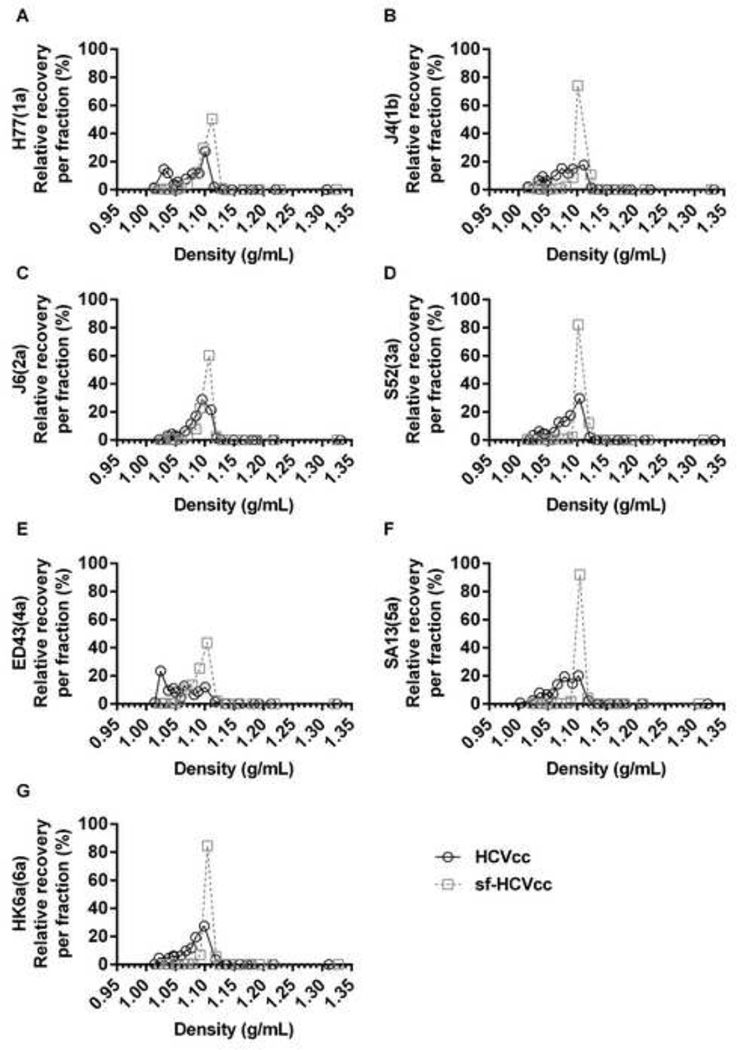
Figure 5. The sf-HCVcc particles of genotype 1–6 displayed an altered density profile with a single infectivity peak.
Of the virus stocks described in Figure 2, 10mL HCVcc supernatant taken from the last harvest of DMEM + 10% FBS culture supernatant (black line) or 10mL sf-HCVcc supernatant taken after 48 hours of AEM culture (grey, dotted line) was concentrated and layered on top of a pre-formed 10–40% iodixanol gradient and subjected to ultracentrifugation as described in Materials and Methods. Fractions were collected from the bottom of the gradients and analyzed by infectivity titration and by density determination as described in Materials and Methods. The HCV Core-E2 sequences of all virus stocks used were determined by direct sequencing. Compared to the plasmid sequence, sf-H77(1a) and H77(1a) had acquired the previously described amino acid change Y361H [11], estimated to be present in 50% of viral genomes. The sf-J4(1b) had acquired amino acid changes T578A and D584G, estimated to be present in the majority of viral genomes. Relative recovery per fraction (%) was calculated by relating the amount of infectious virus detected in each fraction to the total amount of infectious virus collected, and is plotted against the density determined for each fraction.