Abstract
PromoSer is a web service that provides an easy and efficient approach to the batch retrieval of a large number of proximal promoters. Since its introduction last year, it has undergone continued development and expansion. At the core, there have been improvements in the filtering of the raw mRNA/EST sequences upon which all predictions are built, improvements in the alignments clustering and transcription start site prediction algorithms, and improvements in the backing database for increased performance. At the user interface level, improvements include enhanced functionality and user options, better integration with other resources on the web and a new visualization tool. PromoSer now also supports queries using a SOAP-based interface and XML-based responses. The service is publicly available at http://biowulf.bu.edu/zlab/PromoSer.
INTRODUCTION
The recent completion of the sequencing of the human genome (1,2) and the increased availability of many more near-complete genomes has paved the way for new approaches to insights into the study of living organisms. Important progress has also been made regarding the process of acquiring static snapshots of the dynamic transcriptional activity inside living cells using microarrays, and methods for the analysis of such data. Linking transcriptional activity profiles to genomic control regions provides further insights into the inner workings of eukaryotic transcription control mechanisms.
Many transcription control elements are within regions close to the Transcription Start Sites (TSSs) of expressed sequences, whether they are for coding or non-coding mRNA (3). With microarray experiments generating data for many thousands of transcribed sequences, an efficient method to obtain the proximal promoters of these transcripts becomes highly desirable. PromoSer (4) is a web service that was designed specifically for this purpose and has since its initial release seen a steady increase in usage, reaching an average of over a hundred hits daily.
As a large-scale data integrator, PromoSer is under constant revision to expand its sequence base and update its genomes with the latest releases. In addition to data updates, various parts of PromoSer have benefited from enhancements in accuracy, performance and usability. We report here on a variety of improvements made to PromoSer since its initial release.
DATA COLLECTION AND FILTERING
The current release of PromoSer is based on the finished human genome (July 2003, NCBI build 34), the draft mouse genome (October 2003, NCBI build 30) and the draft rat genome (June 2003, Baylor build HGSC v3.1). All expressed sequence tag (EST), mRNA and RefSeq data available on February 4, 2004 were downloaded from the GenBank (5) ftp site. The Eukaryotic Promoter Database (EPD) (6) release 77 was also downloaded.
A central concept in PromoSer is the prediction of TSSs based on purely experimental transcript data and no computational gene predictions, thus maximizing the confidence in the identified TSSs. This requires PromoSer to be sensitive to all expressed sequence data since some rare transcripts may be observed only once in the entire GenBank collection. The danger of this approach is that it can be led astray by noisy raw data. Yet, GenBank is a general data repository that employs minimal data quality control in its generic categories (such as EST and mRNA). Thus, specially designed GenBank queries are needed to filter out many of the sequences that would seriously degrade the quality of PromoSer predictions. In particular, any sequence with one of the following ‘bad’ words was discarded: synthetic construct, transgenic, chimeric, recombinant and probe.
The initial design of PromoSer assumed users would only search for promoters of a transcribed sequence based on a known GenBank accession number for the sequence. It was found that, for a number of cases, a record of the transcribed sequence was not available, or the user was not aware of it. Instead, a genomic record of the transcript was the only available key to search with. To accommodate such cases, PromoSer now supports a limited set of genomic records as well.
CLUSTERING AND TSS PREDICTION
The total collection of records that PromoSer handles is over 10 million. To efficiently map these onto their corresponding genomes, BLAT (7) was used on a 256-processor Linux cluster. To improve sensitivity, the standalone version of BLAT was used to compare all sequences against each chromosome. The mapping results of every sequence were compared to determine the best alignment, or in some cases where several mappings were nearly equally good, the best alignment and those within 1% of its score were retained.
Furthermore, transcript sequences are canonically supposed to be 5′ to 3′ oriented. This rule is broken in about 50% of the EST records and the annotation of the direction is notoriously unreliable. For reliable orientation identification, which is essential for the clustering, the alignments were checked against the genome to determine the intron orientation based on the GT/AG rule. For unspliced sequences, the original sequence was examined for the presence of the polyA tail or, if not found, a polyA signal. The location of the tail or the signal was used to infer the transcript orientation. The tail length was subtracted to determine a more accurate percentage identity score of an alignment (which is only a part of the scoring scheme used for optimal alignment selection).
In the initial design of PromoSer, alignments were subjected to strict criteria in order to be used. This resulted in the exclusion of a significant fraction of sequences and the inability to locate their promoters. To improve performance and retain accuracy, the filtering process was performed in three stages. First, a low-stringency (80% identity over at least 100 bases) initial filter was applied to select alignments used in the clustering. Once clustered, a second, stringent filter (90% identity for EST and 95% otherwise, and no more than 5 unaligned bases at the start of the sequence) was used to select alignments that may be used to predict the TSS. Finally, a third pass heuristically assigns the alignments that did not pass the initial filter to the smallest cluster that fully overlaps that alignment. Those guessed alignments are marked and reported as being a guess when the user searches for them. With that scheme, PromoSer coverage increases to about 90% of the attempted sequences instead of the 28% reported previously, with improvement mainly in the EST category.
To cluster sequences, all alignments that overlap a genomic region and are transcribed in the same orientation are collected. They are then separated into sub-clusters based on the sharing of transcribed regions. Finally, each sub-cluster is examined to determine if it contains independent subcomponents that are connected through a single EST. If so, the sub-cluster is broken up into its subcomponents. Such ESTs have been observed when the clusters were visualized using the cluster viewer (described below) and they could be artifacts of the EST library.
After clustering, candidate TSSs are identified as the 5′-most position of transcripts passing the stringent filter (see above) plus the 5′-most position in the cluster overall. Sites within 20 bp are grouped and the 5′-most one is retained. Instead of an overall cluster quality score, individual TSSs are now assigned a quality and a support score as follows: A TSS that coincides with an EPD-identified TSS has a quality score of 4. A TSS that comes from a RefSeq sequence is given quality 3, one from an mRNA record a quality of 2 and one from an EST only a quality score of 1. If no evidence supports a site (e.g. the 5′-end of a sequence that is known to be truncated) it gets a score of 0. The support score is the count of sequences that contribute to the TSS prediction. For quality 2 and above, ESTs are excluded from this count.
The cluster is finally annotated based on its largest RefSeq or mRNA sequence. The locations of gaps longer than 500 bases and other clusters upstream are noted as possible boundaries to promoter sequence extraction.
THE USER INTERFACE
The simplicity of PromoSer's interface is one of its biggest advantages, and this is largely maintained from the previous release. The user needs only to provide a list of accession numbers for which promoters are required. A number of options further customize the user's experience and tailor the results to the user's needs. A user may provide a sequence in the FASTA format rather than an accession number, and PromoSer will try to find the clusters that best match the user's sequence. Alternatively, the user can provide a specially formatted list to directly extract batches of genomic sequence.
The lengths of upstream and downstream sequences flanking a TSS are user-specifiable up to certain limits. We provide options that allow the user to
set the minimum quality and support score for the predicted TSSs and associated promoters;
eliminate duplicate promoters and/or suppress guessed alignments or those that map to multiple loci;
control repeat-masking in the returned sequence;
control boundary conditions for sequence extraction based on gaps and upstream clusters; and
control which of several possibilities the user wishes to select when a cluster contains more than one predicted TSS.
The results report from PromoSer provides a list of the promoters found and some information about them. Each promoter found can be clicked to show a detailed view of the cluster containing the clicked entry using the cluster viewer described below. Alternatively the cluster can also be displayed in the UCSC genome browser (8).
VISUALIZATION TOOLS
Two new tools are now provided to visualize the data contained in PromoSer. The chromosome viewer shows a selected chromosome (human, mouse or rat) as one long line wrapped around into a number of rows. Clusters that PromoSer has identified are plotted on this long line. The browser allows the user to visualize much of the cluster-related data with this simple display tool.
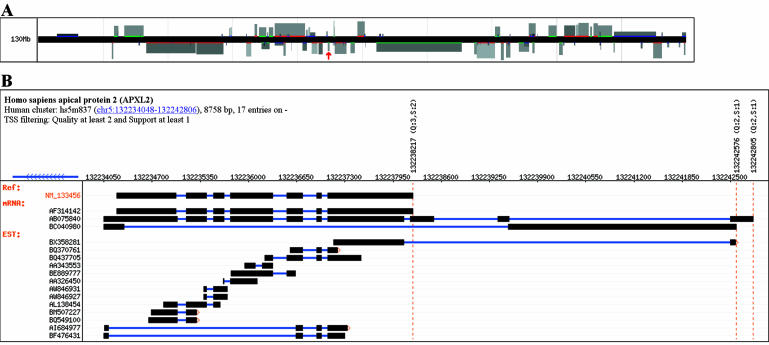
Clusters on the plus strand are shown above the line, while those on the minus strand are shown below the line. The cluster's location and width in the display represent its genomic location and size. The cluster's height represents the number of transcripts in the cluster, and its shade represents the highest quality score of the TSS predictions. Figure 1A shows an example of the chromosome browser display. Blocks can be clicked to show the details of the cluster in the cluster viewer. The chromosome viewer is helpful in understanding the neighborhood of a cluster and how this may affect its regulation.
Figure 1.
(A) A region of human chromosome 5 shown by the Chromosome Viewer of PromoSer. Each block indicates a cluster of transcripts that correspond to a gene. The block height represents the number of transcripts in the cluster, and its shade represents the highest quality score of the TSS predictions. The block pointed to by a red arrow is the human apical protein 2 (APXL2), and is detailed in (B). (B) A Cluster-Viewer display of APXL2 on chromosome 5. Notice the gradual view of the cluster revealed by the aligned ESTs and the three possible alternative transcription start sites indicated by red vertical dashed lines.
The cluster viewer represents the elements comprising a cluster in a track-type display, similar to the one used by the UCSC genome browser (8). Our viewer is specialized and contains additional features to show (as vertical lines across the image) predicted TSSs that are above certain specifiable thresholds. Alignments are shown as connected horizontal blocks representing exons. If an alignment has more than five bases unaligned at its 5′ end, a small arrow appears beside the alignment to indicate the truncation. Tracks can be clicked to link to the corresponding GenBank record. The TSS annotation can also be clicked to obtain a zoom-in view centered on the TSS at single-base resolution. Figure 1B shows an example of the cluster viewer.
The cluster viewer can be accessed from the chromosome viewer or through a home page where a user can enter an accession number to search for and specify thresholds of quality and support scores. In the results report, clicking a promoter entry invokes the cluster viewer containing the cluster that corresponds to the entry.
THE SOAP INTERFACE
Simple Object Access Protocol (SOAP) is emerging as a favored standard for access to various web services in a platform- and language-independent manner (9). It provides easy access to such services in programming and in pipeline-type applications. A fully functional SOAP server for PromoSer that captures most of the web user interface functionality has recently been implemented. The service is described with a Web Services Description Language (WSDL) document accessible at http://biowulf.bu.edu/zlab/promoser/promoser.wsdl. The service works using a job-ticket model, in which a reference ID is immediately returned upon successful request submission. The user can then poll the server to check for the availability of the results for that job ticket. When found, PromoSer will return an XML-formatted results file that contains both the web-based report and the promoters.
COMPARISON TO OTHER PROMOTER-RETRIEVAL METHODS
Several methods have the ability to extract promoter sequences, including EnsMart (10), EZ-Retrieve (11), CONPRO (12) and DBTSS (13). A number of important features set PromoSer apart from these services, summarized as follows.
PromoSer has mechanisms to track (and select) alternative promoters identified for each cluster of transcripts. No other methods currently provide this function. Alternative transcription start sites and the resulting alternative proximal promoters are an emerging genomic regulation paradigm recently recognized to participate in the huge variability of the transcriptome and the proteome relative to the rather limited annotated genome (together with alternative splicing and alternative termination).
PromoSer relies solely on filtered experimental data. No computationally predicted gene models are used in PromoSer's clusters and TSS predictions. Together with its alternative promoter concept, our approach will allow a more robust and comprehensive examination of proximal promoter organization. Both EnsMart and CONPRO rely on predicted gene models.
PromoSer relies on an integrative approach that combines together all transcript sequence data made publicly available (by being deposited into GenBank) for the supported organisms. This data set is ever expanding and PromoSer will continue to be updated to utilize new additions. Many other resources (e.g. DBTSS) rely only on their internally generated data to make TSS predictions and are thus less comprehensive and slower to update than PromoSer.
As PromoSer is mainly aimed toward facilitating promoter extraction, it is able to provide an intuitive, uncluttered and specific user interface.
PromoSer provides a batch extraction service that can be queried using either common sequence identifiers or sequences in the FASTA format, from the same convenient user interface. In contrast, most other resources accept either identifiers (e.g. EZ-Retrieve) or sequences (e.g. CONPRO), but not both. In addition, some are often limited to single queries (e.g. DBTSS), and others require the user to go through multiple screens with many options (e.g. EnsEMBL). PromoSer also supports a SOAP interface, which greatly facilitates access by advanced users and computer scripts. This feature is essential for integrating diverse biological data and applications (9). Currently we are not aware of other SOAP interfaces for batch retrieval of promoter sequences.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Peter M. Haverty for detailed testing of the SOAP interface and many helpful suggestions about the XML output. We thank Brian Pierce for thoroughly proof reading the manuscript. This work has been supported in part by NSF grants DBI-0078194 and MRI DBI-0116574, and NIH grants 1P20GM066401-01 and 1R01HG03110-01.
REFERENCES
- 1.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 2.Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 3.Novina C.D. and Roy,A.L. (1996) Core promoters and transcriptional control. Trends Genet., 12, 351–355. [PubMed] [Google Scholar]
- 4.Halees A.S., Leyfer,D. and Weng,Z. (2003) PromoSer: a large-scale mammalian promoter and transcription start site identification service. Nucleic Acids Res., 31, 3554–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wheeler D.L., Church,D.M., Federhen,S., Lash,A.E., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Sequeira,E., Tatusova,T.A. et al. (2003) Database resources of the National Center for Biotechnology. Nucleic Acids Res., 31, 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmid C.D., Praz,V., Delorenzi,M., Perier,R. and Bucher,P. (2004) The Eukaryotic Promoter Database EPD: the impact of in silico primer extension. Nucleic Acids Res., 32 (Database issue), D82–D85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kent W.J. (2002) BLAT—the BLAST-like alignment tool. Genome Res., 12, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kent W.J., Sugnet,C.W., Furey,T.S., Roskin,K.M., Pringle,T.H., Zahler,A.M. and Haussler,D. (2002) The human genome browser at UCSC. Genome Res., 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stein L. (2002) Creating a bioinformatics nation. Nature, 417, 119–120. [DOI] [PubMed] [Google Scholar]
- 10.Kasprzyk A., Keefe,D., Smedley,D., London,D., Spooner,W., Melsopp,C., Hammond,M., Rocca-Serra,P., Cox,T. and Birney,E. (2004) EnsMart: a generic system for fast and flexible access to biological data. Genome Res., 14, 160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H., Ramanathan,Y., Soteropoulos,P., Recce,M.L. and Tolias,P.P. (2002) EZ-Retrieve: a web-server for batch retrieval of coordinate-specified human DNA sequences and underscoring putative transcription factor-binding sites. Nucleic Acids Res., 30, e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu R. and States,D.J. (2002) Consensus promoter identification in the human genome utilizing expressed gene markers and gene modeling. Genome Res., 12, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki Y., Yamashita,R., Nakai,K. and Sugano,S. (2002) DBTSS: DataBase of human Transcriptional Start Sites and full-length cDNAs. Nucleic Acids Res., 30, 328–331. [DOI] [PMC free article] [PubMed] [Google Scholar]