Abstract
Recently, cell culture systems producing hepatitis C virus particles (HCVcc) were developed. Establishment of serum-free culture conditions is expected to facilitate development of a whole-virus inactivated HCV vaccine. We describe generation of genotype 1–6 serum-free HCVcc (sf-HCVcc) from Huh7.5 hepatoma cells cultured in adenovirus expression medium. Compared to HCVcc, sf-HCVcc showed 0.6 to 2.1 log10 higher infectivity titers (4.7–6.2 log10 Focus Forming Units/mL), possibly due to increased release and specific infectivity of sf-HCVcc. In contrast to HCVcc, sf-HCVcc had a homogeneous single-peak density profile. Entry of sf-HCVcc depended on HCV co-receptors CD81, LDLr, and SR-BI, and clathrin-mediated endocytosis. HCVcc and sf-HCVcc were neutralized similarly by chronic-phase patient sera and by human monoclonal antibodies targeting conformational epitopes. Thus, we developed serum-free culture systems producing high-titer single-density sf-HCVcc, showing similar biological properties as HCVcc. This methodology has the potential to advance HCV vaccine development and to facilitate biophysical studies of HCV.
Keywords: Hepatitis C virus, cell culture system, serum-free, high-titer, biophysical characterization, vaccine development, receptor blocking, neutralization, genotypes, adenovirus expression medium
Introduction
Hepatitis C virus (HCV) is a major public healthcare burden with 3–4 million new infections occurring each year and more than 150 million individuals estimated to be chronically infected worldwide [1]. Many of these individuals develop serious chronic liver diseases such as cirrhosis and hepatocellular carcinoma, making HCV the most frequent cause of liver transplantation [2].
HCV is an enveloped, positive-stranded RNA virus of the genus Hepacivirus within the Flaviviridae family. Due to a high degree of genetic heterogeneity, HCV has been classified in 6 epidemiologically important genotypes and numerous subtypes, differing in approximately 30% and 20% of their nucleotide and amino acid sequence, respectively [3,4]. Genotypes show important clinical and biological differences [5–10]. Serotypes have not been defined; however, different genotypes and subtypes show differential sensitivity to neutralizing antibodies found in sera of chronically infected patients and to monoclonal neutralizing antibodies with therapeutic potential [6,11–14].
The 9.6 kb HCV genome consists of 5’ and 3’ untranslated regions and a single open reading frame encoding structural proteins (Core, E1 and E2), the viroporin p7, and nonstructural proteins (NS2, NS3, NS4A, NS4B, NS5A and NS5B) [4]. The HCV virion is believed to consist of a nucleocapsid of HCV Core proteins containing the genomic RNA, covered by a lipid envelope with the HCV envelope glycoproteins E1 and E2. The HCV life cycle is tightly linked to the hepatic lipid metabolism. During assembly and release, the HCV virion is believed to associate with very-low-density-lipoprotein (VLDL) or VLDL-like structures, creating lipo-viro-particles (LVP) [15,16]. Thus, HCV apparently circulates in infected patients associated to different classes of lipoproteins [16], resulting in a heterogeneous density profile apparent following buoyant density gradient ultracentrifugation [16,17]. Components of the VLDL assembly and secretion pathway, such as apolipoprotein E (ApoE), might be important for the association between HCV and lipoproteins [18].
HCV entry is mediated by several co-receptors, including CD81, the low-density-lipoprotein receptor (LDLr) and the scavenger receptor class B type I (SR-BI) [19]. While HCV is believed to interact directly with CD81 through E2 [20,21], interactions with other receptors, such as LDLr and SR-BI, might occur through lipoprotein components present on the LVP, such as ApoE [22,23], although direct interactions between E2 and SR-BI have also been reported [23,24]. Eventually, HCV is internalized through clathrin-mediated endocytosis [25,26].
There is no vaccine available for HCV. Current standard-of-care, based on pegylated interferon-α2 and ribavirin, has limited efficacy and is associated with severe side effects and contraindications [9]. Even though promising new compounds for treatment of HCV are being developed and licenced [9,10], only a minority of HCV-infected individuals is expected to be diagnosed and treated, mainly due to the asymptomatic nature of infection, economic constraints and contraindications [1]. Thus, an HCV vaccine is needed to control HCV globally. Most successful antiviral vaccines employ inactivated or attenuated whole viral particles as vaccine antigen and depend on the induction of neutralizing antibodies [27,28]. Due to a lack of HCV particle-producing cell culture systems, this approach was not feasible for HCV [29,30].
Only in 2005, the first HCV cell culture system supporting the full viral life cycle was developed, based on the genotype 2a isolate JFH1 and the human hepatoma cell line Huh7 and derived cell lines [31–33]. Subsequently, culture systems producing HCV particles (HCVcc) of the major genotypes were developed using JFH1-based recombinants expressing genotype specific Core, E1, E2, p7 and NS2 [11,12,34–37]. Such particles could serve as antigens in a whole-virus inactivated HCV vaccine primarily aiming at induction of neutralizing antibodies against structural proteins of the major HCV genotypes.
However, HCVcc yields from the developed cell culture systems are relatively low compared to quantities envisioned to be required for vaccine production. Further, as patient derived HCV particles [17], HCVcc showed a heterogeneous density profile [6,32,38,39], making density-based purification and concentration procedures difficult. Also, cell cultures are typically treated with animal-derived trypsin, and growth medium used for production of HCVcc is typically supplemented with fetal bovine serum (FBS) [40]. Vaccine development, as well as other research applications, such as biophysical studies of HCV particle composition, require generation of purified and concentrated HCVcc stocks. This is expected to be facilitated by reducing concentrations of non-HCV proteins such as FBS derived proteins in HCVcc producing cell cultures. Further, use of FBS and animal-derived trypsin increases the risk of contamination with adventitious microbial agents, of relevance for HCV vaccine development [40,41]. Thus, development of methods for production of HCVcc under serum-free conditions is a research focus. At the onset of this study it had been demonstrated that Huh7 cells could be cultured in serum-free medium (RPMI 1640 supplemented with Na2SeO3) without previous adaptation for an extended period of time, and that serum-free cell cultures (DMEM supplemented with Na2SeO3 and lipid rich albumin) allowed replication of HCV [42,43].
In this study, we aimed at developing and characterizing serum-free genotype 1–6 HCVcc particles (sf-HCVcc). From infected Huh7.5 cell cultures maintained in adenovirus expression medium (AEM) without using trypsin, we were able to harvest genotype 1–6 sf-HCVcc containing culture supernatants. The sf-HCVcc particles were characterized by increased infectivity titers and an altered density profile with a single infectivity peak, expected to facilitate density-based purification procedures. However, sf-HCVcc showed similar biological properties as HCVcc regarding routes of viral entry and susceptibility to neutralizing antibodies. Establishment of a robust methodology for generation of high-titer single-density serum-free HCVcc is expected to aid HCV vaccine development.
Materials and Methods
Huh7.5 cell culture and infection with HCV recombinants
Human hepatoma Huh7.5 cells were cultured in culture flasks (Nunc) in DMEM (Gibco/Invitrogen) supplemented with 10% fetal bovine serum (FBS, [Sigma]), penicillin 100 U/mL and streptomycin 100 µg/mL (Gibco/Invitrogen), referred to as DMEM + 10% FBS. Cells were kept sub-confluent and split every 2–3 days. For splitting, cells were washed in PBS (Invitrogen) and detached using trypsin (Sigma-Aldrich). For serum-free cultures, cells were plated in DMEM + 10% FBS. When cells were 80% confluent, DMEM + 10% FBS was removed, cells were washed in PBS and adenovirus expression medium (AEM, [Gibco/Invitrogen]) supplemented with penicillin 100 U/mL and streptomycin 100 µg/mL, referred to as AEM, was added without splitting the cells. Every 2–3 days, supernatant was removed and fresh AEM was added to the cells. Huh7.5 cell cultures were maintained at 37°C and 5% CO2.
Generation of HCVcc virus stocks
For generation of HCVcc and sf-HCVcc virus stocks, Huh7.5 cells cultured in DMEM + 10% FBS at 80% confluency were infected at a multiplicity of infection (MOI) of 0.003 using 1st or 2nd viral passage stocks of the following HCVcc intra- and intergenotypic recombinants: H77C/JFH1V787A,Q1247L(referred to as H77(1a)), J4/JFH1F886L,Q1496L(J4(1b)), J6/JFH1 (J6(2a)), S52/JFH1I787S,K1398Q (S52(3a)), ED43/JFH1T827A,T977S (ED43(4a)), SA13/JFH1A1022G,K1119R (SA13(5a)), and HK6a/JFH1F350S,N417T (HK6a(6a)) [11,12,32,34,35]. The % infection was monitored by HCV-specific immunostaining as described below. For generation of HCVcc virus stocks, cells were maintained in DMEM + 10% FBS; supernatants were harvested every 2–3 days, when cells were split, until % of infected cells declined [34], as detected by immunostaining. High-titer stocks, collected at the peak of viral infection, were used for further experiments. Stocks with relatively low peak titers were concentrated using Amicon 100 kDa centrifugation filters (Millipore). For generation of sf-HCVcc virus stocks, DMEM + 10% FBS cell cultures with 40–80% HCV infected cells were washed with PBS and AEM was added. Cells were maintained in AEM, and supernatants were harvested every 2–3 days, when AEM was exchanged, for up to 29 days. Supernatants were sterile filtered and stored at −80°C. The HCV Core-E2 sequences of all virus stocks used for further experiments were determined by direct sequencing (described below). Sequences were identical to the plasmid sequence unless otherwise indicated in respective figure and table legends.
Evaluation of HCV infected cell cultures
Spread of HCV recombinants in cell cultures was monitored by HCV NS5A immunostaining. Cells plated onto chamber slides (Nunc) the previous day were fixed for 10 minutes in ice-cold acetone (Sigma-Aldrich) and washed twice with PBS and twice with PBS + 0.1% Tween-20 (Sigma-Aldrich). Cells were stained for HCV NS5A using primary anti-NS5A antibody 9E10 [32] at 1:1,000 dilution in PBS + 1% bovine serum albumin (BSA, [Roche Applied Science]) + 0.2% skim milk (PBS/BSK) for two hours at room temperature. Cells were washed twice with PBS and twice with PBS + 0.1% Tween-20, and stained using secondary antibody Alexa Fluor 594-conjugated goat anti-mouse IgG (H+L) (Invitrogen) at 1:500 dilution and Hoechst 33342 (Invitrogen) at 1:1,000 dilution in PBS + 0.1% Tween-20. Cells were washed twice in PBS, before being covered by Fluoromount-G (SouthernBiotech) and a cover-slip.
Culture supernatant infectivity titers were determined as Focus Forming Units (FFU)/mL. Huh7.5 cells, plated at 6,000 cells/well onto poly-D-lysine coated 96-well plates (Nunc) the day before, were infected with serially-diluted supernatants (lowest dilution 1:2). Forty-eight hours after infection, cells were fixed in ice-cold methanol and washed twice with PBS + 0.1% Tween-20 before being incubated with 3% H2O2 for five minutes at room temperature. Cells were washed twice with PBS + 0.1% Tween-20 and HCV NS5A was immunostained with primary anti-NS5A antibody 9E10 at 1:1,000 dilution in PBS/BSK at 4°C. The next day, cells were washed twice with PBS + 0.1% Tween-20 and stained using secondary antibody ECL anti-mouse IgG horseradish peroxidase (HRP)-linked whole antibody (GE Healthcare Amersham) at 1:300 in PBS + 0.1% Tween-20 for 30 minutes at room temperature before being visualized by 30 minutes incubation at room temperature with a DAB substrate kit (Dako). FFU were counted automatically using an ImmunoSpot series 5 UV analyzer (CTL Europe GmbH) with customized software as described previously [44]. Lower limit of detection was calculated for each 96-well plate as the mean of at least 6 negative wells plus 3 standard deviations plus 3. Upper limit of detection was set to 200 FFU/well as this was within the linear range of test dilution series and comparable with manual determinations [45].
For determination of HCV RNA titers in culture supernatant, RNA was extracted from 200 µL supernatant using the Total Nucleic Acid Isolation Kit (Roche Applied Science); titers were determined by TaqMan real-time PCR as previously described [34]. HCV Core titers in culture supernatant were determined using the ARCHITECT HCV Ag assay (Abbott).
Direct sequencing of cell culture-derived HCV
HCV RNA was purified from 200 µL cell culture supernatant using the High Pure Viral Nucleic Acid Kit (Roche Applied Science). Overall, reverse transcription, 1st round PCR and 2nd round nested PCR were carried out as previously described [34]. Primers used to generate cDNA and PCR amplicons spanning the Core-E2 region have been previously reported for H77(1a) and ED43(4a) [35]; J4(1b) and HK6a(6a) [11], J6(2a) and S52(3a) [34]; as well as SA13(5a) [12]. Direct sequencing of amplicons was carried out by Macrogen Europe.
Single-cycle virus production assay in S29 cells
Overall, S29 cell experiments were carried out as previously described [46]. Briefly, 400,000 CD81-deficient S29 cells [47] were plated in 6-well plates 24 hours before transfection. In vitro HCV RNA transcripts of SA13(5a) [12] as well as of positive control (J6(2a)) and of negative control (J6(2a)-GND) [32] were generated using T7 RNA polymerase (Promega) for 2 hours at 37°C, DNAse treated using DNA RQ1 DNAse (Promega) and purified using RNeasy kit (Qiagen). HCV RNA transcripts (2.5 µg) were mixed with 5µL Lipofectamine 2000 (Invitrogen) in 500µL serum-free Opti-MEM (Gibco/Invitrogen). S29 cells were incubated with transfection complexes for 4 hours in Opti-MEM. Following transfection, Opti-MEM was replaced by either DMEM + 10% FBS or AEM. S29 cells were collected at 4, 24, 48 and 72 hours post transfection and prepared for determination of intracellular HCV Core and infectivity titers as previously described [46]. Culture supernatants were collected at 24, 48 and 72 hours post transfection for determination of extracellular HCV Core and infectivity titers. Infectivity titers were determined as described above, while Core titers were determined using the ARCHITECT HCV Ag assay (Abbott).
Equilibrium density gradient ultracentrifugation
Semi-continuous 10–40% iodixanol gradients were prepared by layering 2.5 mL of 40%, 30%, 20% and 10% OptiPrep (iodixanol; Sigma-Aldrich) on top of each other as described previously [6]. HCVcc containing supernatants were either loaded directly on top of the gradient, or concentrated using Amicon 100 kDa centrifugation filters before loading. A final volume of ~250 µL was loaded for all samples. The samples were ultracentrifugated at 151,000 × relative centrifugal force (RCF) for 18 hours at 4°C using a Beckman SW-41 rotor mounted in a Beckman XL-70 ultracentrifuge. After centrifugation, fractions of ~550 µL were collected from the bottom of the tube and 400 µL portions were weighed (model SI-114; Denver Instruments) to determine fraction densities. Fraction infectivity titers were determined as described above. Iodixanol containing fractions were diluted to contain ≤ 10% iodixanol before titration.
Receptor-, endocytosis- and neutralization assays
For receptor blocking assays we used Purified Mouse Anti-Human CD81 primary antibody (JS-81) and Purified Mouse IgG1K isotype control (MOPC-21) (both BD Biosciences); Purified Goat Anti-human LDLr polyclonal antibody (AF2148) and Normal Goat IgG control (AB108C) (both R&D Systems); Anti-SR-BI primary antibody (C16–71) and control antibody (D) were previously described [48]. For HCVcc neutralization, we used chronic-phase serum from patient H taken 29 years after acute infection (H06 [35]) and chronic-phase serum from a genotype 5a infected patient (SA3 [12]) as well as a panel of monoclonal antibodies AR1B and AR2A-5A, which were previously described [13,49]. For ApoE neutralization we used a mouse monoclonal primary antibody (1D7) blocking the ApoE receptor binding site, and mouse IgG1κ (1D1) control antibody previously described [50]. For inhibition of clathrin-mediated endocytosis, we used chlorpromazine hydrochloride (Calbiochem).
Huh7.5 cells were plated at 7,000 cells/well onto poly-D-lysine coated 96-well plates. On the following day, for receptor-blocking assays, antibodies were diluted in DMEM +10 % FBS as specified and added to cells for 1 hour. For chlorpromazine assays, chlorpromazine was diluted in DMEM + 10% FBS as specified and added to cells for 30 minutes. HCVcc was diluted in DMEM + 10% FBS, whereas sf-HCVcc was diluted in AEM with FBS concentration adjusted to 10%. Virus dilutions were added to the cells incubated with blocking antibodies or chlorpromazine. Cell cultures were incubated for an additional 6 hours.
For HCV neutralization assays, chronic-phase HCV sera, or AR1B and AR2A-5A monoclonal antibodies were diluted in DMEM + 10% FBS as specified and mixed with either HCVcc or sf-HCVcc diluted as for receptor blocking assays. Patient serum-virus or antibody-virus mixes were incubated for 1 hour, before being added to cells. Cell cultures were incubated for 6 hours. For ApoE neutralization, 1D7 and 1D1 monoclonal antibodies were diluted in DMEM + 10% FBS as specified and mixed with either HCVcc or sf-HCVcc, diluted in DMEM + 10% FBS. Antibody-virus mixes were incubated for 30 minutes, before being added onto cells. Cell cultures were incubated for 3 hours.
For blocking and neutralization assays, after 3 or 6 hours incubation as indicated above, the cells were washed in PBS and DMEM + 10% FBS was added to all cultures. Cells were incubated and fixed 48 hours post infection in ice-cold methanol and HRP-stained for HCV NS5A as described above. Single HCV NS5A positive cells were counted automatically using an ImmunoSpot series 5 UV analyzer (CTL Europe GmbH) with customized software as described previously [45,51]. The % blocking and neutralization were calculated by relating counts of experimental wells to the mean count of six replicate wells with untreated control virus. For receptor blocking and neutralization assays, following logarithmic transformation of X-values, variable-slope sigmoidal dose-response curves [Y = Bottom+(Top-Bottom)/(1 + 10(Log10EC50−X)×HillSlope)] were fitted to the data using GraphPad Prism 6.0. For receptor blocking and neutralization assays “Bottom” was constrained to “0”. For neutralization assays, “Top” was constrained to “100”, when appropriate, as indicated in Figure legends, and median inhibitory concentrations (IC50) were calculated using GraphPad Prism 6.0. For receptor blocking assays, maximum blocking rates (Bmax), the Y values at the top plateaus of the fitted curves, were calculated using GraphPad Prism 6.0.
HCV immunoprecipitation using anti-ApoE antibody
Immunoprecipitation was done using the ApoE-specific antibody 1D7 and isotype-matched control antibody 1D1 as previously described [52]. Briefly, 50 µl of magnetic-bead slurry was washed in antibody binding buffer (immunoprecipitation kit; Dynabeads Protein G; 100.070D; Invitrogen) and incubated it on a shaker with 5 µg antibody in 50 µl antibody binding buffer for 20 minutes at room temperature. The beads were subsequently washed two times in washing buffer and incubated with 106 IU of the virus in 200 µl of complete medium on a shaker for 1 hour at room temperature. The beads were removed and washed three times in 200 µl of washing buffer prior to elution in 50 µl according to the manufacturer’s instructions. HCV RNA was extracted from the complete eluate and measured in duplicates as previously described [34].
Cell viability and proliferation assays
For determination of Huh7.5 cell viability in DMEM + 10% FBS versus AEM, we plated 6,000 cells per well of poly-D-lysine coated 96-well plates in DMEM + 10% FBS. The following day, medium was removed and cells were incubated in DMEM + 10% FBS or AEM for 48 hours. Then, cell viability was determined using the CellTiter 96® AQueous One Solution Cell Proliferation Assay kit (Promega) according to the manufacturer’s instructions. The % viability was calculated by relating absorbance at 490 nm determined for 10 AEM cultures to the mean absorbance of 10 replicate DMEM + 10% FBS cultures.
For determination of chlorpromazine cytotoxicity, chlorpromazine was diluted in DMEM + 10% FBS as specified and then added to 6,000 Huh 7.5 cells/well, plated the previous day in poly-D-lysine coated 96-well plates. Cells were incubated for 6 hours before chlorpromazine was removed and DMEM + 10% FBS was added. Cell viability was determined 6 and 48 hours post-treatment using the CellTiter 96® AQueous One Solution Cell Proliferation Assay kit. The % viability was calculated by relating absorbance at 490 nm determined for chlorpromazine treated cultures to the mean absorbance of at least three replicate untreated cultures.
For determination of Huh7.5 cell proliferation in DMEM + 10% FBS versus AEM, we used the BrdU cell proliferation kit (Millipore). Cells were plated in poly-d-lysine coated 96-well plates at 2,000 cells/well in DMEM + 10% FBS according to the manufacturer’s instructions. The following day, medium was removed and cells were incubated in DMEM + 10% FBS or AEM for 48 hours. Then, cell proliferation was determined using the BrdU cell proliferation kit according to the manufacturer’s instructions. The % proliferation was calculated by relating absorbance at 450 nm determined for 10 replicate AEM cultures to the mean absorbance of 10 replicate DMEM + 10% FBS cultures.
Flow cytometry
For surface staining of HCV co-receptors we used Phycoerythrin (PE) Mouse Anti-Human CD81 primary antibody (BD Biosciences, JS-81), Anti-mouse LDL R-Phycoerythrin primary antibody (R&D systems, 263123), Purified Mouse Anti-Human CLA-1 (SR-BI) primary antibody (BD Transduction Laboratories, 25/CLA-1) with PE Goat Anti-Mouse Ig secondary antibody (BD Biosciences, polyclonal 550589) and Anti-human Claudin-1 primary antibody (R&D systems, 421203) with PE Goat Anti-Rat Ig secondary antibody (BD Biosciences, polyclonal 550767). Cells were detached by treatment with a 10 mM solution of EDTA in PBS for 10 minutes at 37°C. The cells were washed in PBS and resuspended in FACS buffer (PBS + 1% FBS) and 2.5 × 105 cells/well were plated in a V-bottom 96-well plate. Cells were stained protected from light at 4°C for 1 hour with either α-CD81 (25 µL/well according to the manufacturer’s instructions), α-LDLr (10µg/mL in FACS buffer), α-SR-BI (5µg/mL in FACS buffer) or α-Claudin-1 (5µg/mL in FACS buffer). Total volume in all wells was adjusted to 50 µL using FACS buffer. After incubation, cells were washed in FACS buffer. SR-BI-and claudin-1-stained cells were stained protected from light at 4°C for 20 minutes with secondary antibodies α-mouse Ig (4µg/mL in FACS buffer) or α-rat Ig (2µg/mL in FACS buffer). Cells were washed in FACS buffer and fixed protected from light at room temperature for 15 minutes using CellFix (BD Biosciences). Cells were washed and resuspended in PBS before they were analyzed on a BD FACSCalibur flow cytometer using CellQuest Pro. Data analysis was done using FlowJo flow cytometry analysis software.
Results
Huh7.5 cells cultured in serum-free medium yield higher HCV infectivity titers than conventional cultures
To produce sf-HCVcc, HCV recombinants were cultured in Huh7.5 cells maintained in AEM, a commercially available cell culture medium without animal- or human serum as described in Materials and Methods. Because AEM cultured Huh7.5 cells did not tolerate detachment, AEM was replaced every 2–3 days without splitting the cells. Thus, animal-derived trypsin was not used during the virus production phase. Cultures handled in this manner became over-confluent but could be maintained for at least 29 days (data not shown).
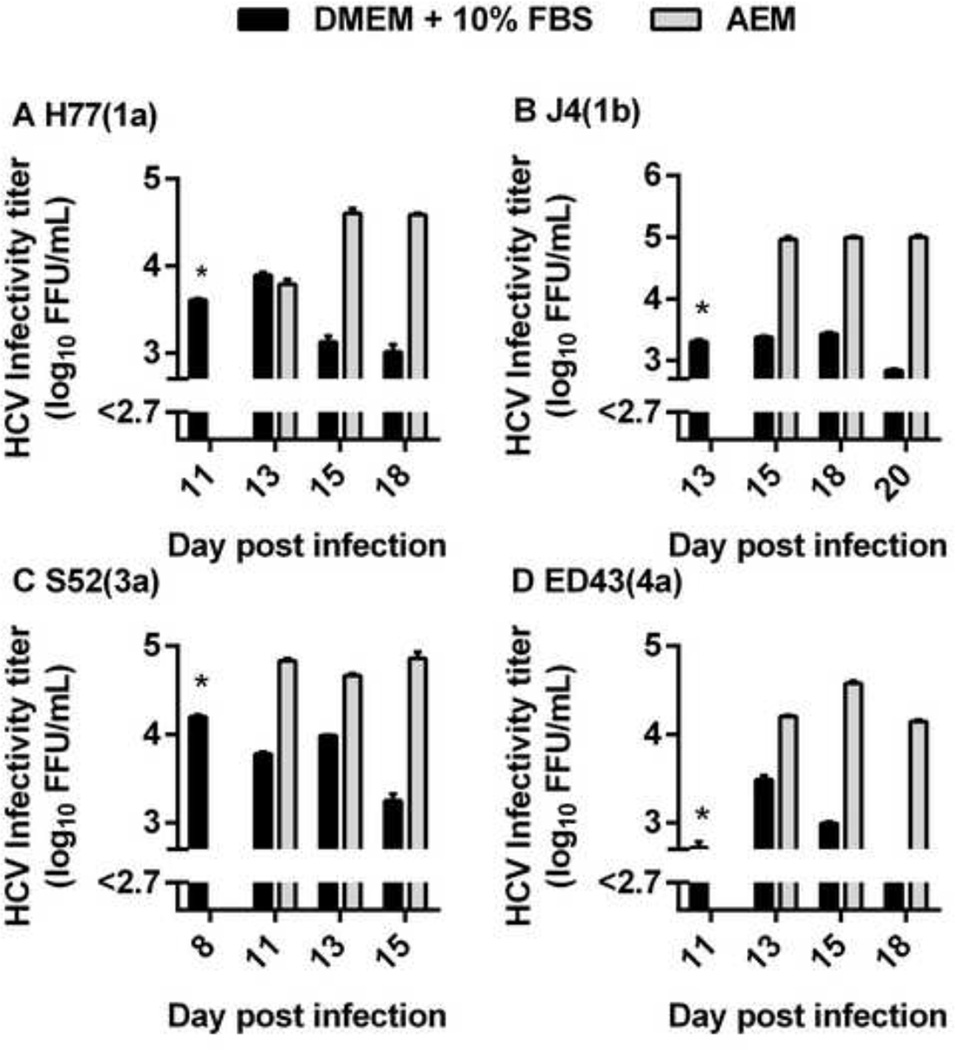
We tested if AEM cultured cells supported production of sf-HCVcc. Huh7.5 cells cultured in DMEM + 10% FBS were initially infected with JFH1-based Core-NS2 recombinants H77(1a), J4(1b), S52(3a) and ED43(4a) [11] (Figure 1). When viral infection had spread to ~80% of culture cells, as determined by immunostaining of HCV NS5A antigen, one replicate culture was maintained in AEM and another in DMEM + 10% FBS. Similar to previous observations [11], HCVcc peak supernatant infectivity titers were 3.4 to 4.2 log10 FFU/ml, followed by a drop in infectivity titers, when virus induced cell death was observed (Figure 1) [34]. For sf-HCVcc, peak infectivity titers were higher than for HCVcc, reaching 4.6 to 5.0 log10 FFU/ml (Figure 1). Also, relatively high infectivity titers were maintained for a longer period for sf-HCVcc than for HCVcc.
Figure 1. Serum-free Huh7.5 cell cultures produced high-titer sf-HCVcc.
Huh7.5 cells were infected with (A) H77(1a), (B) J4(1b), (C) S52(3a) and (D) ED43(4a) JFH1-based Core-NS2 recombinants in DMEM + 10% FBS for 18 hours. Cells were split into two replicate DMEM + 10% FBS cultures. When ~80% of culture cells were infected, as determined by HCV NS5A immunostaining, one replicate culture was maintained in DMEM + 10% FBS (black bars), while the other replicate culture was maintained in AEM (grey bars). At the indicated day post infection, supernatants were collected and DMEM + 10% FBS cultures were split, while fresh medium was added to AEM cultures as described in Materials and Methods. Supernatant HCVcc infectivity titers are shown as means of 3 replicates with standard error of the mean (SEM). The lower limit of detection in the experiments shown was up to 2.7 log10 FFU/ml, indicated by y-axis break. *, at this time point, the experiment consisted only of replicate cultures maintained in DMEM + 10% FBS; only one culture was titrated.
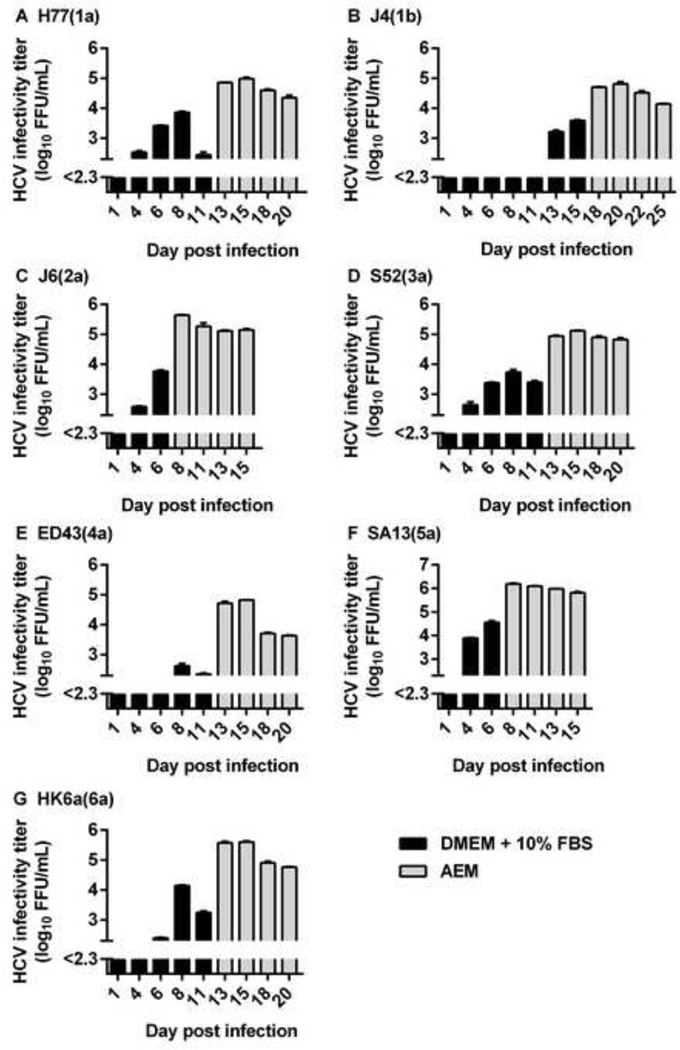
We next aimed at producing sf-HCVcc virus stocks of prototype strains of the six major HCV genotypes and epidemiologically important subtype 1b for further characterization. We infected Huh7.5 cells cultured in DMEM + 10% FBS with Core-NS2 recombinants indicated in Figure 2. DMEM + 10% FBS was replaced by AEM, when 40–80% of culture cells were infected (Figure 2). From these cultures, high-titer sf-HCVcc virus stocks were harvested at four consecutive time points, before cultures were closed. Peak infectivity titers between 4.7 and 6.2 log10 FFU/ml were observed for sf-HCVcc, with sf-J4(1b) and sf-ED43(4a) showing the lowest and sf-SA13(5a) showing the highest titers (Figure 2 and Table 1). Thus, sf-HCVcc stocks showed 0.6 to 2.1 log10 FFU/mL increased infectivity titers compared to previously described HCVcc reference stocks (Table 1) [11]. HCV RNA and Core titers for sf-HCVcc and HCVcc reference stocks were similar (Table 1). Thus, genotype 1–6 sf-HCVcc showed increased specific infectivities compared to HCVcc reference stocks (Table 1). This increase in specific infectivity was most pronounced for recombinants with comparatively low infectivity titers. Thus, based on RNA titers, specific infectivity was 20-fold increased for sf-J4(1b) and 40-fold increased for sf-ED43(4a) compared to their HCVcc counterparts (Table 1). This resulted in differences in specific infectivity between sf-HCVcc of different genotypes being smaller than those between HCVcc of different genotypes. For sf-HCVcc, based on RNA titers, specific infectivities were between 1/40 FFU/IU and 1/631 FFU/IU, while HCVcc showed specific infectivities between 1/398 FFU/IU and 1/12,589 FFU/IU (Table 1).
Figure 2. Generation of genotype 1–6 sf-HCVcc virus stocks.
Huh7.5 cells were infected with the indicated viruses in DMEM + 10% FBS at an MOI of 0.003 for 6 hours. On day 6 to 15 post infection, depending on the growth kinetics of the respective virus, when 40–80% of culture cells were infected, as determined by HCV NS5A immunostaining, DMEM + 10% FBS was replaced by AEM. At the indicated day post infection, supernatant was collected and fresh AEM was added to the cells. Supernatant HCVcc infectivity titers are shown as means of 3 replicates with standard error of the mean (SEM). Black bars represent DMEM + 10% FBS supernatant HCVcc infectivity titers; grey bars represent AEM supernatant sf-HCVcc infectivity titers. The lower limit of detection in the experiments shown was up to 2.3 log10 FFU/ml, indicated by y-axis break.
Table 1.
Characteristics of genotype 1–6 sf-HCVcc virus stocks compared to HCVcc reference stocks
| Peak HCV infectivity titerb log10FFU/mL |
Peak HCV RNA titerc log10IU/mL |
Peak HCV Core titerd log10 pmol Core/mL |
Specific infectivitye FFU/IU |
Specific infectivityf FFU/pmol Core |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Isolate (genotype)a | sf-HCVcc | HCVcc | sf-HCVcc | HCVcc | sf-HCVcc | HCVcc | sf-HCVcc | HCVcc | sf-HCVcc | HCVcc |
| H77(1a) | 5.0 | 4.3 | 7.6 | 7.5 | 5.7 | 5.4 | 1/316 | 1/1,585 | 1/4.8 | 1/13.5 |
| J4(1b) | 4.7 | 3.2 | 7.5 | 7.3 | 5.6 | 5.1 | 1/631 | 1/12,589 | 1/8.1 | 1/79.8 |
| J6(2a) | 5.6 | 5.0 | 7.6 | 7.6 | 5.5 | 5.5 | 1/100 | 1/398 | 1/0.8 | 1/3.4 |
| S52(3a) | 4.9 | 4.3 | 7.4 | 7.2 | 5.5 | 5.4 | 1/316 | 1/794 | 1/3.8 | 1/12.5 |
| ED43(4a) | 4.7 | 3.6 | 7.1 | 7.6 | 5.0 | 5.4 | 1/251 | 1/10,000 | 1/2.0 | 1/58.0 |
| SA13(5a) | 6.2 | 4.1g | 7.8 | 7.0 | 5.5 | 4.9 | 1/40 | 1/794 | 1/0.2 | 1/6.6 |
| HK6a(6a) | 5.6 | 4.0 | 7.7 | 7.7 | 5.6 | 4.8 | 1/126 | 1/1,000 | 1/0.9 | 1/5.9 |
Serum-free cultures were infected and maintained as described in Materials and Methods (Figure 2). For sf-HCVcc, supernatant HCV infectivity titers, Core antigen, and RNA titers were determined, and specific infectivities was calculated. Representative peak infectivity titers as well as Core and RNA titers from the same sample are shown. Core-E2 sequences were determined by direct sequence analysis as described in Materials and Methods. For sf-J6(2a), sf-S52(3a), sf-ED43(4a), sf-SA13(5a) and sf-HK6a(6a), Core-E2 sequences were identical to the plasmid sequence. The sf-H77(1a) had acquired the previously described amino acid change Y361H, estimated to be present in 50% of viral genomes; this change was also present in the H77(1a) HCVcc stock shown in this table [11]. The sf-J4(1b) had acquired amino acid changes T578A and D584G, estimated to be present in the majority of viral genomes. For HCVcc, characteristics of references stocks (HCVcc ref. stocks) are reproduced from Gottwein et al. 2009 [11].
Isolate and genotype of Core-NS2 of the used JFH1-based recombinants is indicated. Recombinants are further described in Materials and Methods.
For sf-HCVcc, supernatant infectivity titers were determined as FFU/mL by a cell culture-based titration assay as described in Materials and Methods. Values are means of three replicates. Iw HCVcc, values are reproduced from [11].
For sf-HCVcc, supernatant RNA titers were determined in the samples, for which infectivity titers are given, as IU/mL by Taq-Man PCR as described in Materials and Methods. Values are means of two replicates. For HCVcc, values are reproduced from [11].
Core titers were determined in the samples for which infectivity titers are given, as pmol/mL using the ARCHITECT HCV Ag assay (Abbott).
For sf-HCVcc, specific infectivity was calculated as FFU/IU by dividing supernatant infectivity titers with the corresponding RNA titers. For HCVcc, values were adapted from [11] to FFU/IU by dividing supernatant infectivity titers with the corresponding RNA titers.
Specific infectivity was calculated as FFU/pmol Core by dividing supernatant infectivity titers with the corresponding Core titers.
The peak infectivity titer of this SA13(5a) reference stock was lower than what we typically observe. Typically, peak titers for SA13(5a) are ~5 log10 FFU/mL (Figure 3A).
Higher infectivity titers of sf-HCVcc might be due to increased viral release and specific infectivity
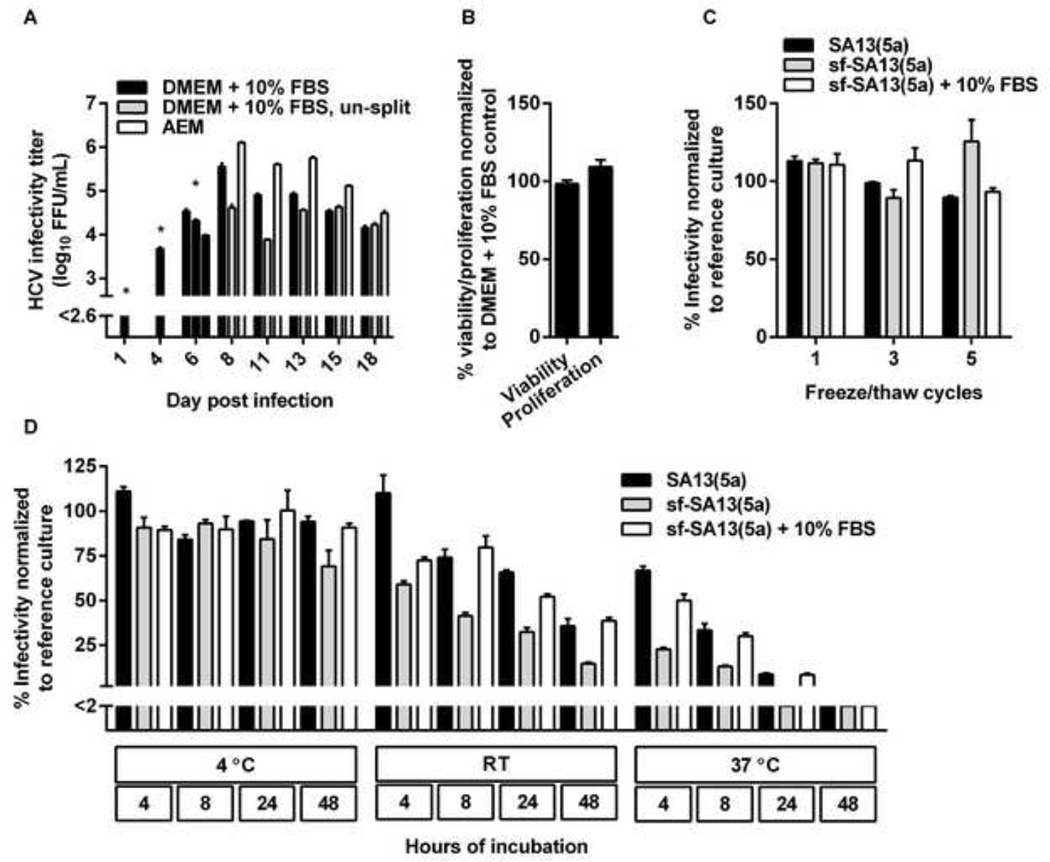
We next aimed at determining if the observed differences in infectivity titers were due to the fact that DMEM + 10% FBS cultures were split at regular intervals, while AEM cultures were kept over-confluent. Huh7.5 cell cultures were infected with SA13(5a) and (i) maintained in DMEM + 10% FBS and split every 2–3 days, (ii) maintained in DMEM + 10% FBS without splitting, or (iii) maintained in AEM without splitting (Figure 3A). The DMEM + 10% FBS culture yielded a peak infectivity titer of 5.6 log10 FFU/ml, while the AEM culture yielded a peak infectivity titer of 6.1 log10 FFU/ml (Figure 3A). However, the DMEM + 10% FBS culture maintained without splitting reached only 4.6 log10 FFU/mL (Figure 3A). This suggested that the high infectivity titers observed for the AEM cultures were not due to reduced stress related to avoiding cell culture splitting.
Figure 3. Increased infectivity titers of serum-free cell cultures were not due to reduced cell-splitting, changes in cell viability or proliferation, or increased viral stability.
(A) AEM cultures produced higher infectivity titers than DMEM + 10% FBS cultures handled similarly and DMEM + 10% FBS control cultures. Huh7.5 cells were infected with the SA13(5a) JFH1-based Core-NS2 recombinant in DMEM + 10% FBS for 3 hours at an MOI of 0.003. On day 4 post-infection, cells were split into three replicate cultures. Following day 6 post infection, when ~80% of culture cells were infected, as determined by HCV NS5A immunostaining, one replicate culture was maintained in DMEM + 10% FBS and split every 2–3 days, another replicate culture was maintained in DMEM + 10% FBS without being split, while the third replicate culture was maintained in AEM without being split. At the indicated days post infection, supernatants were collected. Supernatant HCVcc infectivity titers are shown as means of 3 replicates with standard error of the mean (SEM). The lower limit of detection in the experiment shown was up to 2.6 log10 FFU/ml, indicated by y-axis break. * The culture originally infected with SA13(5a) was first split into three replicates at day 4 post infection; thus, on day 1 and 4 post infection only one infectivity titer is shown. On day 6, DMEM + 10% FBS supernatants were harvested from the three replicate cultures; subsequently cultures were maintained in the different growth media indicated. (B) AEM cultures showed similar viability and proliferation as DMEM + 10% FBS cultures. Cell viability or proliferation of Huh7.5 cells cultured for 48 hours in AEM versus DMEM + 10% FBS was determined as described in Materials and Methods. The % viability/proliferation was calculated by relating absorbance at 490 nm (viability) or 450 nm (proliferation) determined for AEM cultures to the mean absorbance of 10 replicate DMEM + 10% FBS cultures. Bars represent the means of 10 replicates with SEM. (C) sf-HCVcc and HCVcc showed similar freeze-thaw stability. SA13(5a) diluted 1:100 in DMEM + 10% FBS or sf-SA13(5a) diluted 1:100 in either AEM or AEM + 10% FBS were exposed to up to 5 freeze/thaw cycles. Samples were thawn at room temperature and frozen at −80°C. After the indicated number of cycles, infectivity titers were determined as described in Materials and Methods. The % infectivity was calculated by relating the infectivity titer of each sample to the mean titer of a reference sample of the same stock, which had been stored at −80°C. Bars represent the means of three replicates with SEM. (D) sf-HCVcc showed decreased stability under temperature stress. SA13(5a) diluted 1:100 in DMEM + 10% FBS or sf-SA13(5a) diluted 1:100 in either AEM or AEM + 10% FBS were incubated at 4°C, room temperature (RT) or 37°C for 4 to 48 hours as indicated. Infectivity titers were determined as described in Materials and Methods. The % infectivity was calculated by relating the infectivity titer of each sample to the mean titer of a reference sample of the same stock, which had been stored at −80°C. Bars represent the means of three replicates with SEM. The lower limit of detection in the experiment shown was up to 2%, indicated by y-axis break.
We subsequently investigated cell viability and proliferation of cells cultured in AEM versus DMEM + 10% FBS. After 48 hours of culture in AEM, when increased infectivity titers were observed, cell viability and proliferation of AEM cultures was similar to that of DMEM + 10% FBS cultures (Figure 3B). Thus, changes in cell viability or proliferation did not explain the increased infectivity titers observed.
We further investigated, whether sf-HCVcc were more stable than HCVcc, which might contribute to the observed increase in infectivity titers. Up to 5 freeze/thaw cycles did not results in major decrease in infectivity or differences in infectivity for SA13(5a), sf-SA13(5a) or sf-SA13(5a) supplemented with 10% FBS (Figure 3C). Incubation for 48 hours at 4°C resulted in a minor decrease in infectivity of sf-SA13(5a) compared to SA13(5a) and sf-SA13(5a) supplemented with 10% FBS (Figure 3D). Incubation for 4 to 48 hours at room temperature and 37°C resulted in a gradual decrease in infectivity; this decrease was more pronounced for sf-SA13(5a) than for SA13(5a), and partially rescued by addition of 10% FBS to sf-SA13(5a) (Figure 3D). Thus, FBS might result in stabilization of HCVcc. However, apparently, increased infectivity titers were not caused by increased stability of sf-HCVcc.
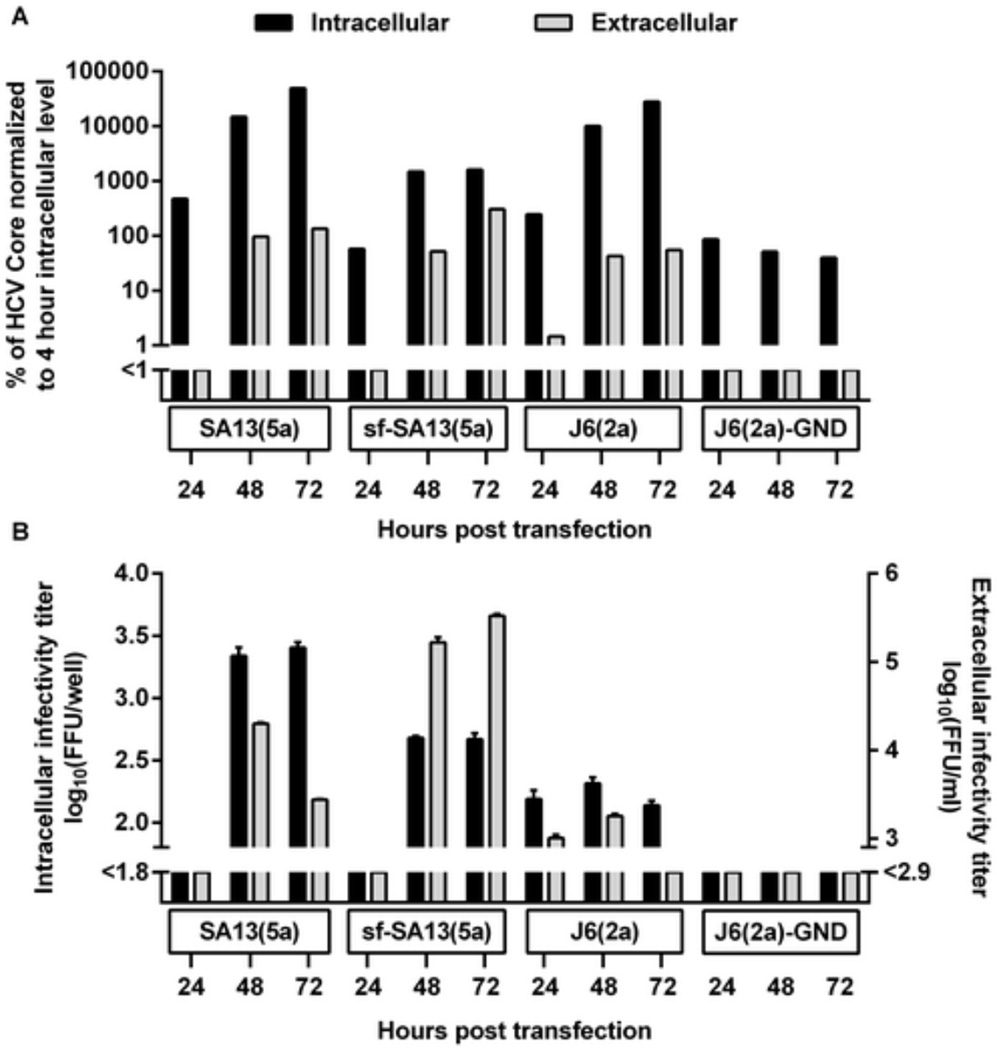
In order to investigate whether certain steps of the viral life cycle were affected by serum-free culture conditions, we carried out single-cycle virus production assays using CD81-deficient S29 cells, which are derived from Huh7.5 cells [47]. Following transfection with SA13(5a) HCV RNA, AEM cultures showed an ~1 log decrease in intracellular HCV Core and infectivity titers compared to DMEM + 10% FBS cultures, indicating a decrease in viral replication/translation (Figure 4A and B). In contrast, AEM and DMEM + 10% FBS cultures showed similar extracellular Core titers, but AEM cultures had a ~1 log increase in extracellular infectivity titers compared to DMEM + 10% FBS cultures. These findings suggest that AEM culture resulted in increase in viral release and that sf-HCVcc had increased specific infectivity compared to HCVcc. Furthermore, for HCVcc, the peak extracellular infectivity titer was observed 48 hours post transfection followed by a decrease at 72 hours post transfection, while for sf-HCVcc high titers were observed at both time points (Figure 4B). This is in agreement with the prolonged peak of infection observed in Huh7.5 cells (Figures 1–Figure 3).
Figure 4. Serum-free culture decreased viral replication/translation but enhanced viral release and specific infectivity.
S29 cells were transfected with SA13(5a) as well as positive control (J6(2a)) and negative control (J6(2a)-GND) HCV RNA transcripts as described in Materials and Methods. (A) Intracellular (black bars) and extracellular (grey bars) Core levels were determined 24, 48 and 72 hours post transfection. Core levels were normalized to intracellular Core levels measured 4 hours post transfection. (B) Intracellular (black bars) and extracellular (grey bars) infectivity titers were determined 24, 48 and 72 hours post transfection. Infectivity titers are shown as the means (FFU/well) of three replicates with SEM. The lower limits of detection are indicated by y-axis breaks.
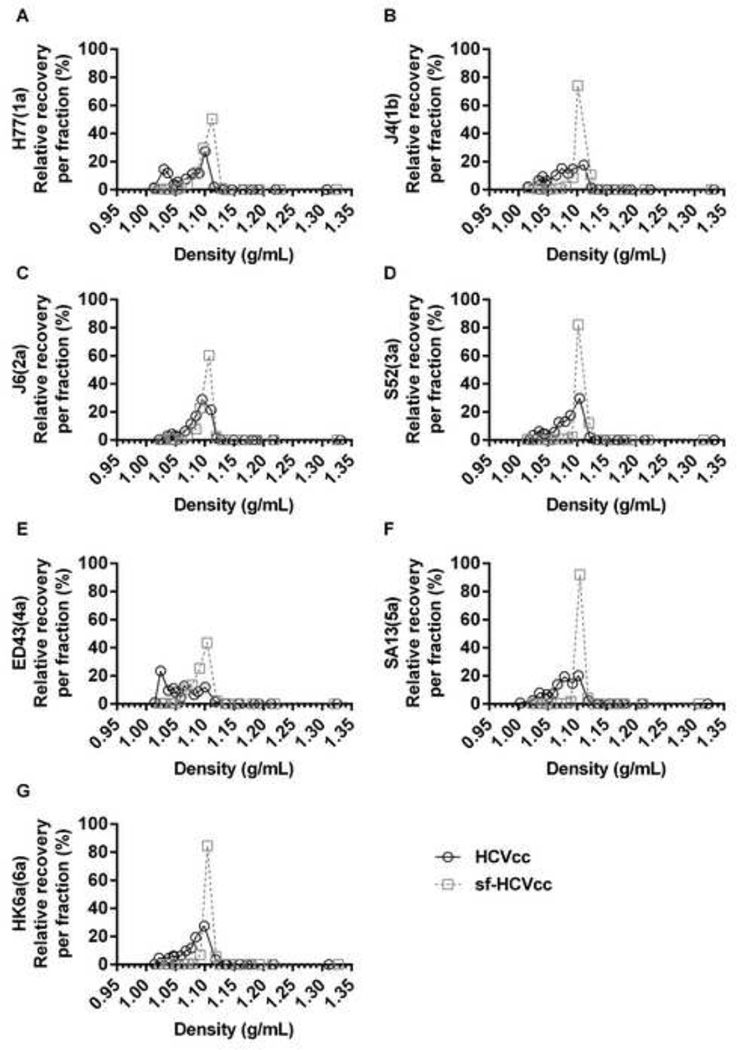
Infectious sf-HCVcc particles displayed a homogeneous density profile
To investigate their biophysical properties we subjected sf-HCVcc (Figure 2) to equilibrium buoyant density ultracentrifugation on iodixanol gradients. As described previously for genotype 2a, 3a, 5a and 6a HCVcc [6,32,38,39], we observed that infectious genotype 1–6 HCVcc particles constituted heterogeneous virus populations with buoyant densities between 1.01–1.10 g/mL (Figure 5). Interestingly, for genotype 1–6 sf-HCVcc harvested after 48 hours of cell culture with AEM, we detected up to 92% of recovered infectious sf-HCVcc at a single density of ~1.10 g/mL, following iodixanol gradient ultracentrifugation. Furthermore, between 71% and 97% of recovered infectious sf-HCVcc could be collected in three fractions with densities between 1.09 g/mL and 1.12 g/mL (Figure 5). Thus, infectious genotype 1–6 sf-HCVcc constituted a more homogeneous virus population compared to their HCVcc counterparts.
Figure 5. The sf-HCVcc particles of genotype 1–6 displayed an altered density profile with a single infectivity peak.
Of the virus stocks described in Figure 2, 10mL HCVcc supernatant taken from the last harvest of DMEM + 10% FBS culture supernatant (black line) or 10mL sf-HCVcc supernatant taken after 48 hours of AEM culture (grey, dotted line) was concentrated and layered on top of a pre-formed 10–40% iodixanol gradient and subjected to ultracentrifugation as described in Materials and Methods. Fractions were collected from the bottom of the gradients and analyzed by infectivity titration and by density determination as described in Materials and Methods. The HCV Core-E2 sequences of all virus stocks used were determined by direct sequencing. Compared to the plasmid sequence, sf-H77(1a) and H77(1a) had acquired the previously described amino acid change Y361H [11], estimated to be present in 50% of viral genomes. The sf-J4(1b) had acquired amino acid changes T578A and D584G, estimated to be present in the majority of viral genomes. Relative recovery per fraction (%) was calculated by relating the amount of infectious virus detected in each fraction to the total amount of infectious virus collected, and is plotted against the density determined for each fraction.
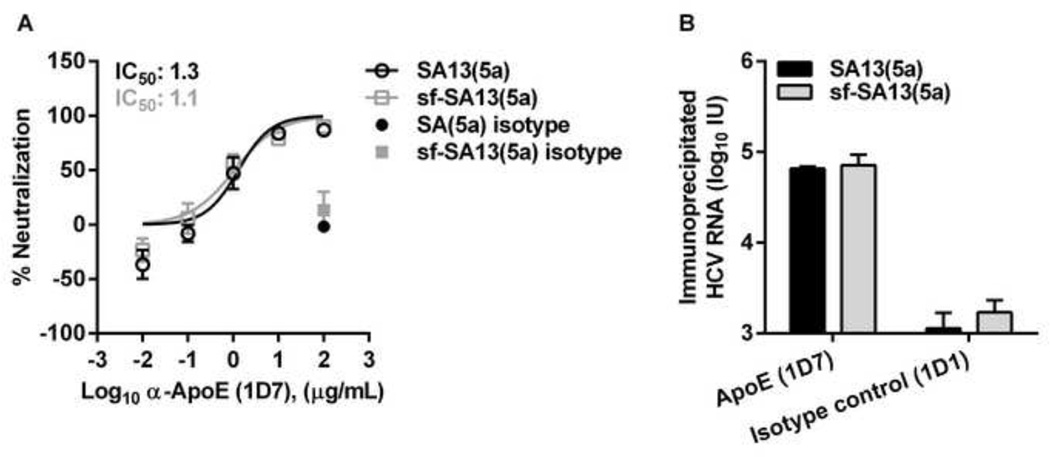
Infectious sf-HCVcc particles were apparently associated with ApoE
The altered density profile of sf-HCVcc might indicate an altered association with lipoproteins. To investigate if sf-HCVcc was associated with ApoE, a key component of HCV associated lipoproteins [15,16], we carried out neutralization assays using a monoclonal antibody directed against ApoE [18,50]. Due to limited availability of this antibody, we studied only the genotype 5a recombinant. SA13(5a) and sf-SA13(5a) showed a similar concentration-dependent response with median inhibitory concentrations (IC50) of 1.3 µg/mL for SA13(5a) and 1.1 µg/mL for sf-SA13(5a), and almost complete neutralization achieved at the highest α-ApoE concentrations (Figure 6A), suggesting that genotype 5a HCVcc and sf-HCVcc showed similar association with ApoE. These data were confirmed by neutralizing SA13(5a) and sf-SA13(5a) using Anti-Apolipoprotein E antibody (ab24139, abcam). Using this polyclonal rabbit IgG, complete neutralization, as well as 50% neutralization, was observed at similar dilutions for SA13(5a) and sf-SA13(5a) (data not shown).
Figure 6. HCVcc and sf-HCVcc showed similar association with ApoE.
(A) Monoclonal α-ApoE antibody (1D7) and control mouse IgG1κ (1D1) were diluted in DMEM + 10% FBS to the indicated concentrations. SA13(5a) (black bars) and sf-SA13(5a) (grey bars) were diluted in DMEM + 10% FBS and incubated with dilutions of α-ApoE or mouse IgG1κ for 30 minutes at 37°C. The virus-antibody mixes were added to Huh7.5 cells, plated the previous day in poly-D-lysine coated 96 well plates. After 3 hours of incubation, virus-antibody mixes were removed and DMEM + 10% FBS was added. Cells were fixed 48 hours post infection and stained, and the number of single HCV NS5A positive cells per well was determined by automated counting as described in Materials and Methods. The % neutralization was calculated by relating counts of experimental wells to the mean count of six replicate wells with untreated control virus. Data points are means of three replicates with SEM (error bars). Following logarithmic transformation of X-values, variable-slope sigmoidal dose-response curves were fitted [Y = Bottom+(Top-Bottom)/(1 + 10(Log10EC50-X)×HillSlope)]. “Bottom” was constrained to “0” for all curves. “Top” was constrained to “100”. (B) Immunoprecipitation was carried out on 106 IU HCV RNA of SA13(5a) and sf-SA13(5a), using monoclonal α-ApoE (1D7) and control mouse IgG1κ (1D1) as described in Materials and Methods. Amounts of HCV RNA (IU) were determined in the immunoprecipitated fractions using TaqMan PCR as described in Materials and Methods. RNA titers are shown as the mean of two replicates with SEM.
We further carried out immunoprecipitation of SA13(5a) and sf-SA13(5a) with the ApoE specific monoclonal antibody also used for neutralization experiments in Figure 6A [18,50]. We observed no major differences in the amount of viral RNA precipitated by this antibody (Figure 6B). Collectively, these data indicate that HCVcc and sf-HCVcc showed similar association with ApoE.
Establishment of the use of sf-HCVcc in biological assays
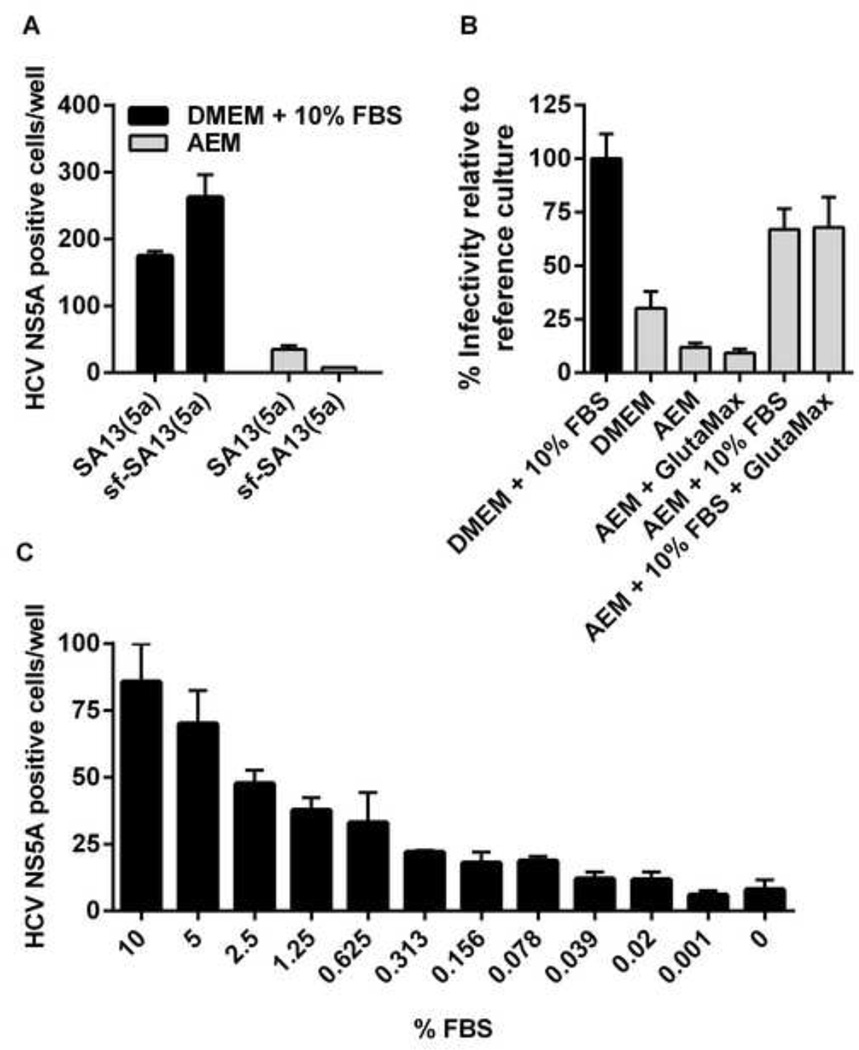
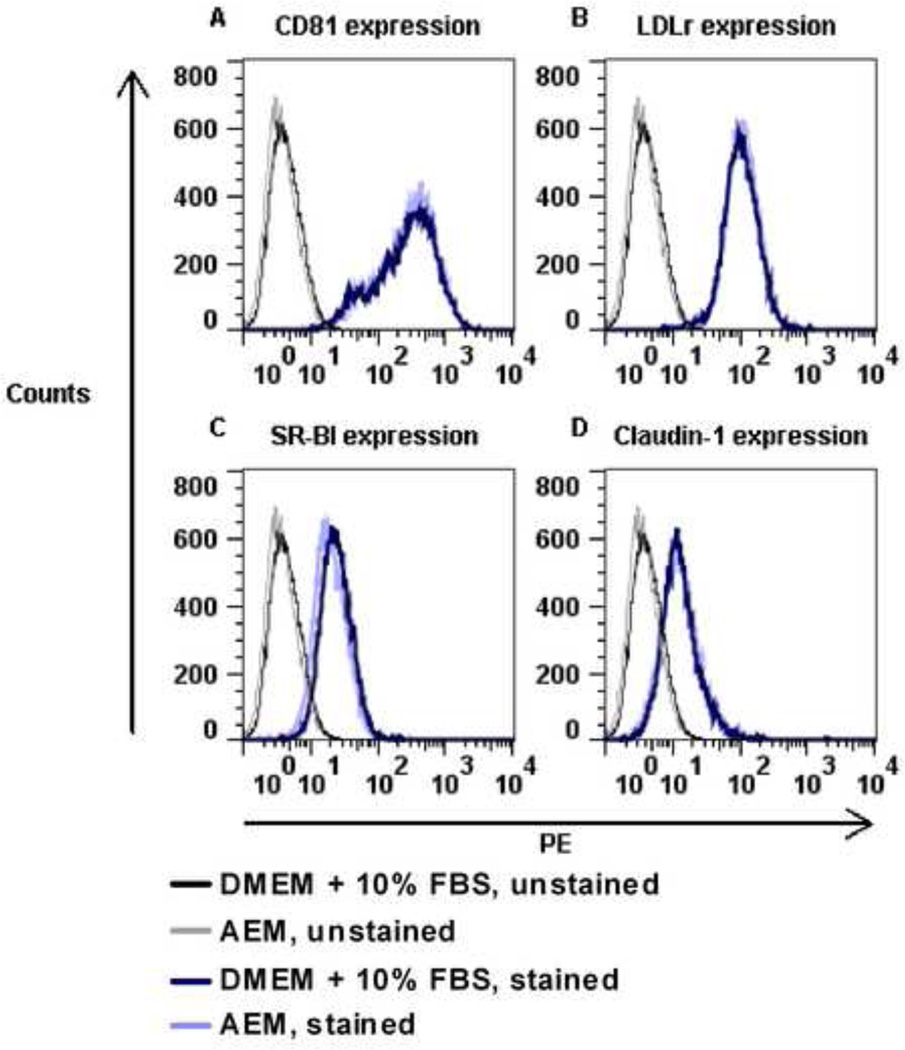
To further characterize sf-HCVcc, we aimed at studying routes of sf-HCVcc entry and sf-HCVcc sensitivity to neutralizing antibodies. To avoid potential in vitro association of sf-HCVcc with FBS components such as lipoproteins [53], we aimed at replacing DMEM + 10% FBS, typically used in such assays, by AEM during the viral infection step. However, using AEM, in initial experiments with genotype 5a and 2a viruses, we found greatly reduced infectivity for sf-SA13(5a) and sf-J6(2a). Interestingly, SA13(5a) and J6(2a) infectivity was also reduced when these viruses were diluted in AEM prior to infection (Figure 7A and data not shown). This loss of infectivity was not due to down-regulation of important HCV co-receptors on AEM cultured Huh7.5 cells, since expression of CD81, LDLr, SR-BI and claudin-1 was similar in Huh7.5 cells cultured for three hours in either DMEM + 10% FBS or AEM, as determined by flow cytometry (Figure 8).
Figure 7. FBS enhances infectivity of both HCVcc and sf-HCVcc.
(A-C) Huh7.5 cells seeded in poly-D-lysine coated 96-well plates the previous day, were incubated with (A) SA13(5a) and sf-SA13(5a) diluted in DMEM + 10% FBS (black bars) or AEM (grey bars), (B) sf-SA13(5a) diluted in different media with supplements as indicated or (C) sf-SA13(5a) diluted in AEM supplemented with different concentrations of FBS. % FBS in growth medium indicates the final FBS concentration. (A–C) Cells were incubated with virus mixes for 3 hours. After incubation, fresh DMEM + 10% FBS was added to all wells. Cells were incubated for 48 hours before they were fixed, stained and the number of single HCV NS5A positive cells per well was determined by automated counting as described in Materials and Methods. Error bars represent SEM of triplicates. For (B), the mean infectivity (HCV NS5A positive cells/well) of triplicate wells of the reference culture (DMEM + 10% FBS, black bar) was set to 100%. The number of HCV NS5A positive cells / experimental well was related to this mean to calculate % infectivity relative to the reference culture.
Figure 8. Huh7.5 cells cultured in DMEM + 10% FBS or AEM showed similar surface expression of HCV co-receptors.
Huh7.5 cells were incubated for 3 hours in DMEM + 10% FBS or AEM and subsequently prepared for flow cytometry analysis as described in Materials and Methods. Cell surface expression of HCV co-receptors was determined using antibodies against (A) CD81, (B) LDL-r, (C) SR-BI and (D) claudin-1 as described in Materials and Methods. PE signals were recorded on a BD FACSCalibur flow cytometer. Histograms show the co-receptor surface expression in cells cultured in DMEM + 10% FBS (dark blue) or AEM (light blue) compared to unstained cells (black and grey, respectively).
To determine how infectivity of sf-HCVcc in AEM could be rescued, we diluted sf-SA13(5a) in DMEM + 10% FBS (reference culture), DMEM, AEM, or AEM supplemented with 10% FBS and/or GlutaMax (a glutamine supplement present in DMEM + 10% FBS culture medium but not in AEM). For sf-SA13(5a) diluted in AEM, infectivity was only 12% of infectivity of the reference culture (Figure 7B). While supplementing AEM with GlutaMax did not influence infectivity, supplementing AEM with 10% FBS or 10% FBS and GlutaMax increased infectivity of sf-SA13(5a) to 67% and 68% respectively, compared to the reference culture (Figure 7B). When sf-SA13(5a) was diluted in DMEM without FBS (GlutaMax only), infectivity was only 30% compared to the reference culture (Figure 7B). Apparently other FBS components than lipoproteins and lipoprotein-associated factors mediated this enhancement of infectivity, because supplementing AEM with either VLDL, low density lipoprotein, high density lipoprotein, ApoB, ApoCI, ApoE or water-soluble cholesterol did not rescue infectivity, while lipoprotein deficient FBS partly restored infectivity (data not shown). Furthermore, we observed that sf-SA13(5a) infectivity correlated with the % of FBS in AEM (Figure 7C). These findings suggested that yet undefined factors in FBS culture medium supplement were crucial for infectivity of HCVcc and sf-HCVcc. Therefore, we investigated if sf-HCVcc was able to associate in vitro with FBS components, leading to alteration of the observed sf-HCVcc density profile. We incubated sf-SA13(5a) with different media and serum concentrations in the absence of cells. Such incubations did not affect the density profile of sf-SA13(5a) (Figure 9), suggesting that association between sf-HCVcc and serum components did not occur to an extent that influenced the previously observed density profile (Figure 5). Thus, it was feasible to carry out further biological studies of sf-HCVcc in AEM supplemented with 10% FBS.
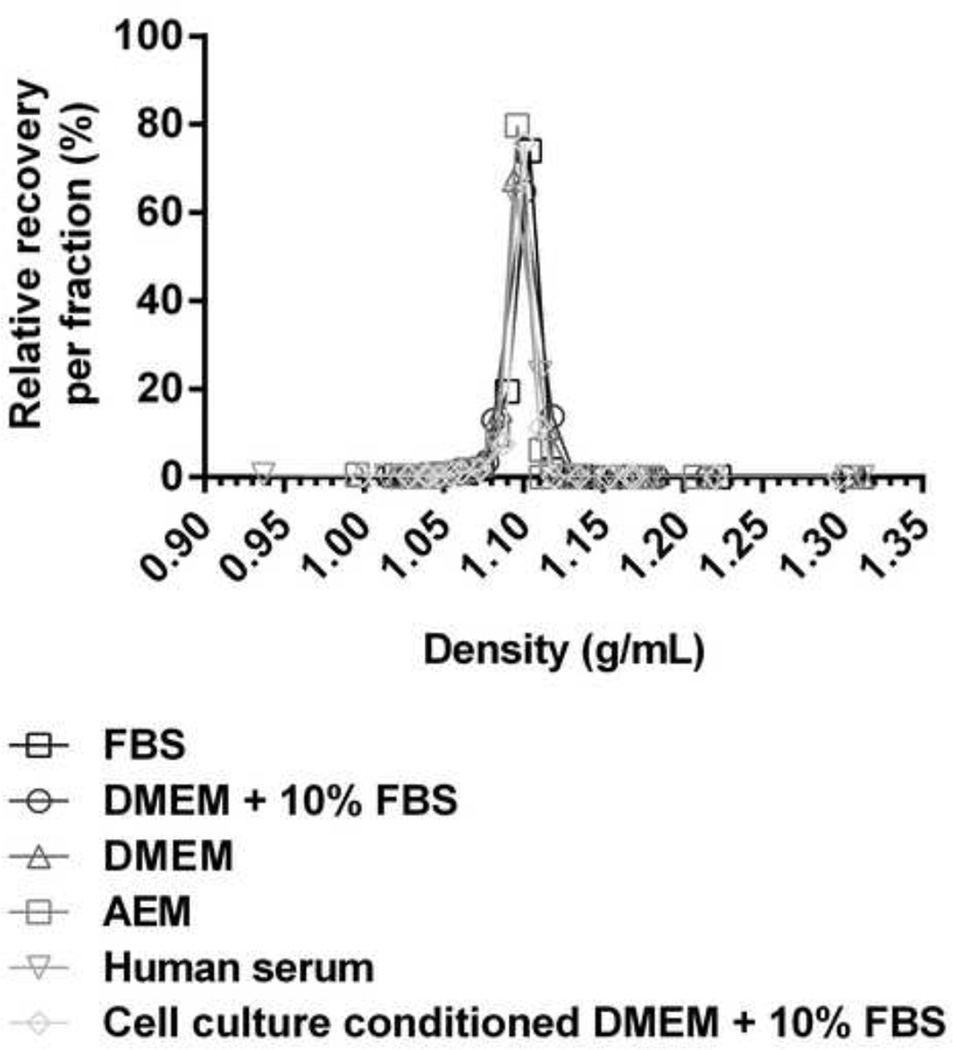
Figure 9. The sf-HCVcc density profile was maintained afterin vitro incubation with serum.
The sf-SA13(5a) recombinant was mixed 1:1 with either 100% FBS, DMEM + 10% FBS, DMEM, AEM, 100% human serum or sterile filtered cell culture conditioned medium (DMEM + 10% FBS harvested after 48 hours culture on naïve Huh7.5 cells) and incubated for 6 hours at 37°C. Mixes were layered on top of a pre-formed 10–40% iodixanol gradient, and subjected to ultracentrifugation as described in Materials and Methods. Fractions were collected from the bottom of the gradients and analyzed by infectivity titration and by density determination as described in Materials and Methods. Relative recovery per fraction (%) was calculated by relating the amount of infectious virus detected in each fraction to the total amount of infectious virus collected, and is plotted against the density determined for each fraction.
Entry of sf-HCVcc depended on HCV co-receptors CD81, LDLr and SR-BI as well as on clathrin-mediated endocytosis
To investigate if sf-HCVcc differed from HCVcc regarding entry into the host cell, we first studied HCV co-receptors LDLr and SR-BI, which might interact with lipoprotein components on the LVP [22,23], as well as CD81, supposed to directly interact with E2 [20,21].
When blocking CD81, for genotype 1–6 HCVcc, maximum blocking rates (Bmax) of ~100% were observed at the highest α-CD81 concentrations; for these viruses, similar blocking rates were previously observed [11]. Genotype 1–6 sf-HCVcc showed similar concentration-dependent sensitivity towards CD81 blocking as their HCVcc counterparts (Figure 10, left column).
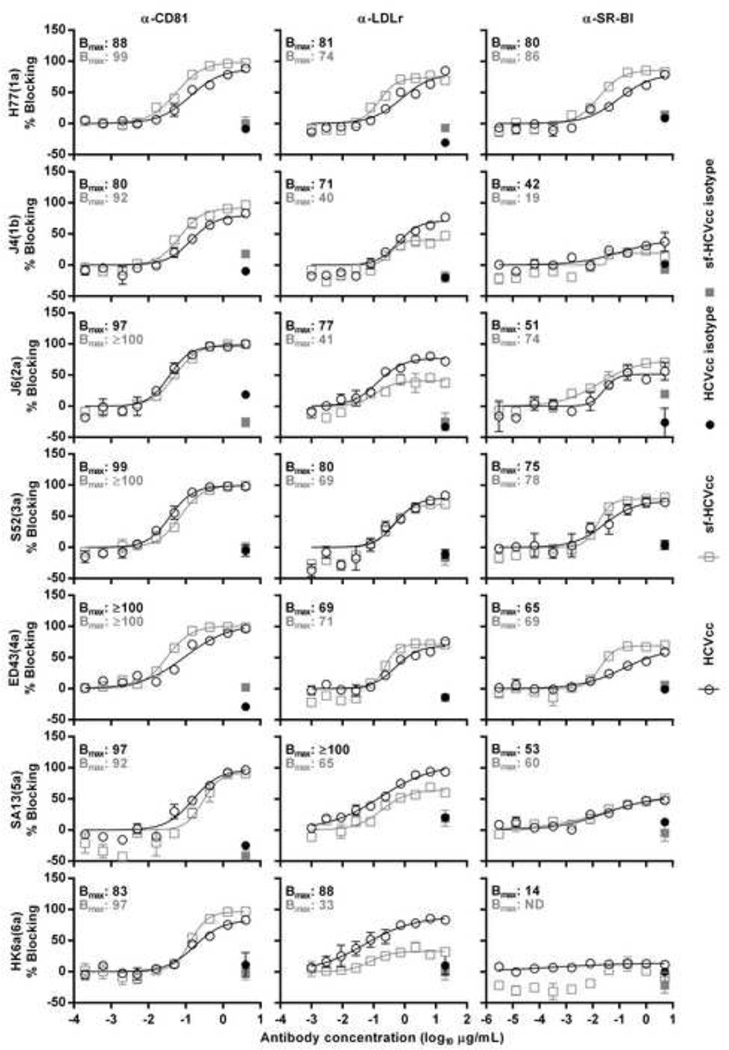
Figure 10. Effect of co-receptor blocking on HCVcc and sf-HCVcc entry.
α-CD81 (left column), α-LDLr (middle column) or α-SR-BI (right column) was diluted in DMEM + 10% FBS to the indicated concentrations. Specified antibody (open symbols) or control antibody (closed symbols) dilutions were added to Huh7.5 cells, plated the previous day onto poly-D-lysine coated 96-well plates and incubated for 1 hour. HCVcc (black circles) were diluted in DMEM + 10% FBS and sf-HCVcc (grey squares) were diluted in AEM + 10% FBS and added to cultures. After 6 hours incubation, antibody-virus mixes were removed and DMEM + 10% FBS was added. Cells were fixed 48 hours post infection and stained, and the number of single HCV NS5A positive cells per well was determined by automated counting as described in Materials and Methods. The HCV Core-E2 sequences of all virus stocks used were determined by direct sequencing. Sequences were identical for HCVcc and sf-HCVcc of the same recombinant. Compared to the plasmid sequence, H77(1a) viruses had acquired amino acid change I348S and J4(1b) had acquired amino acid change V710L, both estimated to be present in the majority of viral genomes. The % blocking was calculated by relating counts of experimental wells to the mean count of six replicate wells with untreated control virus. Data points are means of three replicates with SEM (error bars). Following logarithmic transformation of X-values, variable-slope sigmoidal dose-response curves were fitted [Y = Bottom+(Top-Bottom)/(1 + 10(Log10EC50-X)×HillSlope)]. “Bottom” was constrained to “0”. Bmax values, the Y values at the top plateaus of the fitted curves, are shown for HCVcc (black) and sf-HCVcc (grey). No curve could be fitted to data points obtained for sf-HK6a(6a) in SR-BI blocking experiments. ND, not determinable.
When blocking LDLr, we found concentration-dependent blocking for genotype 1–6 HCVcc. Bmax values were between 69% for ED43(4a) and 100% for SA13(5a) at the highest concentrations of α-LDLr, suggesting genotype/isolate-specific differences in dependency on LDLr (Figure 10, middle column). Genotype 1–6 sf-HCVcc could also be blocked in a concentration-dependent manner, with Bmax values between 33% for sf-HK6a(6a) and 74% for sf-H77(1a) at the highest concentrations of α-LDLr. While sf-H77(1a), sf-S52(3a) and sf-ED43(4a) showed similar Bmax values as their HCVcc counterparts, sf-J4(1b), sf-J6(2a), sf-SA13(5a) and sf-HK6a(6a) showed 31–55% lower Bmax values than their HCVcc counterparts, suggesting that sf-HCVcc of certain genotypes had lower dependency on LDLr than their HCVcc counterparts.
Blocking of SR-BI had only limited effect on entry of J4(1b) and HK6a(6a) HCVcc with Bmax values <50% (Figure 10, right column). For HCVcc of other genotypes we observed concentration-dependent blocking with Bmax values between 51% for J6(2a) and 80% for H77(1a) at the highest concentrations of α-SR-BI. Thus, sensitivity to SR-BI blocking apparently depended on the genotype/isolate. Blocking of SR-BI also had limited effect on entry of sf-J4(1b) and sf-HK6a(6a). Entry of sf-HCVcc of other genotypes was blocked in a concentration-dependent manner, with Bmax values between 60% for sf-SA13(5a) and 86% for sf-H77(1a). Bmax values were similar between HCVcc and sf-HCVcc of the same genotype. Thus, overall, HCVcc and sf-HCVcc of the same genotype showed similar sensitivity to SR-BI blocking.
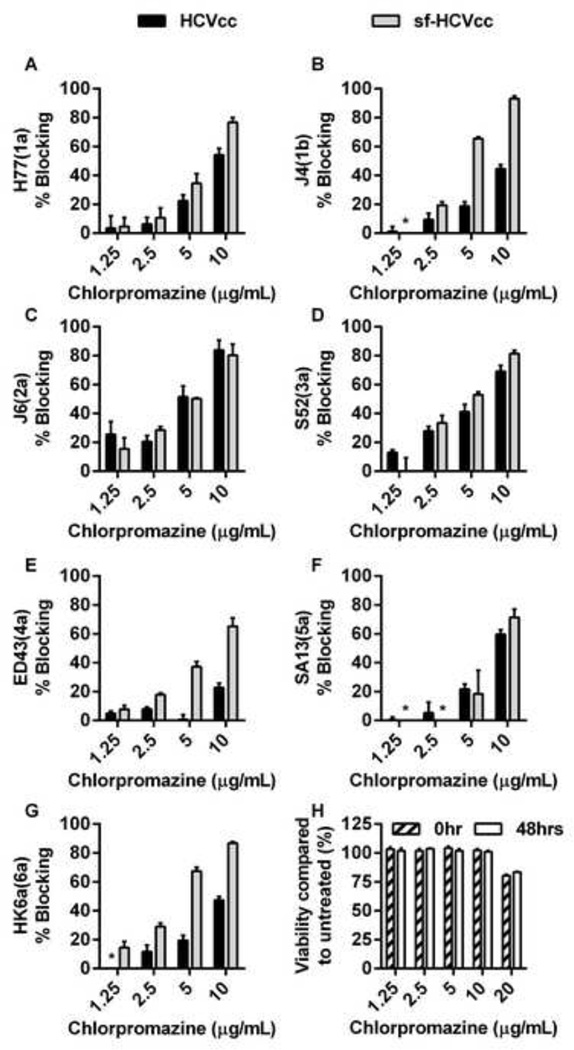
Finally, we studied dependency of genotype 1–6 sf-HCVcc on clathrin-mediated endocytosis. When pre-treating cells with chlorpromazine, we observed concentration-dependent blocking rates of up to 93%, suggesting that both sf-HCVcc and HCVcc depended on clathrin-mediated endocytosis (Figures 11 A–G). We were not able to achieve 100% blocking for any of the recombinants at 10 µg/mL chlorpromazine, the highest concentration not resulting in cytotoxic effects (Figure 11H). Interestingly, most sf-HCVcc were slightly more sensitive to chlorpromazine treatment than their HCVcc counterparts. This difference was greatest for J4(1b), ED43(4a) and HK6a(6a). For J6(2a) and SA13(5a) no obvious difference was observed, while H77(1a) and S52(3a) showed relatively small differences. This suggested that dependency on clathrin-mediated endocytosis might be slightly greater for sf-HCVcc of most genotypes/isolates than for HCVcc.
Figure 11. Effect of chlorpromazine treatment on HCVcc and sf-HCVcc entry.
(A-G) Chlorpromazine was diluted in DMEM + 10% FBS to the concentrations indicated and then added to Huh7.5 cells, plated the previous day onto poly-D-lysine coated 96-well plates, and incubated for 30 minutes. HCVcc (black bars) were diluted in DMEM + 10% FBS and sf-HCVcc (grey bars) were diluted in AEM + 10% FBS and added to cultures. After 6 hours incubation, chlorpromazine-virus mixes were removed and DMEM + 10% FBS was added. Cells were fixed 48 hours post infection and stained, and the number of single HCV NS5A positive cells per well was determined by automated counting as described in Materials and Methods. The HCV Core-E2 sequences of all virus stocks used were determined by direct sequencing. Sequences were identical for HCVcc and sf-HCVcc of the same recombinant. Compared to the plasmid sequence, H77(1a) viruses had acquired amino acid change I348S and J4(1b) had acquired amino acid change V710L, both estimated to be present in the majority of viral genomes. The % blocking was calculated by relating counts of experimental wells to the mean count of six replicate wells with untreated control virus. Data points are means of three replicates with SEM (error bars). (H) Chlorpromazine was diluted in DMEM + 10% FBS to the concentrations indicated and then added to Huh 7.5 cells, plated the previous day in poly-D-lysine coated 96-well plates. Cells were incubated for 6 hours before chlorpromazine was removed and DMEM + 10% FBS was added. A cell viability assay was carried out on cells incubated for 6 hours with chlorpromazine and on control cultures as described in Materials and Methods (0 hrs post treatment; dashed bars). An additional cell viability assay was carried out on chlorpromazine treated- and control cultures 48 hours post treatment (white bars). The % viability was calculated by relating absorbance at 490 nm determined for chlorpromazine treated cultures to the mean absorbance of three replicate untreated cultures. Bars represent the means of three replicates with SEM. *, values <0.
In conclusion, these data suggest that, overall, entry of HCVcc and sf-HCVcc relied on CD81, LDLr and SR-BI HCV co-receptors as well as on clathrin-mediated endocytosis, with exception of genotype 1b and 6a particles, which could not be blocked by α-SR-BI. However, we detected minor differences for recombinants of different HCV genotypes and for HCVcc versus sf-HCVcc regarding dependency on certain receptors and clathrin-mediated endocytosis.
Chronic-phase patient sera and monoclonal antibodies against conformational epitopes in E1E2 and E2 neutralize sf-HCVcc
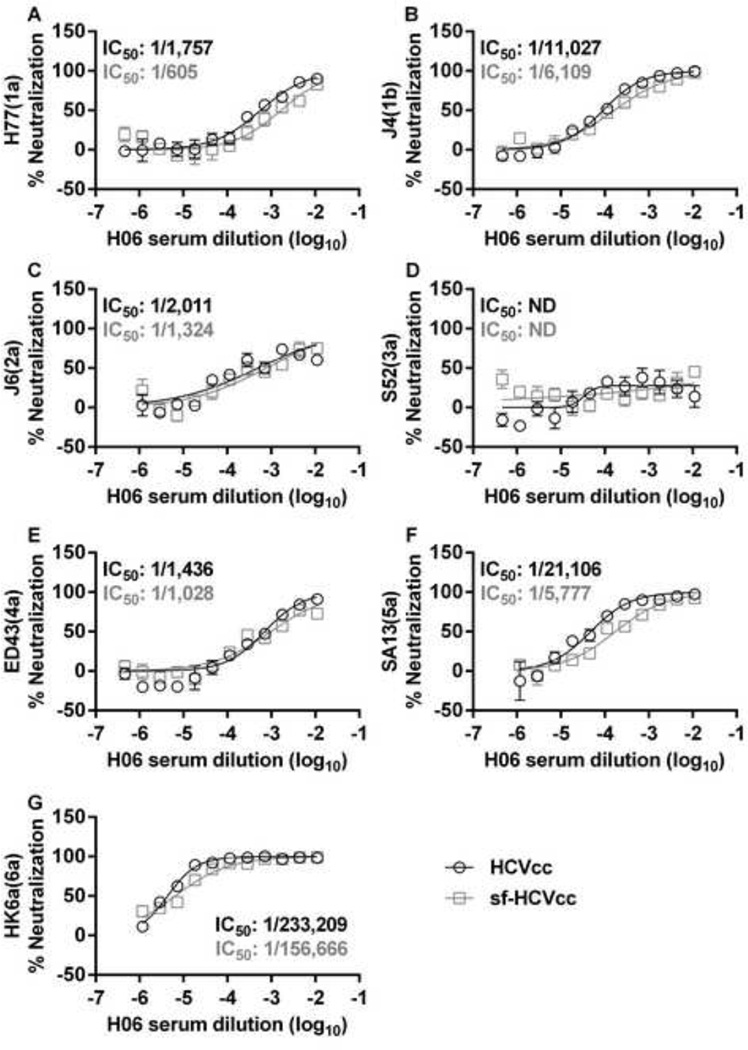
To investigate if there were differences between HCVcc and sf-HCVcc in sensitivity to neutralizing antibodies, we first did neutralization of genotype 1–6 viruses using serum from genotype 1a infected Patient H, taken 29 years after acute infection (H06 [35]). For HCVcc, as previously described, S52(3a) was the least sensitive to neutralization with H06 (Figure 12D) [6,11,35]. For HCVcc of other genotypes we observed dilution-dependent neutralization with IC50 values ranging from 1/1,436 to 1/233,209 and relatively high neutralization rates by high concentrations of H06 serum (Figure 12). For genotype 1–6 sf-HCVcc, we found similar neutralization patterns as for their HCVcc counterparts. For all sf-HCVcc except S52(3a) we observed dilution dependent neutralization with IC50 values ranging from 1/605 to 1/156,666 and relatively high neutralization rates by high concentrations of H06 serum (Figure 12). Thus, sf-HCVcc particles showed similar susceptibility to neutralizing antibodies in chronic phase patient serum as HCVcc.
Figure 12. HCVcc and sf-HCVcc show similar sensitivity to neutralization with genotype 1a chronic-phase patient serum.
Genotype 1a serum H06 was diluted in DMEM + 10% FBS as indicated. HCVcc (black circles) were diluted in DMEM + 10% FBS and sf-HCVcc (grey squares) were diluted in AEM + 10% FBS, mixed with H06 serum dilutions and incubated 1 hour at 37°C. Virus-serum mixes were added to Huh7.5 cells, plated the previous day onto poly-D-lysine coated 96 well plates. After 6 hours incubation, virus-serum mixes were removed and DMEM + 10% FBS was added. Cells were fixed 48 hours post infection and stained, and the number of single HCV NS5A positive cells per well was determined by automated counting as described in Materials and Methods. The HCV Core-E2 sequences of all virus stocks used were determined by direct sequencing. Sequences were identical for HCVcc and sf-HCVcc of the same recombinant. Compared to the plasmid sequence, H77(1a) viruses had acquired amino acid change I348S and J4(1b) had acquired amino acid change V710L, both estimated to be present in the majority of viral genomes. The % neutralization was calculated by relating counts of experimental wells to the mean count of six replicate wells with untreated control virus. Data points are means of three replicates with SEM (error bars). Following logarithmic transformation of X-values, variable-slope sigmoidal dose-response curves were fitted [Y = Bottom+(Top-Bottom)/(1 + 10(Log10EC50-X)×HillSlope)]. “Bottom” was constrained to “0” for all curves. “Top” was constrained to “100” for all curves in all panels except D; for these curves, median inhibitory concentrations (IC50) were calculated (black for HCVcc and grey for sf-HCVcc). ND, not determinable.
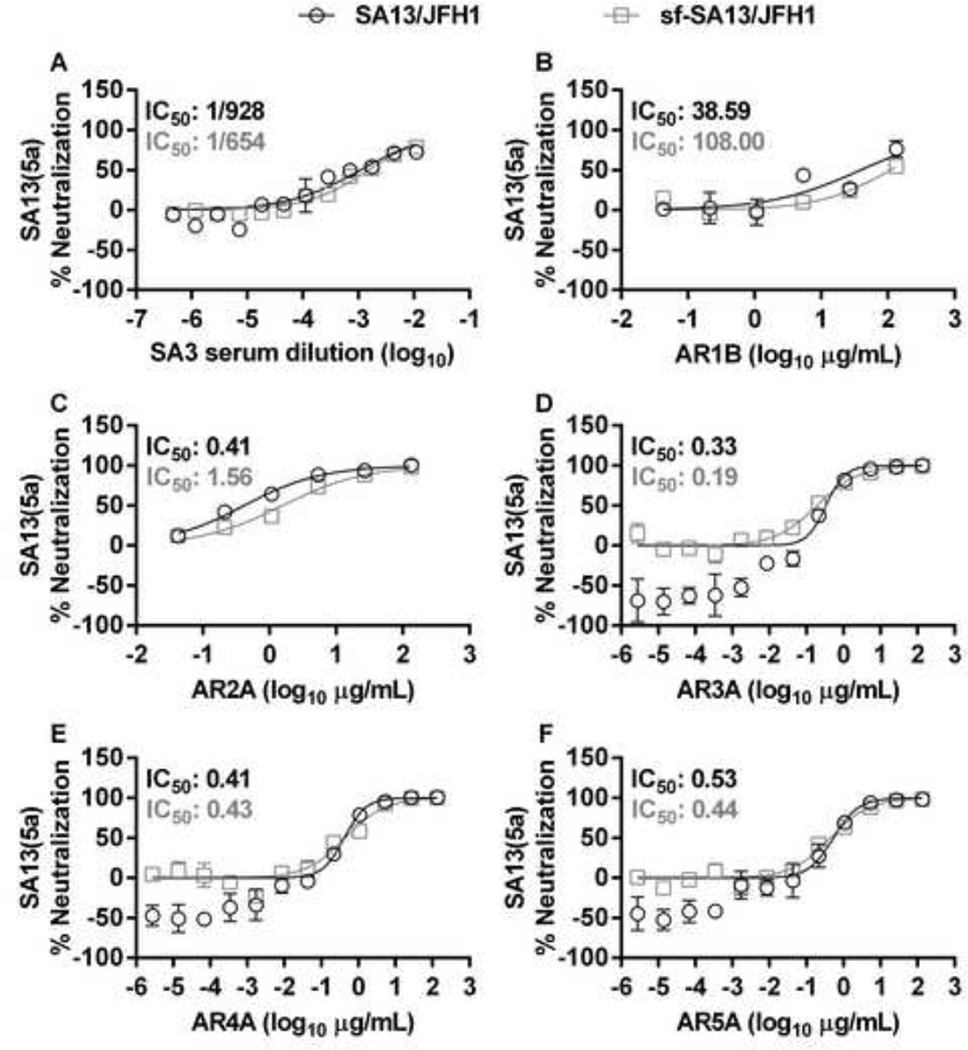
To confirm these observations, we next neutralized SA13(5a) and sf-SA13(5a) with a genotype 5a chronic-phase patient serum (SA3 [12]). These viruses showed similar neutralization profiles, with IC50 values of 1/928 for SA13(5a) and 1/654 for sf-SA13(5a) as well as high neutralization rates by high concentrations of SA3 serum (Figure 13A).
Figure 13. SA13(5a) and sf-SA13(5a) show similar susceptibility to genotype 5a patient serum and human monoclonal antibodies.
(A) Genotype 5a chronic phase serum SA3 or (B-F) monoclonal antibodies AR1B and AR2A-5A were diluted in DMEM + 10% FBS as indicated. HCVcc (black circles) were diluted in DMEM + 10% FBS and sf-HCVcc (grey squares) were diluted in AEM + 10% FBS, mixed with SA3 serum, AR1B or AR2A-5A antibody dilutions and incubated 1 hour at 37°C. Virus-serum or virus-antibody mixes were added to Huh7.5 cells, plated the previous day in poly-D-lysine coated 96 well plates. After 6 hours incubation, virus-serum or virus-antibody mixes were removed and DMEM + 10% FBS was added. Cells were fixed 48 hours post infection and stained, and the number of single HCV NS5A positive cells per well was determined by automated counting as described in Materials and Methods. The % neutralization was calculated by relating counts of experimental wells to the mean count of six replicate wells with untreated control virus. Data points are means of three replicates with SEM (error bars). Following logarithmic transformation of X-values, variable-slope sigmoidal dose-response curves were fitted [Y = Bottom+(Top-Bottom)/(1 + 10(Log10EC50-X)×HillSlope)]. “Bottom” was constrained to “0” for all curves. “Top” was constrained to “100” for all curves. Median inhibitory concentrations (IC50) were calculated (black for HCVcc and grey for sf-HCVcc).
Finally, we tested a panel of monoclonal antibodies (AR1B and AR2A-5A) targeting defined conformational epitopes in E1E2 and E2 [13,49], against SA13(5a) and sf-SA13(5a). AR1B was the least efficient for both viruses without reaching a top plateau (Figure 13B). AR2A-5A neutralized SA13(5a) and sf-SA13(5a) with similar concentration-response profiles, with IC50 values ranging from 0.33 to 0.53 µg/mL for SA13(5a) and 0.19 to 1.56 µg/mL for sf-SA13(5a); for both viruses complete neutralization was observed at high antibody concentrations (Figure 13C–F). This further supports that sf-HCVcc show similar sensitivity to neutralizing antibodies as HCVcc. In addition, these findings suggest that sf-HCVcc and HCVcc do not show major differences regarding conformation of E1 and E2.
Discussion
In this study, we describe the generation and characterization of genotype 1–6 serum-free HCVcc particles, using AEM to culture infected Huh7.5 hepatoma cells. Compared to HCVcc, sf-HCVcc showed similar biological properties but increased infectivity titers and a homogenous single-peak density profile. These unique characteristics, as well as the reduced concentration of non-HCV proteins in serum-free culture supernatants, are expected to facilitate generation of purified and concentrated virus stocks, required for vaccine development and biophysical studies of HCV particle composition [54–57]. Further, the developed serum-free culture conditions might reduce the risk of contamination with adventitious microbial agents in future vaccine antigen preparations [40,41].
Efficient production of HCVcc has primarily been achieved in the continuous hepatoma cell line Huh7 and derived cell lines, such as Huh7.5 cells [29,30]. Due to their increased permissiveness to infection with recombinant HCV, Huh7.5 cells were previously used for cell culture adaptation and growth of HCV genotype 1–6 recombinants used in this study [11,12,32,34,35]. According to WHO recommendations, a wide range of continuous cell lines are now considered as suitable substrates for production of various medicinal substances if certain requirements are met [40]. These requirements include use of well-characterized cell banks, use of suitable manufacturing procedures aiming at a high degree of purification of the end product, and thorough characterization of the end product [40]. Thus, Huh7.5 cells could potentially be characterized to comply with these recommendations, allowing their use for vaccine development. Huh7 derived cell lines have typically been subjected to long-term passage using serum-containing growth medium and animal-derived trypsin. According to WHO recommendations, animal derived products should be reduced or eliminated from cell cultures used for production of medicinal substances due to risk of contamination with adventitious microbial agents [40]. In this study, we describe a method for production of sf- HCVcc, thus avoiding the use of trypsin and bovine serum during the virus production phase. To further reduce presence of animal-derived components in sf-HCVcc producing cell culture, it might be possible to culture Huh7.5 cells in serum-free medium [42], prior to sf-HCVcc production. Alternatively, based on recently generated knowledge on host-factors required for HCV infection, it might be possible to engineer cell lines already approved for vaccine development with susceptibility to HCV infection [58–63]. However, this might be a cumbersome process and might require re-approval of the modified cell-line. Of note, most genotype 1–6 recombinants used in this study [11,12,32,34,35] contained adaptive mutations conferring efficient growth in Huh7.5 cells; for H77(1a), J4(1b) and HK6a(6a) cell culture adaptive mutations localized to the envelope proteins. In future studies, it will be of relevance to develop a panel of genotype 1–6 recombinants without envelope mutations, thus not differing from naturally occurring isolates.
Recently, proof-of-concept for immunogenicity of genotype 2a HCVcc was obtained, since immunization of mice resulted in induction of HCV neutralizing antibodies [41,57,64]. This underlines the potential of inactivated HCVcc particles as future vaccine antigens. However, HCVcc used for immunizations were grown in cell culture medium supplemented with 2% FBS [57], even though the authors had previously reported development of serum-free cultures for genotype 2a recombinants JFH1 and J6/JFH1, using growth medium DMEM/F-12 supplemented with Insulin-Transferrin-Selenium-X [65]. In contrast to our study, infectivity titers and specific infectivity of 2a virus from such serum-free cultures were apparently only equal to or lower than titers of viruses from serum-supplemented cultures [65]. In addition, serum-free 2a HCVcc showed a similar density profile as 2a HCVcc derived from serum-containing cell culture, following sucrose gradient ultracentrifugation [65]. These differences between previously produced serum-free 2a HCVcc [65] and sf-HCVcc described in this study are most likely due to the different culture media used and/or other differences in experimental conditions.
We describe establishment of serum-free cell cultures producing HCV particles of prototype strains of genotypes 1–6 with favourable biophysical- and biological characteristics (Figures 2 and 5 and Table 1) [11,66]. Supernatant infectivity titers of sf-HCVcc were 0.6 to 2.1 log10 FFU/ml higher than titers of HCVcc (Table 1 and Figure 2) [11]. Of the panel of previously developed HCVcc recombinants [11], SA13(5a) showed the highest infectivity titers (~5 log10 FFU/ml). Infectivity titers of >6 log10 FFU/ml, as observed for sf-SA13(5a) (Table 1 and Figures 2F and 3A), are among the highest infectivity titers reported to date for cell culture grown HCV [47,67–71]. Furthermore, recombinants with relatively low infectivity titers, such as J4(1b) and ED43(4a) [11,35], yielded significantly increased infectivity titers, when grown under serum-free conditions (Table 1 and Figures 2B and E). Genotype 1b is considered to be the most prevalent genotype worldwide and in certain countries, such as Egypt, genotype 4a has a prevalence of up to 15%; thus genotype 4a sf-HCVcc might prove an important antigen for vaccine trials [3,72]. The reason why serum-free culture conditions resulted in increased infectivity titers remains to be fully elucidated. Our studies indicated that increased infectivity in AEM cultures was not due to (i) avoiding stress related to cell splitting (Figure 3A), (ii) changes in Huh7.5 cell viability or proliferation (Figure 3B) or (iii) increased stability of sf-HCVcc (Figures 3C and D). While in S29 cells, serum-free culture conditions resulted in reduction of viral replication/translation, they increased viral release and specific infectivity, possibly contributing to the higher infectivity titers observed (Figure 4).
Specific infectivities were generally higher for sf-HCVcc than for HCVcc as observed in both Huh7.5 cell cultures (Table 1) and S29 cell cultures (Figure 4). This is in line with previous reports that HCVcc fractions with the highest specific infectivity had a buoyant density between 1.09–1.10 g/mL [32], similar to the density of the majority of infectious sf-HCVcc particles (Figure 5). Higher specific infectivity might be due to absence of serum, which might have non-specific neutralizing or inhibitory activity; alternatively, less immature viral particles might be produced using the developed culture conditions.
Furthermore, in our serum-free cultures, supernatants with high infectivity titers could typically be harvested for a prolonged period of time compared to DMEM + 10% FBS cultures (Figures 1–4). For example, for SA13(5a), high-titer supernatants could typically be harvested at 2–3 subsequent time points in DMEM + 10% FBS cultures, whereas in serum-free cultures, high-titer supernatants could be harvested at 4–6 subsequent time points (Figures 2F and 3A). This further increased the yield of infectious virus that could be harvested from serum-free cultures.
In contrast to HCVcc, sf-HCVcc displayed a homogeneous density distribution with a single peak of infectious virus at densities of ~1.10 g/mL, following iodixanol gradient ultracentrifugation (Figure 5). We believe that the density profile of sf-HCVcc might allow more effective density-based purification and concentration using ultracentrifugation and gel chromatography, since a single fraction, containing the majority of infectious virus, can be collected. Density changes were previously observed for HCVcc without hypervariable region 1 (HVR1) [6,73], HCVcc with a specific E2 mutation [54,74,75] and for HCV recovered from HCVcc-infected chimpanzees and uPA-SCID mice engrafted with human liver cells [38]. These density changes were suggested to be due to differences in lipoprotein association [6,38,73–75]. We showed that sf-SA13(5a) could be neutralized as efficiently as its HCVcc counterpart by a monoclonal α-ApoE antibody and polyclonal α-ApoE IgG (Figure 6A and data not shown). Further, immunoprecipitation of sf-SA13(5a) and SA13(5a) showed similar efficacy (Figure 6B). These data indicate that HCVcc and sf-HCVcc do not show major differences in association to ApoE, and thus possibly to lipoproteins [76]. Therefore, further studies will be required to elucidate the cause for the observed density shift (Figure 5). Preliminary studies indicated a decrease in intracellular lipid content in serum-free cultures (data not shown). However, determination of expression levels of genes involved in lipid production or of lipid/lipoprotein composition of HCVcc versus sf-HCVcc [55] was considered outside the scope of this study.
Compared to HCVcc, HCVcc without HVR1, displaying a similar density distribution as sf-HCVcc, were less susceptible to blocking of SR-BI and more susceptible to neutralizing antibodies [6,52,73]. Furthermore, previously described serum-free HCVcc were more susceptible to blocking of CD81 and SR-BI and to neutralization by a monoclonal antibody targeting E2 (AP33) [65]. Of note, in this study biological assays were carried out in AEM supplemented with FBS, which was required for viral infection, but did apparently not alter composition of sf-HCVcc LPV (Figures 7 and 9). When blocking CD81 and SR-BI, we did not observe major differences between sf-HCVcc and HCVcc (Figure 10). Even though sf-HCVcc of certain genotypes showed slightly lower dependency on LDLr and slightly higher dependency on clathrin mediated endocytosis than their HCVcc counterparts (Figures 10 and 11), overall our findings suggested that sf-HCVcc relied on similar routes of entry as HCVcc. In the future, it will be of interest to further investigate the small differences observed for LDLr usage and dependency on clathrin mediated endocytosis using different blocking antibodies and alternative methods of inhibition such as RNA interference.
We further confirmed previous results showing that HCVcc of genotype 1–6 showed similar dependency on CD81 [11]. Previously, we reported that blocking SR-BI had a similar effect on genotype 1–6 HCVcc entry [11]. However, in this study and another recent study by our group [52], using a different blocking antibody, we found differential sensitivity of genotype 1–6 HCVcc to SR-BI blocking. Single E2 mutations in culture adapted JFH1(2a) were reported to cause reduced dependency on SR-BI [71,75]. Further studies are required to elucidate if the E2 mutations present in HK6a(6a) and J4(1b) HCVcc virus stocks mediated reduced dependency on SR-BI. For genotype 1–6 HCVcc, we also observed small but consistent differences regarding dependency on LDLr (Figure 10). Whereas dependency on LDLr for entry has been shown for JFH1(2a) [22,77] and was recently shown for H77(1a), J6(2a) and S52(3a) HCVcc [52], this study is the first to show dependency on LDLr for J4(1b), ED43(4a), SA13(5a) and HK6a(6a) HCVcc. In future studies, also involving recombinants of additional isolates of each genotype, it will be of interest to investigate if different genotypes, subtypes or isolates differ regarding receptor usage.
Compared to HCVcc, sf-HCVcc showed similar sensitivity to neutralization by chronic phase patient sera and human monoclonal antibodies targeting conformational epitopes in E1E2 and E2 (Figures 12 and 13). These results suggest that sf-HCVcc resemble HCVcc regarding epitope exposure and conformation, of importance for vaccine development using sf-HCVcc as antigen.
In conclusion, we have established a method allowing for robust production of genotype 1–6 sf-HCVcc with favourable biological and biophysical characteristics. Serum-free culture apparently reduced viral replication/translation but enhanced viral release and specific infectivity. Sf-HCVcc had increased infectivity titers compared to HCVcc, and compared to serum-free HCVcc reported previously, thus contributing to an increased yield of infectious virus from infected cell cultures. Furthermore, sf-HCVcc displayed a homogeneous density distribution. Together with a reduced concentration of non-HCV proteins in supernatants from serum-free cultures, these features are expected to facilitate viral purification and concentration required for vaccine production and morphological analysis of HCV particles. Biologically, sf-HCVcc particles resembled their HCVcc counterparts regarding association to ApoE, routes of viral entry and sensitivity to neutralizing antibodies. Thus, sf-HCVcc particles could prove important as antigens in a prophylactic HCV vaccine against all six epidemiologically important HCV genotypes. To this aim future studies are required, focussing on establishment of large-scale sf-HCVcc production as well as efficient purification, concentration and inactivation. Finally, it will be of great interest to test immunogenicity of genotype 1–6 sf-HCVcc in small animal models.
Highlights.
We describe production of hepatitis C virus (HCV) from serum free cell culture (sf-HCVcc).
The sf-HCVcc has higher infectivity titers than HCV from conventional cultures (HCVcc).
Compared to HCVcc, sf-HCVcc displays an altered single-peak density profile.
The sf-HCVcc displays similar biological properties as HCVcc.
The sf-HCVcc might facilitate development of a whole virus inactivated HCV vaccine.
Acknowledgements
We are grateful to Lubna Ghanem for technical assistance, to Lotte Mikkelsen and Anna-Louise Sørensen for general laboratory support, to Andrea Galli and Stéphanie Serre for scientific discussions and to Jens Ole Nielsen, Bjarne Ørskov Lindhardt, Ove Andersen and Kristian Schønning (all Copenhagen University Hospital, Hvidovre) for their support of the project, as well as to Robert Milne and Anna Toma (Ottawa Heart Institute, Canada) and Charles Rice (Rockefeller University, US) for providing reagents.
This work was supported by A.P. Møller and Chastine Mc-Kinney Møllers Foundation for Medical Sciences (JMG, JB); Hvidovre Hospital, Research foundation (CKM); Proof-of-Concept Grant (JMG, JB); The Danish Cancer Society (JMG, JB); The Danish Council for Independent Research, Medical Sciences (JB); The Lundbeck Foundation (JPR, JMG, JB); and The Novo Nordisk Foundation (JMG, JB); and by Ph.D. stipends from Faculty of Health and Medical Sciences, University of Copenhagen (CKM, TBJ, JPR) and an individual DFF-postdoctoral grant from the Danish Council for Independent Research (JPR). ML is supported by the United States National Institutes of Health grant number AI79031.
Abbreviations
- AEM
Adenovirus expression medium
- ApoB, ApoCI, ApoE
apolipoprotein B, CI, E
- BSA
bovine serum albumin
- HCVcc
cell culture produced hepatitis C virus
- FBS
fetal bovine serum
- FFU
Focus Forming Units
- HCV
hepatitis C virus
- HRP
horseradish peroxidase
- HVR1
hypervariable region 1
- LVP
lipo-viro-particle
- LDL
low-density-lipoprotein
- LDLr
low-density-lipoprotein receptor
- Bmax
maximum blocking rate
- IC50
median inhibitory concentration
- MOI
multiplicity of infection
- NS
nonstructural protein
- ND
not determinable
- BSK
PBS + 1%BSA +0.2% skim milk
- PE
phycoerythrin
- RCF
relative centrifugal force
- RT
room temperature
- SR-BI
scavenger receptor class B type I
- sf-HCVcc
serum-free cell culture produced hepatitis C virus
- SEM
standard error of the mean
- VLDL
very-low-density-lipoprotein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference list
- 1.Fauvelle C, Lepiller Q, Felmlee DJ, Fofana I, Habersetzer F, Stoll-Keller F, et al. Hepatitis C virus vaccines – Progress and perspectives. Microb. Pathog. 2013;58:66–72. doi: 10.1016/j.micpath.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Alter HJ, Seeff LB. Recovery, Persistence, and Sequelae in Hepatitis C Virus Infection: A Perspective on Long-Term Outcome. Semin. Liver Dis. 2000;20:0017–0036. doi: 10.1055/s-2000-9505. [DOI] [PubMed] [Google Scholar]
- 3.Simmonds P, Bukh J, Combet C, Deléage G, Enomoto N, Feinstone S, et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42:962–973. doi: 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- 4.Gottwein JM, Bukh J. Chapter 2 Cutting the Gordian Knot-Development and Biological Relevance of Hepatitis C Virus Cell Culture Systems. Adv. Virus Res. 2008;71:51–133. doi: 10.1016/S0065-3527(08)00002-X. [DOI] [PubMed] [Google Scholar]
- 5.Amoroso P, Rapicetta M, Tosti ME, Mele A, Spada E, Buonocore S, et al. Correlation between virus genotype and chronicity rate in acute hepatitis C. J. Hepatol. 1998;28:939–944. doi: 10.1016/s0168-8278(98)80340-1. [DOI] [PubMed] [Google Scholar]
- 6.Prentoe J, Jensen TB, Meuleman P, Serre SBN, Scheel TKH, Leroux-Roels G, et al. Hypervariable Region 1 Differentially Impacts Viability of Hepatitis C Virus Strains of Genotypes 1 to 6 and Impairs Virus Neutralization. J. Virol. 2011;85:2224–2234. doi: 10.1128/JVI.01594-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheel TKH, Prentoe J, Carlsen THR, Mikkelsen LS, Gottwein JM, Bukh J. Analysis of Functional Differences between Hepatitis C Virus NS5A of Genotypes 1–7 in Infectious Cell Culture Systems. PLoS Pathog. 2012;8:e1002696. doi: 10.1371/journal.ppat.1002696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cross TJS, Rashid MM, Berry PA, Harrison PM. The importance of steatosis in chronic hepatitis C infection and its management: A review. Hepatol. Res. 2010;40:237–247. doi: 10.1111/j.1872-034X.2010.00626.x. [DOI] [PubMed] [Google Scholar]
- 9.Sarrazin C, Hézode C, Zeuzem S, Pawlotsky J-M. Antiviral strategies in hepatitis C virus infection. J. Hepatol. 2012;56:S88–S100. doi: 10.1016/S0168-8278(12)60010-5. [DOI] [PubMed] [Google Scholar]
- 10.Scheel TKH, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat. Med. 2013;19:837–849. doi: 10.1038/nm.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottwein JM, Scheel TK, Jensen TB, Lademann JB, Prentoe JC, Knudsen ML, et al. Development and characterization of hepatitis C virus genotype 1–7 cell culture systems: role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology. 2009;49:364–377. doi: 10.1002/hep.22673. [DOI] [PubMed] [Google Scholar]
- 12.Jensen TB, Gottwein JM, Scheel TKH, Hoegh AM, Eugen-Olsen J, Bukh J. Highly Efficient JFH1-Based Cell-Culture System for Hepatitis C Virus Genotype 5a: Failure of Homologous Neutralizing-Antibody Treatment to Control Infection. J. Infect. Dis. 2008;198:1756–1765. doi: 10.1086/593021. [DOI] [PubMed] [Google Scholar]
- 13.Giang E, Dorner M, Prentoe JC, Dreux M, Evans MJ, Bukh J, et al. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc. Natl. Acad. Sci. 2012;109:6205–6210. doi: 10.1073/pnas.1114927109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meunier J-C, Engle RE, Faulk K, Zhao M, Bartosch B, Alter H, et al. Evidence for cross-genotype neutralization of hepatitis C virus pseudo-particles and enhancement of infectivity by apolipoprotein C1. Proc. Natl. Acad. Sci. U. S. A. 2005;102:4560–4565. doi: 10.1073/pnas.0501275102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Popescu C-I, Dubuisson J. Role of lipid metabolism in hepatitis C virus assembly and entry. Biol. Cell. 2010;102:63–74. doi: 10.1042/BC20090125. [DOI] [PubMed] [Google Scholar]
- 16.Felmlee D, Hafirassou M, Lefevre M, Baumert T, Schuster C. Hepatitis C Virus, Cholesterol and Lipoproteins — Impact for the Viral Life Cycle and Pathogenesis of Liver Disease. Viruses. 2013;5:1292–1324. doi: 10.3390/v5051292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nielsen SU, Bassendine MF, Burt AD, Martin C, Pumeechockchai W, Toms GL. Association between hepatitis C virus and very-low-density lipoprotein (VLDL)/LDL analyzed in iodixanol density gradients. J. Virol. 2006;80:2418–2428. doi: 10.1128/JVI.80.5.2418-2428.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang K-S, Jiang J, Cai Z, Luo G. Human Apolipoprotein E Is Required for Infectivity and Production of Hepatitis C Virus in Cell Culture. J. Virol. 2007;81:13783–13793. doi: 10.1128/JVI.01091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ploss A, Evans MJ. Hepatitis C virus host cell entry. Curr. Opin. Virol. 2012;2:14–19. doi: 10.1016/j.coviro.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, et al. Binding of Hepatitis C Virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 21.McKeating JA, Zhang LQ, Logvinoff C, Flint M, Zhang J, Yu J, et al. Diverse Hepatitis C Virus Glycoproteins Mediate Viral Infection in a CD81-Dependent Manner. J. Virol. 2004;78:8496–8505. doi: 10.1128/JVI.78.16.8496-8505.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Owen DM, Huang H, Ye J, Gale M., Jr Apolipoprotein E on hepatitis C virion facilitates infection through interaction with low-density lipoprotein receptor. Virology. 2009;394:99–108. doi: 10.1016/j.virol.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thi VLD, Granier C, Zeisel MB, Guerin M, Mancip J, Granio O, et al. Characterization of hepatitis C virus particle sub-populations reveals multiple usage of the scavenger receptor BI for entry steps. J. Biol. Chem. 2012;287:31242–31257. doi: 10.1074/jbc.M112.365924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, et al. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 2002;21:5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blanchard E, Belouzard S, Goueslain L, Wakita T, Dubuisson J, Wychowski C, et al. Hepatitis C Virus Entry Depends on Clathrin-Mediated Endocytosis. J. Virol. 2006;80:6964–6972. doi: 10.1128/JVI.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meertens L, Bertaux C, Dragic T. Hepatitis C Virus Entry Requires a Critical ostinternalization Step and Delivery to Early Endosomes via Clathrin-Coated Vesicles. J. Virol. 2006;80:11571–11578. doi: 10.1128/JVI.01717-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plotkin SA, Plotkin SA. Correlates of Vaccine-Induced Immunity. Clin. Infect. Dis. 2008;47:401–409. doi: 10.1086/589862. [DOI] [PubMed] [Google Scholar]
- 28.Plotkin SA, Plotkin SL. The development of vaccines: how the past led to the future. Nat. Rev. Microbiol. 2011;9:889–893. doi: 10.1038/nrmicro2668. [DOI] [PubMed] [Google Scholar]
- 29.Steinmann E, Pietschmann T. Cell Culture Systems for Hepatitis C Virus. Hepat. C Virus Mol. Virol. Antivir. Ther. 2013:17–48. doi: 10.1007/978-3-642-27340-7_2. [DOI] [PubMed] [Google Scholar]
- 30.Lohmann V, Bartenschlager R. On the history of hepatitis C virus cell culture systems. J. Med. Chem. 2014;57:1627–1642. doi: 10.1021/jm401401n. [DOI] [PubMed] [Google Scholar]
- 31.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat.Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindenbach BD, Evans MJ, Syder AJ, Wölk B, Tellinghuisen TL, Liu CC, et al. Complete Replication of Hepatitis C Virus in Cell Culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 33.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, et al. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gottwein JM, Scheel TK, Hoegh AM, Lademann JB, Eugen-Olsen J, Lisby G, et al. Robust hepatitis C genotype 3a cell culture releasing adapted intergenotypic 3a/2a (S52/JFH1) viruses. Gastroenterology. 2007;133:1614–1626. doi: 10.1053/j.gastro.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 35.Scheel TK, Gottwein JM, Jensen TB, Prentoe JC, Hoegh AM, Alter HJ, et al. Development of JFH1-based cell culture systems for hepatitis C virus genotype 4a and evidence for cross-genotype neutralization. Proc.Natl.Acad.Sci.U.S.A. 2008;105:997–1002. doi: 10.1073/pnas.0711044105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pietschmann T, Kaul A, Koutsoudakis G, Shavinskaya A, Kallis S, Steinmann E, et al. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. 2006;103:7408–7413. doi: 10.1073/pnas.0504877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yi M, Ma Y, Yates J, Lemon SM. Compensatory Mutations in E1, p7, NS2, and NS3 Enhance Yields of Cell Culture-Infectious Intergenotypic Chimeric Hepatitis C Virus. J. Virol. 2007;81:629–638. doi: 10.1128/JVI.01890-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindenbach BD, Meuleman P, Ploss A, Vanwolleghem T, Syder AJ, McKeating JA, et al. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc.Natl.Acad.Sci.U.S.A. 2006;103:3805–3809. doi: 10.1073/pnas.0511218103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gastaminza P, Kapadia SB, Chisari FV. Differential Biophysical Properties of Infectious Intracellular and Secreted Hepatitis C Virus Particles. J. Virol. 2006;80:11074–11081. doi: 10.1128/JVI.01150-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.WHO Requirements for the Use of Animal Cells asin vitroSubstrates for the Production of Biologicals (Requirements for Biological Susbstances No. 50) Biologicals. 1998;26:175–193. doi: 10.1006/biol.1998.0153. [DOI] [PubMed] [Google Scholar]
- 41.Houghton M, Law JL, Tyrrell DL. An Inactivated Hepatitis C Virus Vaccine on the Horizon? Gastroenterology. 2013;145:285–288. doi: 10.1053/j.gastro.2013.06.029. [DOI] [PubMed] [Google Scholar]
- 42.Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Growth of Human Hepatoma Cell Lines with Differentiated Functions in Chemically Defined Medium. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- 43.Abe K, Ikeda M, Ariumi Y, Dansako H, Kato N. Serum-free cell culture system supplemented with lipid-rich albumin for hepatitis C virus (strain O of genotype 1b) replication. Virus Res. 2007;125:162–168. doi: 10.1016/j.virusres.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 44.Gottwein JM, Scheel TKH, Callendret B, Li Y-P, Eccleston HB, Engle RE, et al. Novel Infectious cDNA Clones of Hepatitis C Virus Genotype 3a (Strain S52) and 4a (Strain ED43): Genetic Analyses and In Vivo Pathogenesis Studies. J. Virol. 2010;84:5277–5293. doi: 10.1128/JVI.02667-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheel TKH, Gottwein JM, Mikkelsen LS, Jensen TB, Bukh J. Recombinant HCV Variants With NS5A From Genotypes 1–7 Have Different Sensitivities to an NS5A Inhibitor but Not Interferon-α. Gastroenterology. 2011;140:1032–1042. doi: 10.1053/j.gastro.2010.11.036. [DOI] [PubMed] [Google Scholar]
- 46.Serre SBN, Krarup HB, Bukh J, Gottwein JM. Identification of Alpha Interferon-Induced Envelope Mutations of Hepatitis C Virus In Vitro Associated with Increased Viral Fitness and Interferon Resistance. J. Virol. 2013;87:12776–12793. doi: 10.1128/JVI.00901-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russell RS, Meunier J-C, Takikawa S, Faulk K, Engle RE, Bukh J, et al. Advantages of a single-cycle production assay to study cell culture-adaptive mutations of hepatitis C virus. Proc. Natl. Acad. Sci. 2008;105:4370–4375. doi: 10.1073/pnas.0800422105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Catanese MT, Graziani R, von Hahn T, Moreau M, Huby T, Paonessa G, et al. High-Avidity Monoclonal Antibodies against the Human Scavenger Class B Type I Receptor Efficiently Block Hepatitis C Virus Infection in the Presence of High-Density Lipoprotein. J. Virol. 2007;81:8063–8071. doi: 10.1128/JVI.00193-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Law M, Maruyama T, Lewis J, Giang E, Tarr AW, Stamataki Z, et al. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat. Med. 2008;14:25–27. doi: 10.1038/nm1698. [DOI] [PubMed] [Google Scholar]
- 50.Weisgraber KH, Innerarity TL, Harder KJ, Mahley RW, Milne RW, Marcel YL, et al. The receptor-binding domain of human apolipoprotein E. Monoclonal antibody inhibition of binding. J. Biol. Chem. 1983;258:12348–12354. [PubMed] [Google Scholar]
- 51.Gottwein JM, Scheel TKH, Jensen TB, Ghanem L, Bukh J. Differential Efficacy of Protease Inhibitors Against HCV Genotypes 2a, 3a, 5a, and 6a NS3/4A Protease Recombinant Viruses. Gastroenterology. 2011;141:1067–1079. doi: 10.1053/j.gastro.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 52.Prentoe J, Serre SBN, Ramirez S, Nicosia A, Gottwein JM, Bukh J. Hypervariable Region 1 Deletion and Required Adaptive Envelope Mutations Confer Decreased Dependency on Scavenger Receptor Class B Type I and Low-Density Lipoprotein Receptor for Hepatitis C Virus. J. Virol. 2014;88:1725–1739. doi: 10.1128/JVI.02017-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wünschmann S, Müller HM, Stipp CS, Hemler ME, Stapleton JT. In Vitro Interaction between Hepatitis C Virus (HCV) Envelope Glycoprotein E2 and Serum Lipoproteins (LPs) Results in Enhanced Cellular Binding of Both HCV E2 and LPs. J. Infect. Dis. 2006;194:1058–1067. doi: 10.1086/507647. [DOI] [PubMed] [Google Scholar]
- 54.Gastaminza P, Dryden KA, Boyd B, Wood MR, Law M, Yeager M, et al. Ultrastructural and Biophysical Characterization of Hepatitis C Virus Particles Produced in Cell Culture. J. Virol. 2010;84:10999–11009. doi: 10.1128/JVI.00526-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Merz A, Long G, Hiet M-S, Brugger B, Chlanda P, Andre P, et al. Biochemical and Morphological Properties of Hepatitis C Virus Particles and Determination of Their Lipidome. J. Biol. Chem. 2010;286:3018–3032. doi: 10.1074/jbc.M110.175018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Catanese MT, Uryu K, Kopp M, Edwards TJ, Andrus L, Rice WJ, et al. Ultrastructural analysis of hepatitis C virus particles. Proc. Natl. Acad. Sci. 2013;110:9505–9510. doi: 10.1073/pnas.1307527110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Akazawa D, Moriyama M, Yokokawa H, Omi N, Watanabe N, Date T, et al. Neutralizing Antibodies Induced by Cell Culture-Derived Hepatitis C Virus Protect Against Infection in Mice. Gastroenterology. 2013;145:447–455. doi: 10.1053/j.gastro.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 58.Narbus CM, Israelow B, Sourisseau M, Michta ML, Hopcraft SE, Zeiner GM, et al. HepG2 Cells Expressing MicroRNA miR-122 Support the Entire Hepatitis C Virus Life Cycle. J. Virol. 2011;85:12087–12092. doi: 10.1128/JVI.05843-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Costa DD, Turek M, Felmlee DJ, Girardi E, Pfeffer S, Long G, et al. Reconstitution of the Entire Hepatitis C Virus Life Cycle in Nonhepatic Cells. J. Virol. 2012;86:11919–11925. doi: 10.1128/JVI.01066-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vogt A, Scull MA, Friling T, Horwitz JA, Donovan BM, Dorner M, et al. Recapitulation of the hepatitis C virus life-cycle in engineered murine cell lines. Virology. 2013;444:1–11. doi: 10.1016/j.virol.2013.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frentzen, Anggakusuma A, Gürlevik E, Hueging K, Knocke S, Ginkel C, et al. Cell entry, efficient RNA replication, and production of infectious hepatitis C virus progeny in mouse liver-derived cells. Hepatology. 2014;59:78–88. doi: 10.1002/hep.26626. [DOI] [PubMed] [Google Scholar]
- 62.Kambara H, Fukuhara T, Shiokawa M, Ono C, Ohara Y, Kamitani W, et al. Establishment of a Novel Permissive Cell Line for the Propagation of Hepatitis C Virus by Expression of MicroRNA miR122. J. Virol. 2012;86:1382–1393. doi: 10.1128/JVI.06242-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sourisseau M, Goldman O, He W, Gori JL, Kiem H, Gouon-Evans V, et al. Hepatic Cells Derived From Induced Pluripotent Stem Cells of Pigtail Macaques Support Hepatitis C Virus Infection. Gastroenterology. 2013;145:966–969. doi: 10.1053/j.gastro.2013.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gottwein JM, Bukh J. Viral hepatitis: Cell-culture-derived HCV—a promising vaccine antigen. Nat. Rev. Gastroenterol. Hepatol. 2013;10:508–509. doi: 10.1038/nrgastro.2013.136. [DOI] [PubMed] [Google Scholar]
- 65.Akazawa D, Morikawa K, Omi N, Takahashi H, Nakamura N, Mochizuki H, et al. Production and characterization of HCV particles from serum-free culture. Vaccine. 2011;29:4821–4828. doi: 10.1016/j.vaccine.2011.04.069. [DOI] [PubMed] [Google Scholar]
- 66.Bukh J, Meuleman P, Tellier R, Engle RE, Feinstone SM, Eder G, et al. Challenge pools of hepatitis C virus genotypes 1–6 prototype strains: replication fitness and pathogenicity in chimpanzees and human liver-chimeric mouse models. J.Infect.Dis. 2010;201:1381–1389. doi: 10.1086/651579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu S, Xiao L, Nelson C, Hagedorn C. A Cell Culture Adapted HCV JFH1 Variant That Increases Viral Titers and Permits the Production of High Titer Infectious Chimeric Reporter Viruses. PLoS ONE. 2012;7:e44965. doi: 10.1371/journal.pone.0044965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pokrovskii MV, Bush CO, Beran RKF, Robinson MF, Cheng G, Tirunagari N, et al. Novel Mutations in a Tissue Culture-Adapted Hepatitis C Virus Strain Improve Infectious-Virus Stability and Markedly Enhance Infection Kinetics. J. Virol. 2011;85:3978–3985. doi: 10.1128/JVI.01760-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Horwitz JA, Dorner M, Friling T, Donovan BM, Vogt A, Loureiro J, et al. Expression of heterologous proteins flanked by NS3-4A cleavage sites within the hepatitis C virus polyprotein. Virology. 2013;439:23–33. doi: 10.1016/j.virol.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jiang J, Luo G. Cell Culture-Adaptive Mutations Promote Viral Protein-Protein Interactions and Morphogenesis of Infectious Hepatitis C Virus. J. Virol. 2012;86:8987–8997. doi: 10.1128/JVI.00004-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dhillon S, Witteveldt J, Gatherer D, Owsianka AM, Zeisel MB, Zahid MN, et al. Mutations within a Conserved Region of the Hepatitis C Virus E2 Glycoprotein That Influence Virus-Receptor Interactions and Sensitivity to Neutralizing Antibodies. J. Virol. 2010;84:5494–5507. doi: 10.1128/JVI.02153-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Abdo AA, Lee SS. Management of hepatitis C virus genotype 4. J. Gastroenterol. Hepatol. 2004;19:1233–1239. doi: 10.1111/j.1440-1746.2003.03325.x. [DOI] [PubMed] [Google Scholar]
- 73.Bankwitz D, Steinmann E, Bitzegeio J, Ciesek S, Friesland M, Herrmann E, et al. Hepatitis C Virus Hypervariable Region 1 Modulates Receptor Interactions, Conceals the CD81 Binding Site, and Protects Conserved Neutralizing Epitopes. J. Virol. 2010;84:5751–5763. doi: 10.1128/JVI.02200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhong J, Gastaminza P, Chung J, Stamataki Z, Isogawa M, Cheng G, et al. Persistent Hepatitis C Virus Infection In Vitro: Coevolution of Virus and Host. J. Virol. 2006;80:11082–11093. doi: 10.1128/JVI.01307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grove J, Nielsen S, Zhong J, Bassendine MF, Drummer HE, Balfe P, et al. Identification of a Residue in Hepatitis C Virus E2 Glycoprotein That Determines Scavenger Receptor BI and CD81 Receptor Dependency and Sensitivity to Neutralizing Antibodies. J. Virol. 2008;82:12020–12029. doi: 10.1128/JVI.01569-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yamamoto M, Aizaki H, Fukasawa M, Teraoka T, Miyamura T, Wakita T, et al. Structural Requirements of Virion-Associated Cholesterol for Infectivity, Buoyant Density and Apolipoprotein Association of Hepatitis C Virus. J. Gen. Virol. 2011;92:2082–2087. doi: 10.1099/vir.0.032391-0. [DOI] [PubMed] [Google Scholar]
- 77.Albecka A, Belouzard S, Beeck D, Op A, Descamps V, Goueslain L, et al. Role of low-density lipoprotein receptor in the hepatitis C virus life cycle, Role of low-density lipoprotein receptor in the hepatitis C virus life cycle. Hepatol. Hepatol. 2012;55:998–1007. doi: 10.1002/hep.25501. [DOI] [PubMed] [Google Scholar]