Abstract
A web application for inferring potentially stabilizing non-bonding interactions in macromolecular structures from input atomic coordinate data is described. The core software, called Monster, comprises a PERL wrapper that takes advantage of scripts developed in-house as well as established software in the public domain to validate atomic coordinate files, identify interacting residues and assign the nature of these interactions. The results are assembled and presented in an intuitive and interactive graphical format. Potential applications of Monster range from mining and validating experimentally determined structures to guiding functional analysis. Non-commercial users can perform Monster analysis free of charge at http://monster.northwestern.edu.
INTRODUCTION
Normal function of biological macromolecules relies on sequence-directed folding into defined three-dimensional (3D) shapes. Although reliable a priori prediction of sequence elements essential for proper folding and function remains elusive at present, high-resolution nuclear magnetic resonance (NMR) and crystal structures provide important clues. Indeed, essential residues are usually recognized by virtue of their involvement in potentially stabilizing intra- and/or inter-molecular non-bonding interactions in these structures, which then provides a rational basis for hypothesis generation and testing.
Contemporary atomic coordinate data mining efforts aimed at identifying residue-specific interactions rely on a variety of software programs, each optimized for the discovery of a particular type of non-bonding interaction. Software programs for the comprehensive description of interactions for macromolecular complexes have also been described (1–4). However, either these are restricted to certain types of complex (e.g. protein–ligand, protein–nucleic acid) or the results are available in a pre-computed format. Here we describe a web application that integrates multiple, commonly used software programs for comprehensive and interactive mining of the structures of macromolecules and macromolecular complexes. We envision the tool being especially useful for structural biologists who perform these types of analysis upon completion of a successful structure determination effort and prior to depositing coordinates in a public database. The tool is also intended for those biologists who may be interested in interrogating macromolecular structures for functional analyses, but who otherwise may not be experienced in analyzing the information using 3D molecular graphics programs.
METHODS
Broadly, Monster analyzes atomic coordinate files (ACFs) in three discrete steps: (i) parsing of the ACF, (ii) identification and classification of interactions and (iii) collation and output of results. Parsing includes interpretation of the ACF and incorporates user-specified definitions of interacting polypeptide segments (Figure 1). An option to generate coordinates for missing hydrogen atoms or to replace existing hydrogen atoms in the input ACF is provided. In either case, protonation of the structure is performed using the program WHAT IF (5). In the second step, Monster farms out the calculation of contact surface areas and hydrogen bond detection to the MSMS (6) and HBPLUS programs, respectively (7). To identify hydrophobic and electrostatic interactions, Monster uses a simple classification scheme of hydrophobic, positive, negative or neutral for every atom of standard amino acids and nucleotides. Monster reports a hydrophobic contact if the distance between two hydrophobic atoms is less than 5 Å. Similarly, an interaction is deemed electrostatic if a pair of positively and negatively charged atoms is within 7 Å (8,9). These cutoffs can be altered by the user while viewing the results (see below). In the final step, Monster integrates the results from these scripts with those from various other programs (see above) to produce a list of all non-covalent interactions (henceforth also referred to as ‘bonds’) and the contact surface areas in ASCII and XML file formats. An ACF corresponding to the user-specified interacting polypeptide segments in RCSB PDB format (10) is also generated.
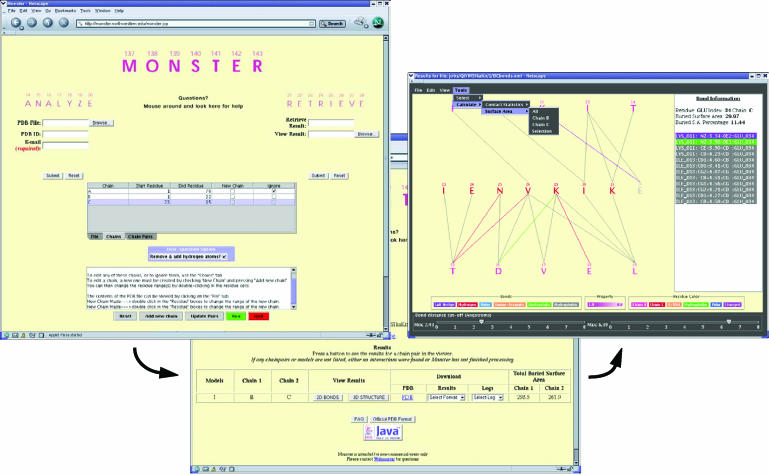
Figure 1.
Screenshots of the ACF parser (left), results retriever (center) and bonds viewer (right) of Monster.
IMPLEMENTATION AND SCOPE
The core software for Monster was developed using PERL. The WHAT IF, MSMS and HBPLUS programs were compiled to run on a Linux platform. The Monster web interface consists of JSP and HTML pages. An Apache Httpd is the primary web server, which runs in parallel to an Apache TomCat server for compiling the JSP pages. Two Java Swing (version 1.4.2) applets were implemented, one that serves as the front-end for interactive user input (designated the ‘ACF parser’) and another for viewing the results in an interactive graphical format (designated the ‘bonds viewer’; Figure 1). The interacting polypeptide segments defined by the user can be viewed in 3D with the Java molecular viewer WebMol (11).
The ACF parser gives the user complete control over the pair of polypeptide chains, or segments thereof, to be used for analysis. Although Monster was originally developed for analyzing inter-molecular interactions, we have provided additional options that permit analysis of intra-molecular interactions as well. This is accomplished by duplicating chains and specifying interacting residue ranges within the parser. For the new or less experienced user, a Help utility and several illustrative examples within the Monster documentation have been provided. The bonds viewer interprets XML files generated by Monster and depicts pairwise interactions as solid lines between residues on a canvas. Present functionality includes the provision to turn on or off different bond types, interactive adjustment of the display to a specified range of bond lengths and the ability to rank residues by color based on bond density (i.e. the number of bonds formed) and contributions to the contact surface area. The latter feature permits the straightforward identification of important residues and is likely to be especially useful to molecular biologists seeking to identify and prioritize residues for mutational analysis. Clicking on any residue on the canvas allows the user to view the bond and contact surface area information for that residue in a sidebar (Figure 1). Provisions to color residues by type (defined by the user) and chain, to control which residues are displayed on the canvas and to calculate bond density and contact surface area for selected residue subsets are some of the additional features included in the viewer. Views can be saved as a PostScript file, a format that can be edited using programs such as Adobe Illustrator®.
Monster has been successfully tested on a variety of protein–protein/peptide, protein–nucleic acid and higher-order nucleic acid complexes. Over two dozen complexes have been tested and the results are available online by following the documentation link from the homepage. The PERL wrapper is presently geared toward parsing ACFs that conform to the RCSB PDB format (10). The current version parses all ATOM and MODEL records and HETATM records for water molecules. Thus, ACFs that contain more than one model, such as those from NMR structure determinations, are also readily analyzed. In addition to producing the normal XML file for each model, Monster produces an overview XML file that permits interactive viewing in the bonds viewer of pairwise interactions in each model as well as the frequency of occurrence of every interaction. The latter allows for better mining of interactions in NMR structures since the average or representative structure can fail to capture all the key elements of an ensemble. By default, only those inter-molecular interactions detected in >50% of the structures are presented in the overview file. Since NMR structures are derived from inter-proton NOEs (nuclear Overhauser effects), which bear a resemblance to the pairwise interactions reported by Monster, we envision that the tool could also be used to check for potential NOEs predicted by the calculated structures This represents another approach for validating NMR structures.
FUTURE DIRECTIONS
The current incarnation of Monster represents the first step of a long-term undertaking to develop tools for interrogating databases of high-resolution macromolecular structures. Obligatory future extensions of Monster include the ability to analyze interactions involving atoms in all HETATM records, the ability to handle alternative input ACF formats in addition to the RCSB PDB format (e.g. XML) and the development of a high-throughput version that will permit analysis of multiple ACFs (as in a database). In conclusion, we have created an interactive tool that complements 3D molecular graphics visualization tools for analyzing non-covalent interactions in macromolecular structures. We anticipate that the tool will be useful in enhancing our understanding of the principles of macromolecular structure and function.
Acknowledgments
ACKNOWLEDGEMENTS
Jin Suntivich is thanked for developing the Monster Help utility. This work was supported by a career development award to I.R. from a SPORE grant in Prostate Cancer from NCI (CA90386). W.J.S. was supported by an undergraduate research grant from Northwestern University.
REFERENCES
- 1.Allers J. and Shamoo,Y. (2001) Structure-based analysis of protein–RNA interactions using the program ENTANGLE. J. Mol. Biol., 311, 75–86. [DOI] [PubMed] [Google Scholar]
- 2.Laskowski R.A., Hutchinson,E.G., Michie,A.D., Wallace,A.C., Jones,M.L. and Thornton,J.M. (1997) PDBsum: a web-based database of summaries and analyses of all PDB structures. Trends Biochem. Sci., 22, 488–490. [DOI] [PubMed] [Google Scholar]
- 3.Sobolev V., Sorokine,A., Prilusky,J., Abola,E.E. and Edelman,M. (1999) Automated analysis of interatomic contacts in proteins. Bioinformatics, 15, 327–332. [DOI] [PubMed] [Google Scholar]
- 4.Neshich G., Togawa,R.C., Mancini,A.L., Kuser,P.R., Yamagishi,M.E., Pappas,G.,Jr, Torres,W.V., Fonseca e Campos,T., Ferreira,L.L., Luna,F.M. et al. (2003) STING Millennium: a web-based suite of programs for comprehensive and simultaneous analysis of protein structure and sequence. Nucleic Acids Res., 31, 3386–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vriend G. (1990) WHAT IF: a molecular modeling and drug design program. J. Mol. Graph., 8, 52–56. [DOI] [PubMed] [Google Scholar]
- 6.Sanner M.F., Olson,A.J. and Spehner,J.C. (1996) Reduced surface: an efficient way to compute molecular surfaces. Biopolymers, 38, 305–320. [DOI] [PubMed] [Google Scholar]
- 7.McDonald I.K. and Thornton,J.M. (1994) Satisfying hydrogen bonding potential in proteins. J. Mol. Biol., 238, 777–793. [DOI] [PubMed] [Google Scholar]
- 8.Draper D.E. (1999) Themes in RNA-protein recognition. J. Mol. Biol., 293, 255–270. [DOI] [PubMed] [Google Scholar]
- 9.Sharp K.A., Honig,B. and Harvey,S.C. (1990) Electrical potential of transfer RNAs: codon–anticodon recognition. Biochemistry, 29, 340–346. [DOI] [PubMed] [Google Scholar]
- 10.Westbrook J., Feng,Z., Jain,S., Bhat,T.N., Thanki,N., Ravichandran,V., Gilliland,G.L., Bluhm,W., Weissig,H., Greer,D.S. et al. (2002) The Protein Data Bank: unifying the archive. Nucleic Acids Res., 30, 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walther D. (1997) WebMol—a Java-based PDB viewer. Trends Biochem. Sci., 22, 274–275. [DOI] [PubMed] [Google Scholar]