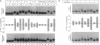Abstract
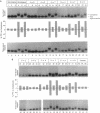
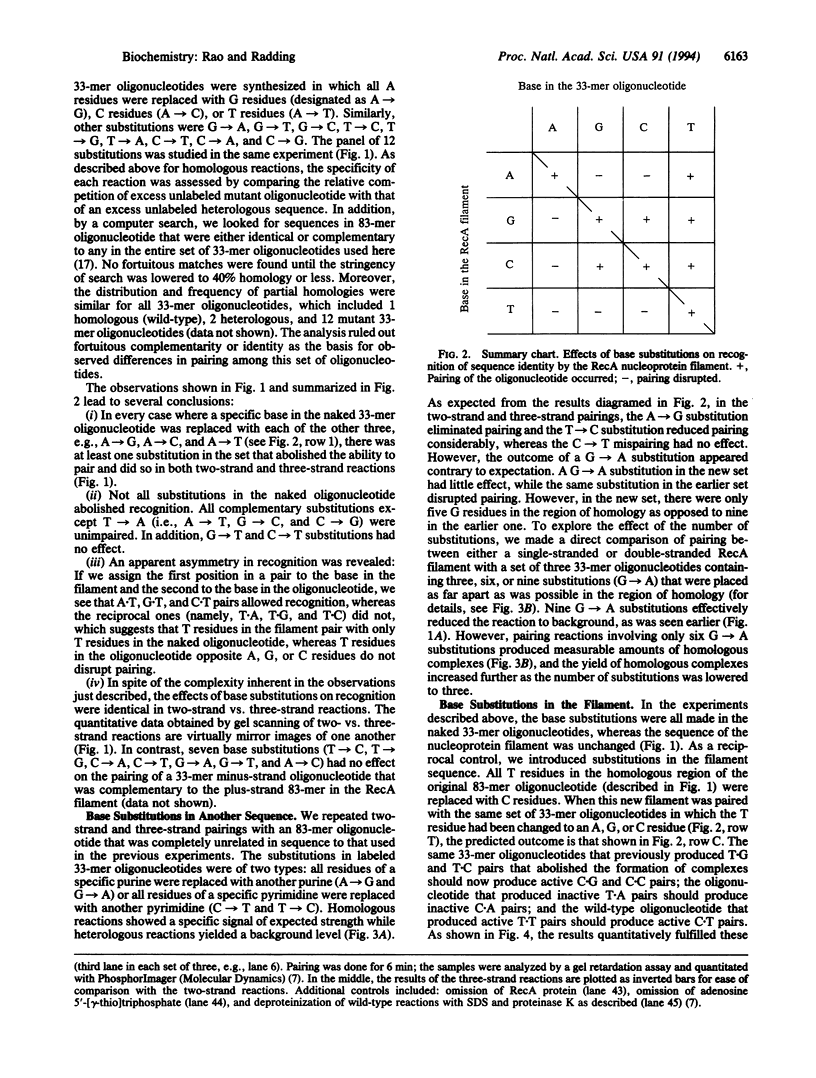
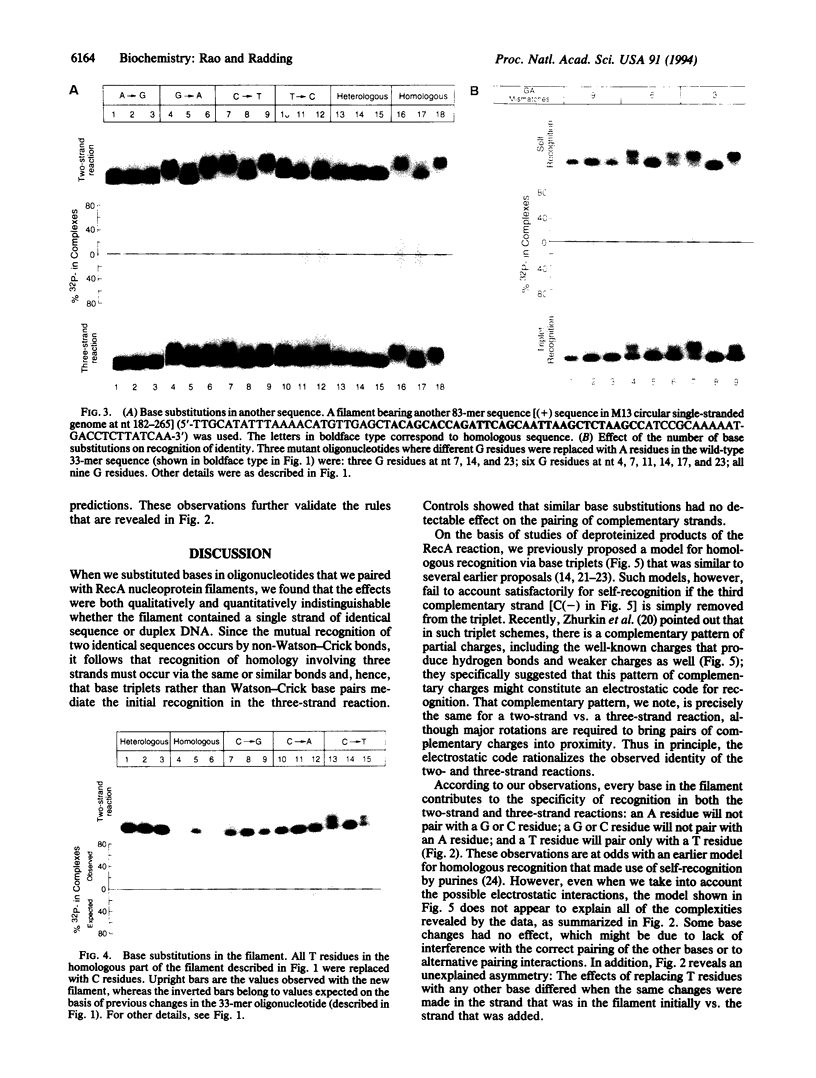
Whereas complementary strands of DNA recognize one another by forming Watson-Crick base pairs, the way in which RecA protein enables a single strand to recognize homology in duplex DNA has remained unknown. Recent experiments, however, have shown that a single plus strand in the RecA filament can recognize an identical plus strand via bonds that, by definition, are non-Watson-Crick. In experiments reported here, base substitutions had the same qualitative and quantitative effects on the pairing of two identical strands in the RecA filament as on the recognition of duplex DNA by a third strand, indicating that similar non-Watson-Crick interactions govern both reactions.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beal P. A., Dervan P. B. Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science. 1991 Mar 15;251(4999):1360–1363. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- Bishop D. K., Park D., Xu L., Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992 May 1;69(3):439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Chiu S. K., Rao B. J., Story R. M., Radding C. M. Interactions of three strands in joints made by RecA protein. Biochemistry. 1993 Dec 7;32(48):13146–13155. doi: 10.1021/bi00211a025. [DOI] [PubMed] [Google Scholar]
- Flory J., Tsang S. S., Muniyappa K. Isolation and visualization of active presynaptic filaments of recA protein and single-stranded DNA. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7026–7030. doi: 10.1073/pnas.81.22.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanvey J. C., Shimizu M., Wells R. D. Intramolecular DNA triplexes in supercoiled plasmids. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6292–6296. doi: 10.1073/pnas.85.17.6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh P., Camerini-Otero C. S., Camerini-Otero R. D. Pairing of homologous DNA sequences by proteins: evidence for three-stranded DNA. Genes Dev. 1990 Nov;4(11):1951–1963. doi: 10.1101/gad.4.11.1951. [DOI] [PubMed] [Google Scholar]
- Kurumizaka H., Ikawa S., Ikeya T., Ogawa T., Shibata T. A chimeric RecA protein exhibits altered double-stranded DNA binding. J Biol Chem. 1994 Jan 28;269(4):3068–3075. [PubMed] [Google Scholar]
- Lacks S. Integration efficiency and genetic recombination in pneumococcal transformation. Genetics. 1966 Jan;53(1):207–235. doi: 10.1093/genetics/53.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T., Yu X., Shinohara A., Egelman E. H. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993 Mar 26;259(5103):1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- Praseuth D., Perrouault L., Le Doan T., Chassignol M., Thuong N., Hélène C. Sequence-specific binding and photocrosslinking of alpha and beta oligodeoxynucleotides to the major groove of DNA via triple-helix formation. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1349–1353. doi: 10.1073/pnas.85.5.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radding C. M. Helical interactions in homologous pairing and strand exchange driven by RecA protein. J Biol Chem. 1991 Mar 25;266(9):5355–5358. [PubMed] [Google Scholar]
- Radding C. M. Homologous recombination: a universal recombination filament. Curr Biol. 1993 Jun 1;3(6):358–360. doi: 10.1016/0960-9822(93)90200-8. [DOI] [PubMed] [Google Scholar]
- Rao B. J., Chiu S. K., Radding C. M. Homologous recognition and triplex formation promoted by RecA protein between duplex oligonucleotides and single-stranded DNA. J Mol Biol. 1993 Jan 20;229(2):328–343. doi: 10.1006/jmbi.1993.1038. [DOI] [PubMed] [Google Scholar]
- Rao B. J., Jwang B., Radding C. M. RecA protein reinitiates strand exchange on isolated protein-free DNA intermediates. An ADP-resistant process. J Mol Biol. 1990 Jun 20;213(4):789–809. doi: 10.1016/S0022-2836(05)80264-5. [DOI] [PubMed] [Google Scholar]
- Rao B. J., Radding C. M. Homologous recognition promoted by RecA protein via non-Watson-Crick bonds between identical DNA strands. Proc Natl Acad Sci U S A. 1993 Jul 15;90(14):6646–6650. doi: 10.1073/pnas.90.14.6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renugopalakrishnan V., Lakshminarayanan A. V., Sasisekharan V. Stereochemistry of nucleic acids and polynucleotides. 3. Electronic charge distribution. Biopolymers. 1971;10(7):1159–1167. doi: 10.1002/bip.360100707. [DOI] [PubMed] [Google Scholar]
- Rippe K., Ramsing N. B., Jovin T. M. Spectroscopic properties and helical stabilities of 25-nt parallel-stranded linear DNA duplexes. Biochemistry. 1989 Nov 28;28(24):9536–9541. doi: 10.1021/bi00450a043. [DOI] [PubMed] [Google Scholar]
- Schwindinger W. F., Warner J. R. DNA sequence analysis on the IBM-PC. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):601–604. doi: 10.1093/nar/12.1part2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T., DasGupta C., Cunningham R. P., Williams J. G., Osber L., Radding C. M. Homologous pairing in genetic recombination. The pairing reaction catalyzed by Escherichia coli recA protein. J Biol Chem. 1981 Jul 25;256(14):7565–7572. [PubMed] [Google Scholar]
- Shinohara A., Ogawa H., Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992 May 1;69(3):457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]