Abstract
We have developed a web tool, PupaSNP Finder (PupaSNP for short), for high-throughput searching for single nucleotide polymorphisms (SNPs) with potential phenotypic effect. PupaSNP takes as its input lists of genes (or generates them from chromosomal coordinates) and retrieves SNPs that could affect the conserved regions that the cellular machinery uses for the correct processing of genes (intron/exon boundaries or exonic splicing enhancers), predicted transcription factor binding sites (TFBS) and changes in amino acids in the proteins. The program uses the mapping of SNPs in the genome provided by Ensembl. Additionally, user-defined SNPs (not yet mapped in the genome) can be easily provided to the program. Also, additional functional information from Gene Ontology, OMIM and homologies in other model organisms is provided. In contrast to other programs already available, which focus only on SNPs with possible effect in the protein, PupaSNP includes SNPs with possible transcriptional effect. PupaSNP will be of significant help in studies of multifactorial disorders, where the use of functional SNPs will increase the sensitivity of identification of the genes responsible for the disease. The PupaSNP web interface is accessible through http://pupasnp.bioinfo.cnio.es.
INTRODUCTION
Single nucleotide polymorphisms (SNPs) are the simplest and most frequent type of DNA sequence variation among individuals and they represent one of the most powerful tools for the analysis of genomes (1). Owing to their widespread distribution, SNPs are particularly valuable as genetic markers in the search for disease susceptibility genes, drug response-determining genes, and so on. In the past decades, linkage analysis has been very successful in the identification of genes responsible for mendelian diseases. Nevertheless, direct application of linkage analysis to the case of complex diseases, in which several genes with weaker genotype–phenotype correlations are involved, has resulted in more modest success (2). Now, it is believed that improved genotyping methods in combination with the proper design strategies could bring the genetics of complex diseases to a point of success comparable to where mendelian genetics now firmly resides (3).
There are examples documented in which alleles of more than one gene contribute to the same disease. It is generally believed that multigenic diseases reflect disruptions in the proteins that participate in a protein complex or a pathway (4). Typically, SNPs have been used as markers; that is, the real determinant of the disease was not the SNP itself but some other mutation in linkage disequilibria with it.
The use of functional SNPs could be an important factor for increasing significantly the sensitivity of association tests. In fact, several complex genetic disorders such as Alzheimer's disease (5) and Crohn's disease (6) have been associated with functional SNPs, lending credence to strategies giving priority to candidate markers based on predictable function. The latest build of NCBI's dbSNP (http://www.ncbi.nlm.nih.gov/SNP/snp_summary.cgi) contains 5 772 564 SNPs, with 2 356 957 of them validated. This means that human variation has been screened to an average resolution of 1 SNP for every 566 nt. There is also curated information on SNPs in HGVbase (7). These figures suggest that the possibility of finding the real determinant of a disease among the characterized SNPs can be seriously considered. In fact, dbSNP build 117 contains 24 483 SNPs located in coding regions that produce amino acid change, affecting a total of 9791 different genes. Several estimate suggest that, overall, only 20% of them could damage the protein (8). Much attention has been focused on the possible phenotypic effects of SNPs that cause amino acid changes. The volume of available information together with the development of more sophisticated methods of protein structure prediction has led to different attempts to relate the effect of amino acid changes to structural distortions and, consequently, possible phenotypic effect. Following this, two main different approaches have been taken: on the one hand is the study of conservation of residues in homologous proteins (9) including more sophisticated approaches taking into account the phylogenetic history (10) and, on the other hand, there is the study of changes in the stability (11,12) and other properties of the protein due to changes of amino acids (8,13).
Nevertheless, there are different ways in which the functionality of a gene product can be affected without requiring a amino acid change in the protein. There is increasing evidence that many human disease genes harbour exonic or non-coding mutations that affect pre-mRNA splicing (14). Alternative splicing produced by mutations in intron/exon junctions, or in distinct binding motifs, such as exonic splicing enhancers (ESEs), to which different proteins involved in splicing bind, is the basis of different diseases. In fact, it has been estimated that 15% of point mutations that result in human genetic diseases cause RNA splicing defects (15). For example, a silent mutation in exon 14 of the APC gene is associated with exon skipping in a Familial Adenomatous Polyposis (FAP) family (16), and there are many more examples [see Table 2 in (14)]. Also, alterations in the level of expression of gene products can cause diseases. Different SNPs are associated with alterations in gene expression (17) and, in some cases, it is known that they alter some regulatory sequence motif. For example, a regulatory polymorphism in the programmed cell death 1 gene (PDCD1), which alters a binding site for the runt-related transcription factor 1 (RUNX1) located in an intronic enhancer, is associated with susceptibility to systemic lupus erythematosus in humans (18). It has also been reported that polymorphisms in the gelatinase A promoter region are associated with diminished transcriptional response to estrogen and genetic fitness (19). A recent large-scale screening over a set of 16 chromosomes, found SNPs in the promoters regions of 35% of the genes, and experimental evidence suggested that around one-third of promoter variants may alter gene expression to a functionally relevant extent (20). Therefore, the inclusion of other possible causes of loss of functionality in gene products, beyond the simple estimation of the possible phenotypic effect of an amino acid change, increases considerably the number of SNPs with potential phenotypic effect to be considered for the design of experiments.
Classical statistical linkage tests need a large number of cases if the number of genes to be tested is high. It has only recently been recognized that reliable identification of genetic variants that affect gene regulation is still a challenge in genomics and is expected to play an important role in the molecular characterization of complex traits (21). Another important consideration when analysing multigenic traits is the information available on the genes. Information allows a more targeted approach, by focusing initially on genes whose functionality is related to the disease studied.
Genome surveys based on the information contained in dbSNP show that there are 361 SNPs mapped in splice sites of introns, 1 387 506 in introns and 242 842 in untranslated regions affecting 336 16 306 and 14 198 genes, respectively. A number of these SNPs could be disease determinants.
With the idea of extracting as much information as possible form SNPs with putative phenotypic effect, we have developed PupaSNP Finder (Putative Phenotypic Alterations caused by SNPs; PupaSNP for short). This tool retrieves all the SNPs present in a set of genes of interest that potentially affect the functionality of the gene product. This list is combined with functional information obtained from Gene Ontology (GO) annotations (22). Genes can be directly retrieved from genomic locations or, alternatively, can be taken from a list provided by the user. This corresponds to two typical problems: (i) traits mapped to a given chromosomal region or (ii) traits associated with a given class of genes (e.g. a signalling pathway). Genome coordinates of genes and SNPs are taken from the Ensembl annotation (23).
METHODS
Finding SNPs with potential phenotypic effect
PupaSNP operates with a collection of entries from dbSNP mapped to the Golden Path genome assembly, as implemented in human section of Ensembl (http://www.ensembl.org). As previously mentioned, PupaSNP uses a list of genes and generates a report in which all the SNPs with possible phenotypic effect are listed. The genes can be selected directly by their location in a region of the genome, or just provided as a list (e.g. genes belonging to a given pathway, involved in a particular biological function). Genomic regions can be selected either by defining a range of chromosome coordinates or by directly choosing the cytoband of interest. The engine finds all the genes located within the specified region as well as their promoter regions using Ensembl APIs. In the case of a user-defined list, Ensembl is used to extract their complete intron/exon structure as well as the promoter regions.
The potential effects on the phenotype taken into account are at both transcriptional and gene product levels. These include alterations in (i) transcription factor binding sites, (ii) intron/exon border consensus sequences, (iii) ESE sequences, which are the binding sites for specific serine/arginine-rich (SR) proteins involved in the splicing machinery (24,25) and (iv) the exons that cause an amino acid change. Additionally, the GO terms (22) associated with the genes can be obtained. This is very useful in the case of looking for genes in a chromosomal region, because it can help to discard genes definitively not involved in the disease studied, based on the annotations.
Transcription factor binding sites
In the search for SNPs with potential phenotypic effect, 10 000 bp upstream of the genes, belonging to the promoter region of each gene in the list, are scanned for the presence of possible transcription factor binding sites (TFBSs). The program Match™ (26), version 1.10, from the Transfac® database (27), version professional 7.3, was used for this purpose. SNPs located within these motifs are considered to have a putative phenotypic effect in the expression of the gene. The options used for the program Match™ were (i) group of matrices: vertebrates, (ii) use high quality matrices only and (iii) cutoff selection for matrix group: to minimize false positives. This cutoff was obtained by exploring the third exon sequences with the weight matrices and was chosen to reduce the number of random putative sites found by the program (26).
Although the scan is done in a region 10 000 bp upstream from the start of the gene, the number of bases to be taken into account in the study is customizable. Obviously, the closer to the start of the gene, the more likely the binding site is to be authentic.
Intron–exon boundaries
Ensembl APIs were used to extract the intron/exon organization of the genes and the corresponding sequences. The two conserved nucleotides at each side of the splicing point, which constitute the splicing signal (14), were then located and all the SNPs altering these signals are recorded.
Exonic splicing enhancers
Mutations that deactivate or activate exonic splicing enhancer sequences may result in exon skipping, malformation, and so on. ESEs also appear to be important in exons that normally undergo alternative splicing. Different classes of ESE consensus motifs have been described, but they are not always easily identified. We have developed a script that scans exon sequences to identify putative ESEs responsive to the human SR proteins SF2/ASF, SC35, SRp40 and SRp55, by using the weight matrices available for them (28). A score is obtained related to the likelihood that the site found is a real ESE. Only ESE sites with scores over the threshold [see (28) and http://exon.cshl.org/ESE/ESEmatrix.html for details] are taken into account in the analysis. Threshold values, above which a score for a given sequence is considered to be significant, are set as the median of the highest score for each sequence in a set of 30 randomly chosen 20 nt sequences (from the starting pool used for functional assays for ESE identification; see http://exon.cshl.org/ESE/ESEmatrix.html). If an SNP disrupts one of these sequences, the new score, corresponding to the mutated sequence, is also calculated. Strong differences between the two score values suggest more drastic effects caused by the SNP.
Changes at amino acid level and functional implications
SNPs that result in a change of amino acid are likely to cause some phenotypic effect and, consequently, are all listed. Since the main purpose of the tool is to cover possible transcriptional effects of the SNPs and there are a number of tools already available for the prediction of phenotypic effects due to mutations in amino acids (see Introduction) PupaSNP only lists them. To help in the identification of possible effects we label SNPs that disrupt any functional motif as listed in Interpro (29), a resource that compiles information on protein families, domains and functional sites. The coordinates of the Interpro motifs within the exons of the genes are extracted from Ensembl and cross-referenced with the SNPs coordinates.
Additional functional information
Since PupaSNP finder works with lists of genes in order to select the best SNP candidates for further use in association analysis, it is very helpful to have functional annotations of the genes. This allows the assignment of priorities based also on the information available on the genes. Information is obtained from (i) Gene Ontology annotations, obtained through the FatiGO engine (30) (available at http://fatigo.bioinfo.cnio.es), (ii) OMIM (Online Mendelian Inheritance in Man), which constitutes a comprehensive, authoritative and timely knowledge base of human genes and genetic disorders (31) and (iii) homologies to other organisms, obtained directly from Ensembl. Gene Ontology is a tree structure (called a directed acyclic graph) in which terms describing three fundamental ontologies (molecular function, biological process and cellular component) have descendants with more detailed descriptions. Thus, descending the hierarchy of GO implies moving towards terms with more detailed descriptions of the ontologies, but, at the same time, there are fewer genes with annotations at such detail. FatiGO works by climbing up the hierarchy to a selected parent level (30) to optimize the number of genes with annotation and the detail of the annotation. Thus, the identification of common parent functions or processes is easier. In this way, the consideration of the SNPs in a functional context can help to understand the potential biological implications of the SNPs and genes studied.
RESULTS
SNPs with possible phenotypic effect
We analysed a total of 24 037 human genes corresponding to the annotations in Ensembl build 34 (version 18.34.1), which contains the mapping of dbSNP 117. By scanning with the Match™ program the 10 000 bp upstream promoter regions of the genes, 2 587 478 transcription factor binding sites, corresponding to 330 different Transfac weight matrices (27), were found. After mapping the SNPs in the promoter regions, 71 444 TFBSs were found to be disrupted by a total of 57 412 SNPs (some SNPs affect more than one TFBS at the same time). A total of 19 010 genes presented at least 1 predicted TFBS disrupted by a SNP, which constitutes a considerable proportion of the total number of genes. The coverage in terms of both SNPs and TFBS predictions was good: only for 54 genes was no single SNP found in the 10 000 bp 5′-upstream region, and only for 2 genes could no predicted TFBS be found (ENSG00000116119, or KV2A_HUMAN, which is the IG KAPPA CHAIN V-II REGION CUM, and ENSG00000174994, or AK057375, which seems to be a DNA binding protein). In a number of cases, SNPs affect overlapping TFBSs, which could have a stronger effect still in the phenotype. There are even 2 SNPs that simultaneously affect 15 TFBSs.
The four conserved bases that define intron–exon boundaries were mutated by 844 SNPs, affecting to a total of 598 genes.
Over eight million ESE motifs were found, covering all the genes studied. A total of 138 746 SNPs were found to disrupt ESE sequences. These SNPs affect a total of 17 312 genes.
These results suggest that, in the search for SNPs with potential phenotypic effects, regulatory SNPs or SNPs affecting splicing should not be neglected.
The web interface
Input data
PupaSNP has been designed for high-throughput screening of functional SNPs. Thus, the input consists of a list of genes. The list can be directly provided as a collection of gene identifiers (Ensembl IDs, or external IDs, which include GenBank, Swissprot/TrEMBL and other gene IDs supported by Ensembl) or can be specified by means of a chromosomal location (cytobands or chromosomal coordinates). In the latter case, PupaSNP extracts all the genes contained in the specified location. Ensembl coordinates are used to extract the genes. Only Ensembl annotated genes, but not predictions, are extracted.
User-defined SNPs
Alternatively, the user can input SNPs not in the database in a very straightforward manner and take advantage of the tools for predicting their potential phenotypic effect. A text file containing the descriptions of the SNPs must be generated. Each line describes one unique SNP with the following tab-delimited data: SNP name, gene (Ensembl ID or external ID), position with respect to the start of the translation and alleles, e.g.
MySNP01 ENSG00000000003 -1830 A/G
MySNP02 ENSG00000157873 421 C/G
This describes two SNPs: the first in the gene ENSG00000000003 (tetraspanin 6, or TSPAN6), 1830 bp away from the transcription start point, with polymorphisms consisting of a change of an A for a G; and the second in gene ENSG00000157873 (tumor necrosis factor receptor-like 2, TNFRSF14), 421 bp within the transcripted region, which corresponds to the first exon of the gene.
The web interface
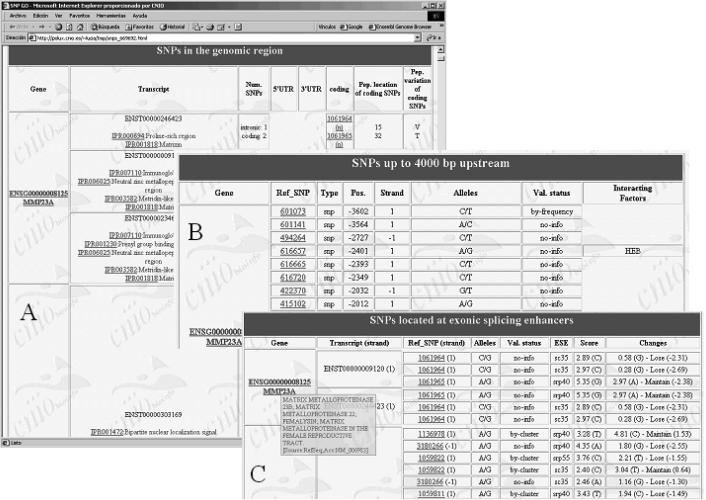
A web interface to PupaSNP is available at http://pupas.bioinfo.cnio.es/. Lists of genes can be defined by chromosome position, which can be specified in terms of cytoband units or in absolute chromosomal position (as mapped in the corresponding Ensembl assembly). The upstream region makes reference to the number of bases upstream in which TFBSs will be searched for (with a upper limit of 10 000 bp). Also, lists of genes can be uploaded or just pasted into the box. PupaSNP finds all the SNPs mapping to locations that might cause a loss of functionality in the genes. Functional information for the genes can also be obtained from OMIM and from Gene Ontology. Information on homologous genes can also be retrieved. Finally, SNPs do not need to be annotated in the genome to be included in the query tool. The user can specify a list of SNPs using a gene as reference. In this way the use of absolute coordinates, which can easily change between assembly versions, is avoided in favour of the use of coordinates relative to genes, which tend to be more stable. Results include SNPs in a the promoter region of the genes, SNPs located at intron boundaries, SNPs located at exonic splicing enhancers and coding SNPs located at Interpro domains. Figure 1 shows part of the results provided by the program for the SNPs with possible phenotypic effect on genes in the p36.33 cytoband of chromosome 1. Figure 1C is especially interesting because it shows how the scores obtained by the motif scanning method can be used to assess the possible impact of the polymorphism on the recognition of the ESE motif by the cellular machinery.
Figure 1.
A selection of results from PupaSNP. (A) List of genes and the corresponding transcripts with the SNPs mapping to the different regions, which include coding and 5′- and 3′-untranslated regions. For coding SNPs, the position within the transcript and the change produced (if any) is reported. (B) SNPs located in the promoter regions (in the example, a limit of 4000 bp was chosen). Disruptions of predicted TFBSs are listed. The validation status of the SNPs (‘no-info’, ‘by-submitter’, ‘by-frequency’, ‘by-cluster’; see dbSNP web page) is also provided. (C) SNPs located at exonic splice enhancers. The scores make reference to the closeness of the site to the motif. If the polymorphism gives a site with a worst score, this would, generally speaking, probably imply worst recognition of the site by the cellular machinery and, consequently, a putative alteration in the normal splicing process. When the cursor is over the gene name, additional information is displayed.
Both the SNPs and the genes found are linked to the Ensembl Genome Browser.
Experimental validation
The validation status of the SNPs is, in some cases, a much more important factor for their selection than their possible functional role. Such information is scarce: 2 359 534 out of 5 798 183 SNPs in dbSNP build 118 have been validated, which constitutes 40%. However, only 160 466 have estimates of population frequencies and only 94 867 have a phenotype associated. To obtain a sense of the reliability of the SNPs annotated with ‘no-info’, a set of SNPs was sought for a list of candidate modifier genes related to a phenotype exhibited by MEN2 (Multiple endocrine neoplasia, type IIA) patients (OMIM, #171400), all of them RET mutation carriers. MEN2 is an autosomal dominant syndrome of multiple endocrine neoplasms, with variable clinical expression even between members of the same family. This fact cannot be explained only by a mutation in a major susceptibility gene, but suggests a role for genetic modifiers, which may also work through quantitative effect.
In most of cases, it was necessary to validate the putative SNPs identified by PupaSNP because there was no information about validation status. To validate SNPs and estimate their allele frequency, 48 non-related individuals from the Spanish population were used. The specific primers used to amplify the fragments of interest by PCR (polymerase chain reaction) were designed using the OLIGO 4.1 program. When possible, the primers were selected and designed to amplify a fragment (200–500 bp) that allowed us to investigate several SNPs at the same time. As a denaturing high-performance liquid chromatograph (dHPLC) system (WAVE, Transgenomics Limited, Crewe, UK) was used for the initial SNP screening, the fragments of interest had a homogeneous GC content across different domains from the DNA fragment to obtain a consistent melting profile. The Navigator software was used for data handling and optimization of the dHPLC system. After normalization, each PCR product that exhibited a change in the chromatogram profile was characterized by sequence analysis. These PCR products were purified using an E.Z.N.A. Cycle-Pure Kit (Omega Bio-tek, USA) according to the manufacturer's instructions, and sequenced using an automatic sequencer ABI PRISM™ 3700 (Applied Biosystems. Perkin Elmer, USA). The reaction was carried out in 4 μl of a Big Dye terminator cycle sequencing Kit (Perkin Elmer, USA), 10 pmol of the sense/antisense primer, 5% DMSO and 6–12 ng of amplified DNA. Although the results obtained here do not pretend to be capable of general extrapolation to the entire database, we have found that 24 out of 28 SNPs assayed proved to be authentic and polymorphic in the Spanish population, which constitutes a good rate.
DISCUSSION
Typically, SNPs have been used as markers to search for the real determinant of a disease in linkage disequilibria with it. As previously mentioned, the use of functional SNPs, which may be the real disease determinants, could be an important factor in increasing the sensitivity of association tests.
Despite the obvious importance that alterations in the regulation, expression level or splicing of genes can have for the phenotype, these have long been ignored in the most common approaches to finding functional SNPs, which have instead focused more on the possible effect of polymorphisms causing amino acid changes. Apart from the databases mentioned above (dbSNP and HGVbase), there are a number of resources available over the net collecting information on phenotypes associated with SNPs, such as The Human Gene Mutation Database (http://www.hgmd.org) at the University of Wales, which classifies SNPs according the lesion they cause (missense substitutions, splice variants, and so on) (32) and PicSNP, a catalogue of non-synonymous SNPs obtained from the human genome assembly (33). However, these are mainly specialized catalogues collecting information on SNPs rather than tools for their selection.
PupaSNP constitutes a tool for selecting SNPs with putative phenotypic effects designed for high-throughput experiments. It deals with lists of genes, instead of focusing on individual genes. In addition, more information on different possible motifs with regulatory function has been included. For example, SNPs in ESE had never previously been included in any catalogue.
Multigenic diseases are generally associated with disruptions in proteins that participate in a protein complex or a pathway (4). The inclusion in PupaSNP of information regarding the participation of genes in signalling cascades or in pathways or in protein complexes will be considered in the near future. Databases containing protein interaction data, such as DIP and BIND (see http://www.hgmp.mrc.ac.uk/GenomeWeb/prot-interaction.html), can be an important source of information to be considered in the search for SNPs affecting multigentic traits.
Despite the fact that PupaSNP is more focused on SNPs with possible effects at transcriptional level, the inclusion of an algorithm for improving the predictions of the effect of SNPs in the proteins, such as FoldX (12), would provide, within the same framework, both types of result.
Minimum SNP set selection allows the user to optimize the number of SNPs required to represent haplotype diversity, thus reducing the cost of genotyping by assaying the minimum number of SNPs required. The inclusion of information on linkage disequilibrium or on haplotype blocks can assist in a more efficient selection of SNPs. Some programs, such as HapScope (34), include information on haplotypes and use them to select minimum subsets of SNPs. Another important issue is the reliability of the SNPs. As previously mentioned, only 40% of the SNPs in dbSNP have been validated, and only for 5% are population frequencies are available. This means that most of the SNPs found in any kind of selection will lack information on their possible presence in the population of interest as a manageable polymorphism. Even though our results suggest a high rate of authenticity, even for the SNPs labeled as ‘no-info’, they must be treated carefully and cannot be directly extrapolated to the entire database. As population frequencies are included in the database, these data could be of interest for use as part of the selection process of SNPs
PupaSNP will be the tool used in the first step of the pipeline for the study of polymorphisms at the Spanish National Genotyping Centre (CeGen). For this reason it has been developed to cope with high-throughput experimental designs. PupaSNP takes as input lists of genes (or generates them from chromosomal coordinates) and provides results which integrate all the information available as well as obtained by means of predictions of SNPs with possible functional consequences.
Acknowledgments
ACKNOWLEDGEMENTS
L.C. and this work are supported by grant PI020919 from the Fondo de Investigaciones Sanitarias. F.A.-S. is supported by grant BIO2001-0068 from Ministerio de Ciencia y Tecnología. This work is also partly supported by a grant from Fundació La Caixa and by the Spanish National Genotyping Centre (CeGen), funded by Genoma España, which is using this program for high-throughput SNP selection.
REFERENCES
- 1.Collins F.S., Green,E.D., Guttmacher,A.E. and Guyer,M.S. (2003) A vision for the future of genomics research. Nature, 422, 835–847. [DOI] [PubMed] [Google Scholar]
- 2.Risch N.J. (2000) Searching for genetic determinants in the new millennium. Nature, 405, 847–856. [DOI] [PubMed] [Google Scholar]
- 3.Botstein D. and Risch,N. (2003) Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nature Genet., 33, 228–237. [DOI] [PubMed] [Google Scholar]
- 4.Badano J.L. and Katsanis,N. (2002) Human genetics and disease: beyond Mendel: an evolving view of human genetic disease transmission. Nature Rev. Genet., 3, 779–789. [DOI] [PubMed] [Google Scholar]
- 5.Strittmatter W.J., Saunders,A.M., Schmechel,D., Pericak-Vance,M., Enghild,J., Salvesen,G.S. and Roses,A.D. (1993) Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer's disease. Proc. Natl Acad. Sci., USA, 90, 1977–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hugot J.P., Chamaillard,M., Zouali,H., Lesage,S., Cezard,J.P., Belaiche,J., Almer,S., Tysk,C., O'Morain,C.A., Gassull,M., Binder,V., Finkel,Y., Cortot,A., Modigliani,R., Laurent-Puig,P., Gower-Rousseau,C., Macry,J., Colombel,J.F., Sahbatou,M. and Thomas,G. (2001) Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature, 411, 599–603. [DOI] [PubMed] [Google Scholar]
- 7.Brookes A.J., Lehvaslaiho,H., Siegfried,M., Boehm,J.G., Yuan,Y.P., Sarkar,C.M., Bork,P. and Ortigao,F. (2000) HGBASE: a database of SNPs and other variations in and around human genes. Nucleic Acids Res., 28, 356–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sunyaev S., Ramensky,V., Koch,I., Lathe,W., Kondrashov,A.S. and Bork,P. (2000) Prediction of deleterious human alleles. Hum. Mol. Genet., 10, 591–597. [DOI] [PubMed] [Google Scholar]
- 9.Ng P.C. and Henikoff,S. (2001) Predicting deleterious amino acid substitutions. Genome Res., 11, 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller M.P. and Kumar,S. (2001) Understanding human disease mutations through the use of interspecific genetic variation. Hum. Mol. Genet., 10, 2319–2328. [DOI] [PubMed] [Google Scholar]
- 11.Chasman D. and Adams,R.M. (2001) Predicting functional consequences of non-synonymous single nucleotide polymorphisms: structure-based assessment of amino acid variation. J. Mol. Biol., 307, 683–706. [DOI] [PubMed] [Google Scholar]
- 12.Guerois R., Nielsen,J.E. and Serrano,L. (2002) Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J. Mol. Biol., 320, 369–387. [DOI] [PubMed] [Google Scholar]
- 13.Ferrer-Costa C., Orozco,M. and de la Cruz,X. (2002) Characterization of disease-associated single amino acid polymorphisms in terms of sequence and structure properties. J. Mol. Biol., 315, 771–786. [DOI] [PubMed] [Google Scholar]
- 14.Cartegni L., Chew,S.L. and Krainer,A.R. (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nature Rev. Genet., 3, 285–298. [DOI] [PubMed] [Google Scholar]
- 15.Krawczak M., Reiss,J. and Cooper,D.N. (1992) The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum. Genet., 90, 41–54. [DOI] [PubMed] [Google Scholar]
- 16.Montera M., Piaggio,F., Marchese,C., Gismondi,V., Stella,A., Resta,N., Varesco,L., Guanti,G. and Mareni,C. (2001) A silent mutation in exon 14 of the APC gene is associated with exon skipping in a FAP family. J. Med. Genet., 38, 863–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan H., Yuan, W., Velculescu,V.E., Vogelstein,B. and Kinzler,K.W. (2002) Allelic variation in human gene expression. Science, 297, 1143. [DOI] [PubMed] [Google Scholar]
- 18.Prokunina L., Castillejo-Lopez,C., Oberg,F., Gunnarsson,I., Berg,L., Magnusson,V., Brookes,A.J., Tentler,D., Kristjansdottir,H., Grondal,G., Bolstad,A.I., Svenungsson,E., Lundberg,I., Sturfelt,G., Jonssen,A., Truedsson,L., Lima,G., Alcocer-Varela,J., Jonsson,R., Gyllensten,U.B., Harley,J.B., Alarcon-Segovia,D., Steinsson,K. and Alarcon-Riquelme,M.E. (2002) A regulatory polymorphism in PDCD1 is associated with susceptibility to systemic lupus erythematosus in humans. Nature Genet., 32, 666–669. [DOI] [PubMed] [Google Scholar]
- 19.Harendza S., Lovett,D.H., Panzer,U., Lukacs,Z., Kuhnl,P. and Stahl,R.A. (2003) Linked common polymorphisms in the gelatinase a promoter are associated with diminished transcriptional response to estrogen and genetic fitness. J. Biol. Chem., 278, 20490–20499. [DOI] [PubMed] [Google Scholar]
- 20.Hoogendoorn B., Coleman,S.L., Guy,C.A., Smith,K., Bowen,T., Buckland,P.R. and O'Donovan,M.C. (2003) Functional analysis of human promoter polymorphisms. Hum. Mol. Genet., 12, 2249–2254. [DOI] [PubMed] [Google Scholar]
- 21.Hudson T.J. (2003) Wanted: regulatory SNPs. Nature Genet., 33, 439–440. [DOI] [PubMed] [Google Scholar]
- 22.Ashburner M., Ball,C.A., Blake,J.A., Botstein,D., Butler,H., Cherry,J.M., Davis,A.P., Dolinski,K., Dwight,S.S., Eppig,J.T., Harris,M.A., Hill,D.P., Issel-Tarver,L., Kasarskis,A., Lewis,S., Matese,J.C., Richardson,J.E., Ringwald,M., Rubin,G.M. and Sherlock, G. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clamp M., Andrews,D., Barker,D., Bevan,P., Cameron,G., Chen,Y., Clark,L., Cox,T., Cuff,J., Curwen,V., Down,T., Durbin,R., Eyras,E., Gilbert,J., Hammond,M., Hubbard,T., Kasprzyk,A., Keefe,D., Lehvaslaiho,H., Iyer,V., Melsopp,C., Mongin,E., Pettett,R., Potter,S., Rust,A., Schmidt,E., Searle,S., Slater,G., Smith,J., Spooner,W., Stabenau,A., Stalker,J., Stupka,E., Ureta-Vidal,A., Vastrik,I. and Birney,E. (2003) Ensembl 2002: accommodating comparative genomics. Nucleic Acids Res., 31, 38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu H.X., Zhang,M. and Krainer, A.R. (1998) Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev., 12, 1998–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaal T.D. and Maniatis,T. (1999) Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol. Cell Biol., 19, 261–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kel A.E., Gößling,E., Reuter,I., Cheremushkin,E., Kel-Margoulis,O.V. and Wingender,E. (2003) MATCHTM: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res., 31, 3576–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wingender E., Chen,X., Hehl,R., Karas,H., Liebich,I., Matys,V., Meinhardt,T., Prüß,M., Reuter,I. and Schacherer,F. (2000) TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res., 28, 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cartegni L., Wang,J., Zhu,Z., Zhang,M.Q. and Krainer,A.R. (2003) ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res., 31, 3568–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mulder N.J., Apweiler,R., Attwood,T.K., Bairoch,A., Barrell,D., Bateman,A., Binns,D., Biswas,M., Bradley,P., Bork,P., Bucher,P., Copley,R.R., Courcelle,E., Das,U., Durbin,R., Falquet,L., Fleischmann,W., Griffiths-Jones,S., Haft,D., Harte,N., Hulo,N., Kahn,D., Kanapin,A., Krestyaninova,M., Lopez,R., Letunic,I., Lonsdale,D., Silventoinen,V., Orchard,S.E., Pagni,M., Peyruc,D., Ponting,C.P., Selengut,J.D., Servant,F., Sigrist,C.J., Vaughan,R. and Zdobnov,E.M. (2003) The InterPro Database brings increased coverage and new features. Nucleic Acids Res., 31, 315–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Al-Shahrour Díaz-Uriarte R. and Dopazo,J. (2004) FatiGO: a web tool for finding significant associations of gene ontology terms with groups of genes. Bioinformatics, 20, 578–580. [DOI] [PubMed] [Google Scholar]
- 31.Hamosh A., Scott,A.F., Amberger,J., Bocchini,C., Valle,D. and McKusick,V.A. (2002) Online Mendelian inheritance in man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids. Res., 30, 52–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stenson P.D., Ball,E.V., Mort,M., Phillips,A.D., Shiel,J.A., Thomas,N.S., Abeysinghe,S., Krawczak,M. and Cooper,D.N. (2003) Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat., 21, 577–581. [DOI] [PubMed] [Google Scholar]
- 33.Chang H. and Fujita,T. (2001) PicSNP: a browsable catalog of nonsynonymous single nucleotide polymorphisms in the human genome. Biochem. Biophys. Res. Commun., 287, 288–291. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J., Rowe,W.L., Struewing,J.P. and Buetow,K.H. (2002) HapScope: a software system for automated and visual analysis of functionally annotated haplotypes. Nucleic Acids Res., 30, 5213–5221. [DOI] [PMC free article] [PubMed] [Google Scholar]