and Bourne,P.E. (1998) Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng., 11, 739–747. [DOI] [PubMed] [Google Scholar]
- 9.Schulz G.E. and Schirmer,R.H. (1979) Principles of protein structure. Springer, New York. [Google Scholar]
- 10.Bennett W. and Huber,R. (1984) Structural and functional aspects of domain motions in proteins. CRC Crit. Rev. Biochem., 15, 291–384. [DOI] [PubMed] [Google Scholar]
- 11.Jacobs D.J., Rader,A.J., Kuhn,L.A. and Thorpe,M.F. (2001) Protein flexibility predictions using graph theory. Proteins, 44, 150–165. [DOI] [PubMed] [Google Scholar]
- 12.Ye Y. and Godzik,A. (2003) Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics, 19, ii246–ii255. [DOI] [PubMed] [Google Scholar]
- 13.Shatsky M., Nussinov,R. and Wolfson,H.J. (2002) Flexible protein alignment and hinge detection. Proteins, 48, 242–256. [DOI] [PubMed] [Google Scholar]
- 14.Gibrat J.F., Madej,T. and Bryant,S.H. (1996) Surprising similarities in structure comparison. Curr. Opin. Struct. Biol., 6, 377–385. [DOI] [PubMed] [Google Scholar]
- 15.Ye Y. and Godzik,A. (2004) Database searching by flexible protein structure alignment. Protein Sci., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murzin A.G., Brenner,S.E., Hubbard,T. and Chothia,C. (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol., 247, 536–540. [DOI] [PubMed] [Google Scholar]