Abstract
We have developed a web server (ProteinDBS) for the life science community to search for similar protein tertiary structures in real time. This system applies computer visualization techniques to extract the predominant visual patterns encoded in two-dimensional distance matrices generated from the three-dimensional coordinates of protein chains. When meaningful contents, represented in a multi-dimensional feature space, have been extracted from distance matrices, an advanced indexing structure, Entropy Balanced Statistical (EBS) k-d tree, is utilized to index the data. Our system is able to return search results in ranked order from a database with 46 075 chains in seconds, exhibiting a reasonably high degree of precision. To our knowledge, this is the first real-time search engine for protein structure comparison. ProteinDBS provides two types of query method: query by Protein Data Bank protein chain ID and by new structures uploaded by users. The system is hosted at http://ProteinDBS.rnet.missouri.edu.
INTRODUCTION
To study the structure–function relationship in proteins, life science researchers have a great need for protein structure retrieval systems for searching for similar three-dimensional (3D) structures. Some concurrent protein structure comparison tools perform structure-to-structure comparison by applying dynamic programming techniques (1–4). The task of such structural alignment is known to be computationally expensive (5).
In recent years, researchers have developed particular approaches to improve both the efficiency and accuracy of structural comparison. Can and Wang (6) applied differential geometry knowledge to protein three-dimensional (3D) structure for extracting signatures such as curvature, torsion and secondary structure type. Camogla et al. (7) built an indexing structure based on secondary structure element triplets using R*-tree. Chionh et al. (8) proposed the SCALE algorithm to compare protein 3D structures based on angle–distance matrices that utilize angles and distances between secondary structure elements. The majority of these works used structural alignment algorithms to find similar chains.
With the advent of new technologies such as synchrotron radiation sources and high-resolution nuclear magnetic resonance (NMR), a great number of new protein structures have been determined in recent years. At February 10, 2004, the primary structural database, the Protein Data Bank (PDB; http://www.rcsb.org/pdb/), contained 24 248 protein structures (∼800 more were added in two months). Given the fact that each protein has either one or multiple polypeptide chains, there were ∼48 000 protein chain structures. Concurrent search engines (through email) often take hours or even days to return a query result. To our knowledge, there is no real-time search engine available to interactively identify similar structures given a new protein tertiary structure. As we expect the number of known protein structures will grow at least at a linear rate, maintaining efficiency and accuracy when finding similar chains in a large database remains a great challenge.
DESCRIPTION AND APPLICATION
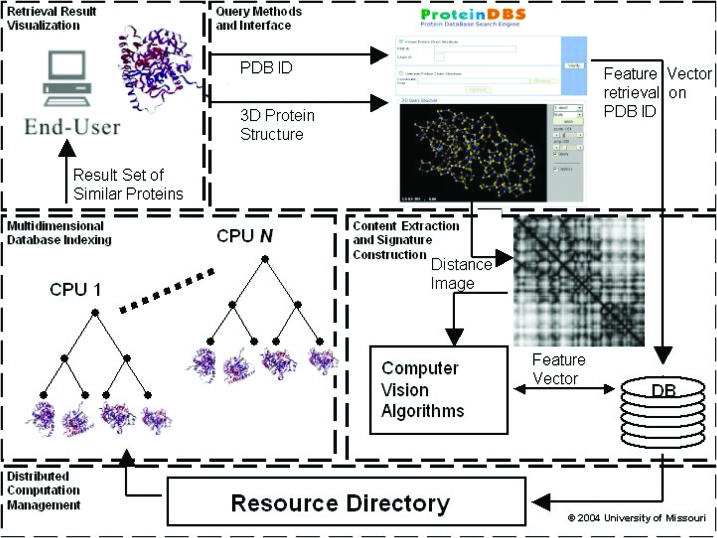
We have implemented ProteinDBS to provide a real-time search engine for the life science community. Figure 1 shows the system architecture, which contains five modules: (i) query methods and interface, (ii) content extraction and signature construction, (iii) multi-dimensional database indexing for protein signatures, (iv) distributed computation management for a large-scale multi-user environment and (v) retrieval results visualization. Owing to page limits in this web server special issue, interested readers are referred to (9) for further discussion of the detailed algorithms. A system tutorial can be viewed at the ProteinDBS website.
Figure 1.
ProteinDBS system diagram. An end-user submits a protein structure encoded in the PDB format or the PDB ID of a protein structure. Given a protein structure PDB file, the interface converts the 3D coordinate structure into a 2D distance matrix image. The application of various computer visualization algorithms produces a protein signature that is used as a query against the database. The protein structure, either computed or retrieved from a relational Database Management System (DBMS) given the PDB ID, is used to determine a similarity-ranked retrieval set from the distributed EBS k-d tree indices. The most similar proteins are then displayed back to end-user.
Query methods and interface
There are two types of query method: query by a new protein structure that is unavailable in the PDB database and query by PDB ID. Using Internet browsers, users can upload a new protein chain structure in PDB format or provide a PDB ID to find similar protein chains in the database.
Content extraction and signature construction
After mapping 3D protein structures into 2D distance matrices (3), the system analyzes the 2D matrices and does further structure comparison based on the patterns in the matrices. Our assumption is that protein chains from the same family should have similar visual patterns in their 2D distance matrices locally and globally. The system applies a suite of computer vision algorithms (10,11) to measure the histograms and textures from the distance matrices. In our current implementation, 23 features are extracted for each protein chain structure. In this 23D feature space, each protein chain structure is represented by a data point.
Multi-dimensional database indexing
The system then uses an advanced tree structure, Entropy Balanced Statistical (EBS) k-d tree (12), to index the distance matrices. By indexing signatures extracted from distance matrices, structural comparison is formulated as a process of finding nearest neighbors in the feature space. Efficiency is achieved by traversing through the EBS k-d tree structure to find similar chains with low computational complexity. In our current setup, the entire PDB database (November 2003 collection) is mapped into 46 075 points.
Distributed computation management
An important aspect of our retrieval system is its distributed nature, more precisely, the distribution of computational tasks. Various architectures involving distributed Java database schemes have been reported in the literature (13). Our system employs a resource directory to manage a collection of distributed index agents. These components handle various tasks including index organization, load balancing, database indexing and retrieval. This architecture allows the distribution of functional abilities and computational demands across various systems. This design makes the system highly expandable in terms of parallelism of indices.
Retrieval results visualization
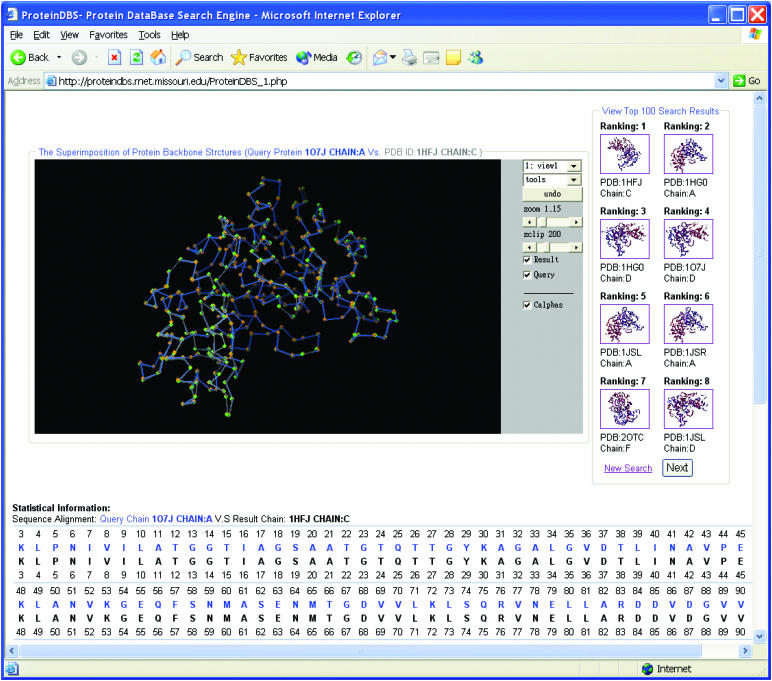
Figure 2 shows the retrieval results for query protein chain 1o7j_A. A set of ranked structures is returned to the user eight at a time. To visualize the quality of search results, a 3D superimposition view of the query structure and the retrieval result is displayed to the user. Figure 2 presents the superimposition view of the query protein chain and the top ranked result, 1hfj_C, generated by clicking on the thumbnail image in the righthand panel. The sequence alignment result is also displayed to the user with root mean square deviation (RMSD) and alignment length values (14).
Figure 2.
ProteinDBS retrieval results visualization. The top left panel shows a superimposed view of a query protein chain and a selected result chain from the top-ranked list (the first ranked protein chain in this figure). Users can check ‘Result’ and/or ‘Query’ to view the retrieved chain and/or the query chain. By clicking on a thumbnail image in the top right panel, users can see an enlarged version of the result superimposed on the query. The lower panel displays the structure alignment results represented by sequences. The RMSD and alignment length, not shown in this figure, are listed immediately after the sequence alignment panel.
Performance evaluation
Two major performance measurements for ProteinDBS, retrieval accuracy and efficiency, have been evaluated using testing data that contained 50 representative groups of protein chains in SCOP (http://scop.mrc-lmb.cam.ac.uk/scop/) release 1.63, with 776 protein chains in total. Each protein chain is used as a query with unknown structure. If the top-ranked results are from the same structural family, we count them as good results. On average, our system exhibits retrieval precision of 84.15% at the 10% recall rate, and 48.54% precision at the 100% recall rate. To query the protein chain with maximum length in the testing set, 566 Cα atoms, it takes 10 s to return ranked search results. These tests were conducted on a Linux server with Quad Xeon III 550 MHz processors with 2 MB cache, 2 GB RAM and a 140 GB SCSI LVD-160 7200 rpm hard drive.
CONCLUSION AND FUTURE WORK
To understand structure–function relationships in the protein universe, biologists are demanding an efficient system to retrieve similar protein chains, especially for new structures that have been discovered recently. It is clear that current structure comparison and retrieval systems are not efficient enough to meet the needs of life science researchers for finding relevant structures for further study. To address this issue, we developed ProteinDBS to provide a real-time protein chain comparison.
To improve retrieval accuracy, we will continue to develop new feature extraction algorithms to handle local-to-global and local-to-local comparisons. Our research plan also includes understanding the relationships between secondary structures and visual patterns in distance matrices and utilizing the EBS k-d tree to achieve rapid classification of unknown protein structures.
To maintain an up-to-date system to serve the life science community, we will populate the database on a monthly basis with newly discovered protein structures released by the PDB.
Acknowledgments
ACKNOWLEDGEMENTS
The authors are grateful to the researchers and groups who made the following software packages and databases available for us to use in ProteinDBS: the Raster3D molecular graphics package (15), which generates ribboned protein images, the Kinemage graphic packages for presenting 3D interactive protein structures, the SCOP database for ground truth testing, and the Protein Data Bank (16) for maintaining the tertiary structures.
REFERENCES
- 1.Needleman S.B. and Wunsch,C.D. (1970) A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol., 48, 443–453. [DOI] [PubMed] [Google Scholar]
- 2.Smith T.F. and Waterman,M.S. (1970) Identification of common molecular subsequences. J. Mol. Biol., 147, 195–197. [DOI] [PubMed] [Google Scholar]
- 3.Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- 4.Shindyalov H.N. and Bourne,P.E. (1998) Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng., 9, 739–747. [DOI] [PubMed] [Google Scholar]
- 5.Godzik A. (1996) The structural alignment between two proteins: is there a unique answer? Protein Sci., 5, 1325–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Can T. and Wang,Y.F. (2003) CTSS: a robust and efficient method for protein structure alignment based on local geometrical and biological features. Proceedings of the IEEE Computer Society Conference on Bioinformatics, August 2003, pp. 169–179. [PubMed] [Google Scholar]
- 7.Camogla O., Kahveci,T. and Singh,A. (2003) PSI: indexing protein structures for fast similarity search. Bioinformatics, 19(Suppl. 1), I81–I83. [DOI] [PubMed] [Google Scholar]
- 8.Chionh C.H., Huang,Z., Tan,K.L., and Yao,Z. (2003) Augmenting SSEs with structural properties for rapid protein structure comparison, Proceedings of the Third IEEE Symposium on Bioinformatics and Bioengineering, pp. 341–348.
- 9.Chi P.H., Scott,G. and Shyu,C.R. (2004) A fast protein structure retrieval system using image-based distance matrices and multidimensional index, Proceedings of the Fourth IEEE Symposium on Bioinformatics and Bioengineering.
- 10.Haralick R.M., Shanmugam,K. and Dinstein,I. (1973) Textural features for image classification. IEEE Trans. Sys. Man Cybernet., SMC-3, 610–621. [Google Scholar]
- 11.Rosenfeld A. and Kak,A.C. (1982) Digital Picture Processing. Academic Press, New York. [Google Scholar]
- 12.Scott G. and Shyu,C.R. (2003) EBS k-d tree—an entropy balanced statistical k-d tree for image databases with ground-truth labels'. Proceedings of the International Conference on Image and Video Retrieval, Urbana-Champaign, IL, 24–25 July, pp. 467–476. [Google Scholar]
- 13.Ro Y., Tsuchida,S., Tamura,M., Nagata,M. and Nakamori,Y. (1999) Remote method invocation based Web database system for global environment models. IEEE SMC'99 Conference Proceedings. pp. 563–568. [Google Scholar]
- 14.Alexandrov N.N. (1996) SARFing the PDB. Protein Eng., 9, 727–732. [DOI] [PubMed] [Google Scholar]
- 15.Merritt E.A. and Bacon,D.J. (1997) Raster3D photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- 16.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]