Abstract
An intense debate concerning the nature and mode of Neolithic transition in Europe has long received much attention. Recent publications of paleogenetic analyses focusing on ancient European farmers from Central Europe or the Iberian Peninsula have greatly contributed to this debate, providing arguments in favor of major migrations accompanying European Neolithization and highlighting noticeable genetic differentiation between farmers associated with two archaeologically defined migration routes: the Danube valley and the Mediterranean Sea. The aim of the present study was to fill a gap with the first paleogenetic data of Neolithic settlers from a region (France) where the two great currents came into both direct and indirect contact with each other. To this end, we analyzed the Gurgy 'Les Noisats' group, an Early/Middle Neolithic necropolis in the southern part of the Paris Basin. Interestingly, the archaeological record from this region highlighted a clear cultural influence from the Danubian cultural sphere but also notes exchanges with the Mediterranean cultural area. To unravel the processes implied in these cultural exchanges, we analyzed 102 individuals and obtained the largest Neolithic mitochondrial gene pool so far (39 HVS-I mitochondrial sequences and haplogroups for 55 individuals) from a single archaeological site from the Early/Middle Neolithic period. Pairwise F ST values, haplogroup frequencies and shared informative haplotypes were calculated and compared with ancient and modern European and Near Eastern populations. These descriptive analyses provided patterns resulting from different evolutionary scenarios; however, the archaeological data available for the region suggest that the Gurgy group was formed through equivalent genetic contributions of farmer descendants from the Danubian and Mediterranean Neolithization waves. However, these results, that would constitute the most ancient genetic evidence of admixture between farmers from both Central and Mediterranean migration routes in the European Neolithization debate, are subject to confirmation through appropriate model-based approaches.
Introduction
The nature and mode of Neolithic expansion in Europe, also referred to as the "Neolithic Revolution", have been highly debated since the beginning of the twentieth century [1]. The Neolithic way of life, i.e., the diffusion of agriculture and farming and potentially the Neolithic people, has been shown to spread from the Near East innovation center according to two major routes of diffusion: the Continental route, following the Danube river, associated with the LBK (LinearBandKeramik) culture in Central Europe; and the Mediterranean route, along the Mediterranean coastlines, associated with the Impressa and Cardium cultures in southern Europe [2,3]. Although traditionally investigated from an archaeological standpoint, this field has substantially benefited from the study of genetics, which has notably contributed to the elaboration of a wide panel of Neolithic diffusion models through the indication of either little genetic contribution from Neolithic farmers to local hunter-gatherers (i.e., cultural diffusion) or various degrees of genetic admixture between expanding farmers and local resident groups of hunters-gatherers (i.e., leapfrog colonization or demic diffusion) [4–6]. Although genetic studies concerning extant European populations generally agree on the contribution of Neolithic farmers to the European gene pool, the degree of this contribution remains highly debated (from 20 to 70% depending on the genetic markers, the methods used and the populations targeted [6–8]). However, recent studies using appropriate model-based approaches, explicitly accounting for drift and admixture, concluded in the congruence between NRY (Non-recombining Region of the Y chromosome) and mtDNA (mitochondrial DNA) data, as both favor the demic diffusion model [9]. These authors also noted a clear decrease in the Neolithic contribution with geographic distance from the Near East (for both molecular markers). This observation suggests that both males and females admixed with the local Palaeolithic populations that inhabited Europe at that time, resulting in the progressive dilution of Near Eastern genes.
Interestingly, with recent advances in molecular biology, ancient DNA (aDNA) data have provided a deeper understanding of the processes involved in the Neolithic diffusion into Europe. Indeed, aDNA data obtained from Early and Middle Neolithic groups can provide direct insights into the gene pool of Neolithic pioneers. Thus, mtDNA data from early Neolithic groups associated with either the Danubian expansion route [10–16] or the Mediterranean route [17–19] have provided substantial evidence for (i) a turnover of mitochondrial genetic diversity with the spread of farmers from the Near East, and (ii) distinct gene pools associated with farmer groups correlated with either the Danubian or Mediterranean expansion waves. If a clear genetic discontinuity of maternal lineages at the advent of farming is regularly highlighted (genetic discontinuity between hunter-gatherers and early farmers mitochondrial lineages [10,14]), the evidence from Middle and Late Neolithic sites in Germany supports the increasing assimilation of female hunter-gatherers [11,20]. Mitochondrial data would therefore suggest a slow but steady increase in hunter-gatherer assimilation following a first period of cohabitation. Nevertheless, the emerging genome data from a few Mesolithic and early farming individuals confirms that early European farmers were primarily of Near Eastern origin and also harbored west European hunter-gatherer-related ancestry [21,22]. Then, more powerful genomic data rather suggest genetic exchange between indigenous hunter-gatherers and Neolithic communities since the advent of farmers' expansion.
The two major migration waves (i.e., Danubian and Mediterranean) may have come into contact in a region that currently encompasses the French territory [2,23]. On the one hand, the archaeological record indicates a clear cultural influence from late LBK in northeastern France, in particular in the Paris Basin (with the LBK-derived cultures RRBP 'Rubané Récent du Bassin Parisien' and VSG 'Villeneuve-Saint-Germain' at the end of the Early Neolithic) [24–26]. On the other hand, archaeologists have observed Mediterranean influences in the material culture found in the Paris Basin [27–30]. Although these archaeological elements demonstrate contacts occurring between Paris Basin and southern France farmer groups, they do not allow for the distinction between pure cultural exchanges versus human group gene flow. By examining the genetic relationships of populations over time, aDNA data can provide insights into the processes involved in these exchanges [14,19]. However, only three mitochondrial sequences are available for Middle Neolithic human remains from Western France [31], and no Early Neolithic aDNA data from the French territory have been published so far.
In this context, the necropolis of Gurgy 'Les Noisats' holds a key position [32]. This site is dated between 5,000 and 4,000 cal. BC, with a main occupation phase ranging from 4,900 to 4,500 cal. BC (i.e., during French Early/Middle Neolithic transition). It is localized along the Yonne River in the southern part of the Paris Basin, in the westernmost part of LBK influence (S1 Fig and S2 Fig). As opposed to the funerary monuments observed during this period in the Paris Basin [33], Gurgy is a monument-lacking necropolis grouping the burials of 128 individuals (S3 Fig). The morphology of burials could permit to draw parallels between Gurgy and western Switzerland funerary practices [34]. The homogeneity characterizing the Gurgy group (related to burial dates, funerary practices or biology; personal data) suggests that a unique population with a common cultural background used the necropolis. We genetically analyzed a total of 102 individuals to describe the maternal lineages comprising this ancient farmer group. The mitochondrial data suggested different evolutionary scenarios to explain the Gurgy gene pool make up. One scenario proposing that Gurgy necropolis reflects the genetic mixing of Neolithic farmers deriving from both Danubian and Mediterranean expansion routes finds particular resonance in archaeological data available for this region.
Material and Methods
Archaeological samples
Gurgy "Les Noisats" is a necropolis site in the southern Paris Basin, on the Yonne department, in Northeast France. Carbon-14 dates of human remains from this site range between 5,000 and 4,000 cal. BC (S1 Table), but the most intensive occupation period ranges from 4,900 to 4,500 cal. BC, i.e., the Early/Middle Neolithic transition in this geographic region (see S1 File for detailed archaeological context). The arrival of the "Neolithic package" in the Paris Basin, from 5,100 BC, has been associated with LBK-derived culture, called Rubané Récent du Bassin Parisien or RRBP [23,35–37]. Around 4,900 BC, i.e., at the end of the Early Neolithic period, a new culture called Villeneuve-Saint-Germain (VSG) developed in the region presenting funerary aspects clearly in the continuity of the Early Neolithic period [26]. Around 4,700 BC monumental funerary structures appeared in the Paris Basin along with the Cerny culture, characterized by two major types of monuments: Structures de Type Passy (STP) and the Malesherbes burials (see S1 File for details). For the period, the Gurgy necropolis corresponds to a third and more inconspicuous funerary profile observed in the region: a monument-lacking necropolis, without monument and any structuring of funerary space [33,38]. The tomb configuration discovered in the site may echo the western Switzerland and the Chamblandes cists [34]. During the same period, on the Southern part of France, the first Impressa settlements give way to Cardial culture around 5,500 BC [39], and from 4,900 BC the Chasséen culture shows an important regional diversity with a large occupation area in Southern France [40].
A total of 134 pits were excavated in the Gurgy necropolis, uncovering 128 individuals. We sampled teeth in the alveolar position, but when these samples were not available, we sampled isolated teeth or bone fragments. We acquired two samples per individual (S1 Table) for a total of 102 individuals. We followed all established aDNA guidelines to reduce contamination from the excavation site to the laboratory (S1 File). To trace the source and time of potential contamination, everyone who was in contact with the samples has been genotyped (S2 Table). Details on the authenticity of the sequences are provided in the Supporting Information. The SRA (Service Régional de l'Archéologie) of the Bourgogne region granted authorization for the Gurgy site archaeological excavation and sampling, with the corresponding site number, 89 198 008.
Ancient DNA extraction
The samples were first submitted to a treatment of bleach and UV radiation. They were then reduced to powder and we followed the procedure of Mendisco et al. [41] to extract the DNA using the 'NucleoSpin Extract II kit' (Macherey-Nagel, Düren, Germany).
SNP analyses
Multiplex assays targeting informative and complementary mitochondrial and Y chromosome SNPs, facilitating the characterization of common European maternal and paternal lineages, were designed with MassArray assay design software (version 4.0). Genotyping reactions were performed using the iPLEX Gold technology (Sequenom, Inc San Diego, CA, USA). The reliability and accuracy of this MALDI-TOF MS-based SNP genotyping technique were particularly adapted to the minute amount of degraded aDNA molecules [41]. A total of 28 mitochondrial SNPs and 10 Y chromosome SNPs were targeted and positive profiles were confirmed at least four times per individual (two genotyping on at least two DNA extracts per sample). The targeted SNPs and the associated primers are detailed in the S3 Table. The hierarchical structure of the targeted SNPs constitutes a powerful internal control for contamination or erroneous results.
HVS-I Amplification and sequencing
We amplified four overlapping fragments [42] to characterize 392 bp (nps 16,009–16,400) of the mtDNA HVS-I control region (see S1 File for details on PCR amplification conditions). At least eight independent PCR reactions were performed per individual (four overlapping fragments—two extractions). All reported mutations were established according to the revised Cambridge Reference Sequence (rCRS) [43,44] and were deduced from the “consensus” among several sequences from multiple amplification products and extracts.
Population groupings
To compare population samples with Gurgy, we distinguished sample groups based on the lower time bound of the Gurgy site occupation period (i.e., 4,000 years BC). We divided ancient hunter-gatherers (HG) and farmers (F) into six groups: European hunter-gatherers anterior to 4,000 BC ('PRE_HG'; N = 41) and European hunter-gatherers dated from after 4,000 BC ('POST_HG'; N = 30), Central European Neolithic farmers anterior to 4,000 BC ('PRE_Central_F'; N = 147) and Central European Neolithic farmers dated from after 4,000 BC ('POST_Central_F'; N = 28), Southern European Neolithic farmers anterior to 4,000 BC ('PRE_South_F'; N = 56) and Southern European Neolithic farmers dated from after 4,000 BC ('POST_South_F'; N = 49) (S4 Table). In addition, we used 20,535 modern HVS-I sequences (between nucleotide positions 16,024 and 16,380) from 78 modern populations from Europe and the Near East (S5 Table).
Statistical Analyses
We used Arlequin version 3.5.1.2 [45] to identify shared mtDNA haplotypes between Gurgy and other ancient and modern groups (S6 Table and S7 Table) and to compute population-specific pairwise genetic distances (F ST). The software R version 3.1.2 (Pumpkin Helmet) was used for Principal Component Analysis (Fig 1 and S4 Fig) and Multidimensional scaling (Fig 2 and S5 Fig). The shared informative haplotype frequencies were used to create a map (S6 Fig) using the software Surfer 12 (Golden Software, Inc.). A median-joining network connecting the mitochondrial sequences anterior to 4,000 BC was constructed for nps 16,056–16,380 using NETWORK 4.6.1.3. (S7 Fig).
Fig 1. Principal Component Analysis (PCA) on the ancient mtDNA dataset.
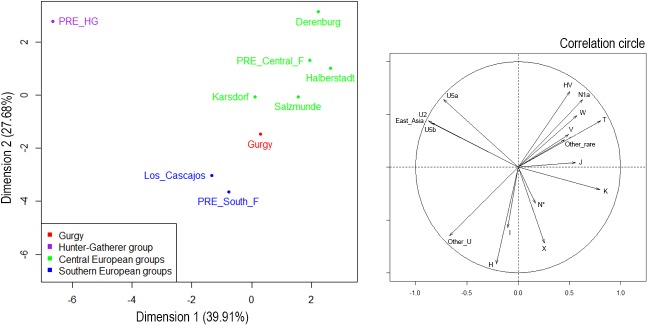
PCA performed with haplogroup frequencies and circle of correlation of the PCA. The ancient samples (S4 Table) included 'PRE_HG' for European Hunter Gatherers anterior to 4,000 BC (N = 41), 'PRE_Central_F' for Central European Neolithic farmers anterior to 4,000 BC (N = 147), and 'PRE_South_F' for Southern European Neolithic farmers anterior to 4,000 BC (N = 56). Some ancient groups anterior to 4,000 BC and sufficiently large enough for comparison at the population level are also shown (Derenburg, Halberstadt, Karsdorf, Salzmünde, from Germany [11,12,14,15]; and Los Cascajos from Spain [19]).
Fig 2. Multidimensional Scaling (MDS) on the ancient mtDNA dataset.
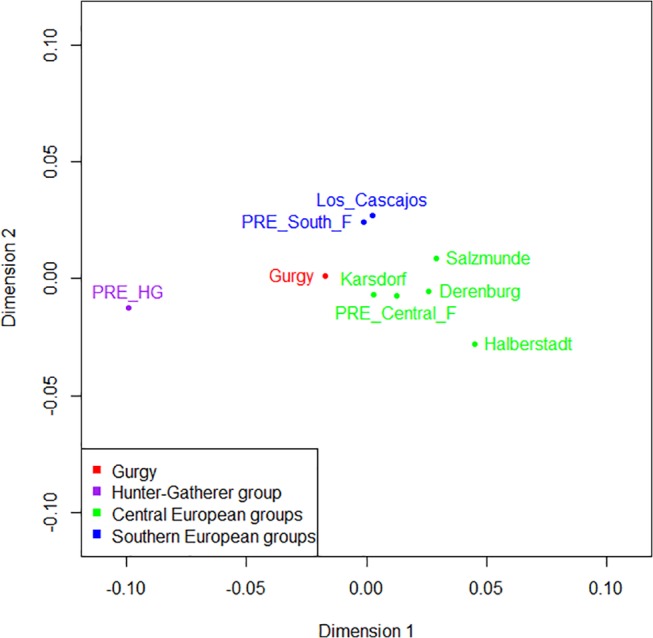
MDS performed on the F ST values between Gurgy and the ancient dataset (same as Fig 1).
Results and Discussion
Gurgy genetic diversity
From the 102 sampled Gurgy human remains, 55 mitochondrial haplogroups were generated (S8 Table), and 39 of those individuals provided HVS-I sequences (nps 16,009–16,400) that met all requested aDNA authenticity criteria (S1 Table, S1 File and S3 Fig) [46]. The sequences were deposited in the GenBank database (http://www.ncbi.nlm.nih.gov/genbank/; accession numbers KP863031-KP863069). To our knowledge, this is the largest mtDNA Neolithic sample recovered from one archaeological site for the Early/Middle Neolithic period. This sample features 27 distinct mitochondrial haplotypes belonging to eight haplogroups (H, K, U, J, N1a, V, X and T). Unfortunately, no sample yielded Y chromosome DNA, suggesting that nuclear DNA might be less well conserved in these samples. Some individuals might be closely maternally related, as five distinct mtDNA haplotypes were shared by more than two individuals (S1 Table). Only three samples sharing a haplotype (K characterized by a 16,224C mutation) originated from spatially close or even superimposed burials (GLN244, GLN245A and GLN245B). Interestingly, the archaeological data obtained for these burials showed the absence of any disturbance between different successively established burials, strongly suggesting the preserved memory of the localization of these areas.
In the last 10 years, an increasing amount of paleogenetic data has been published, providing substantial insights into the genetic relationships between ancient hunter-gatherers, ancient farmers and modern Europeans [10–19,47–62]. We used this published data (detailed in S4 Table) to examine the genetic relationships between samples from Gurgy and other temporally and spatially related aDNA samples.
The Gurgy necropolis genetic diversity (Hd = 0.9447 +/- 0.0284) is similar to the genetic diversity values obtained for Early Neolithic groups from Central Europe (e.g., Derenburg-Meerenstieg II Hd = 0.9567 +/- 0.0238 [14,15] and Halberstadt-Sonntagsfeld Hd = 0.9613 +/- 0.0178 [11,12]) but is slightly higher than that of the Los Cascajos group in Southern Europe (Hd = 0.8234 +/- 0.0670 [19]) (S9 Table). The Gurgy group shows nucleotide diversity (π = 0.010056 +/- 0.005795) intermediate between the values observed for LBK groups and farmers from southwestern Europe (S9 Table). We also observed that the Gurgy genetic diversity is close to the value calculated for the hunter-gatherer group PRE_HG (Hd = 0.9305 +/- 0.0250). This result is striking, as we observe analogous genetic diversity between a hunter-gatherer sample geographically (from Spain to Russia; S4 Table) and chronologically (from 38,000 BP to 4,450 BC; S4 Table) largely dispersed and a local group of Neolithic farmers who buried their dead in the Gurgy necropolis for approximately 500 years. This observation is consistent with population-genomic evidence suggesting lower diversity for hunter-gatherers than for farmers [21,22,63].
Relationship of Gurgy farmers with ancient European groups
To understand of the genetic relationships between the Gurgy group and other temporally and spatially related aDNA samples of hunter-gatherers and farmers, we divided published aDNA data into three groups anterior or contemporaneous to the Gurgy group (i.e., anterior to 4,000 BC, the most recent date obtained for Gurgy sample remains): European hunter-gatherers anterior to 4,000 BC ('PRE_HG'; N = 41), Central European Neolithic farmers anterior to 4,000 BC ('PRE_Central_F'; N = 147) and Southern European Neolithic farmers anterior to 4,000 BC ('PRE_South_F'; N = 56) (Fig 3 and S4 Table). Four common basal mtDNA haplotypes were shared between Gurgy, 'PRE_HG', 'PRE_Central_F' and 'PRE_South_F' groups (S6 Table). Three other haplotypes were shared between 'PRE_Central_F' and 'PRE_South_F' populations, and 20 haplotypes were specific to Gurgy farmers. The number of unique haplotypes encountered in the Gurgy group (similar to the Central and South European farmer groups) must be associated with the still weak aDNA dataset available for ancient European populations. A median-joining network connecting all ancient HVS-I sequences (from hunter-gatherers and farmers anterior to 4,000 BC) is proposed in S7 Fig. The resulting network is not completely satisfactory, as it only considers short HVS-I sequences, as responsible for creating artificial links between different haplogroups. Nevertheless, this network suggests that most of the Gurgy haplotypes are either shared with other ancient European farmers or likely derived from the sequences obtained from Central and South Europe farmers (S7 Fig).
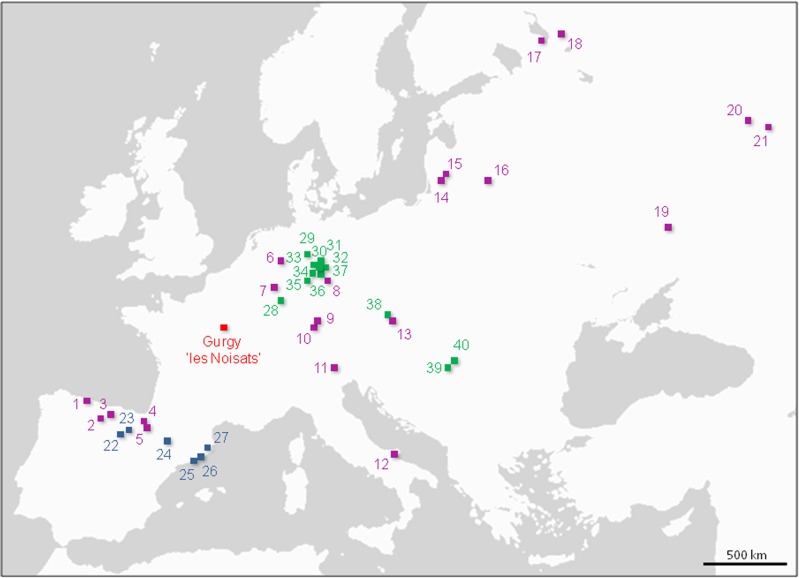
Fig 3. Location of the sites anterior to 4000 cal BC.
PRE_HG (purple): 1) La Braña, 2) La Pasiega, 3) La Chora, 4) Erralla, 5) Aizpea, 6) Oberkassel, 7) Reuland-Loschbour, 8) Bad Durrenberg, 9) Hohleinstein, 10) Hohler Fels, 11) Villabruna, 12) Paglicci, 13) Dolni Vestonice, 14) Spiginas, 15) Donkalnis, 16) Kretuonas, 17) Uznyi Oleni Ostrov, 18) Popovo, 19) Kostenki, 20) Chekalino, 21) Lebyazhinka. PRE_South_F (blue): 22) Los Cascajos, 23) Paternanbidea, 24) Chaves, 25) Can Sadurni, 26) Sant Pau del Camp, 27) Avellaner. PRE_Central_F (green): 28) Flomborn, 29) Wittmar, 30) Oberwiederstedt, 31) Unterwiederstedt, 32) Salzmünde, 33) Derenburg, 34) Halberstadt, 35) Naumburg, 36) Karsdorf, 37) Esperstedt, 38) Vedrovice, 39) Szarvas, 40) Ecsegfalva. References in S4 Table.
Previous work has demonstrated the mtDNA distinctiveness of ancient hunter-gatherers (i.e., 'PRE_HG') compared with Early/Middle Neolithic farmers [10,11,14,19]. This point is clearly illustrated via principal component analysis (PCA) and Multidimensional Scaling (MDS) performed on the ancient groups (Figs 1 and 2). Indeed, the PCA revealed that most of the variation in mtDNA haplogroup frequencies lied between ancient hunter-gatherers (PRE_HG) and farmer groups, including Gurgy (first component, Fig 1). Notably, 'PRE_HG' is characterized by specific haplogroups, such as U2, U4, U5 and several H, which are absent or rare in Gurgy or other farmer groups. Calculation of the pairwise F ST values between ancient populations confirmed an important genetic distance between ancient European hunter-gatherers and Gurgy farmers (0.08016; Fig 2 and Fig 4 and S10 Table). These observations are consistent with the results from Bramanti et al. [10] and Haak et al. [14], who proposed the genetic discontinuity of maternal lineages between local hunter-gatherers and early farmers (a genetic distinction that decreased during later periods through progressive hunter-gatherers assimilation [11]). Notably, the F ST value measured between Gurgy and PRE_HG groups was slightly lower than that measured between other ancient farmers groups (from Central and southern Europe) and PRE_HG. These results are consistent with the study of Rasteiro and Chikhi [9], showing a clear decrease in the Neolithic contribution / higher hunter-gatherer admixture with geographic distance from the Near East. The involvement of hunter-gatherers in the constitution of the Gurgy gene pool requires more powerful model-based approaches for further clarification. However, the important genetic distance measured between Gurgy and hunter-gatherer groups clearly contrasts with the genetic similarities observed between Gurgy and Early Neolithic farmers from Central and Southern Europe.
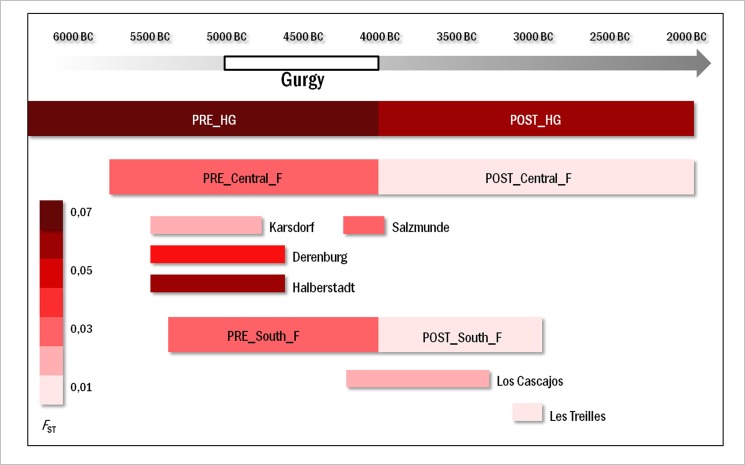
Fig 4. Pairwise F ST distances.
F ST measured between Gurgy and ancient European hunter-gatherers / farmer groups (S4 Table and S10 Table). 'PRE_HG' for European hunter-gatherers anterior to 4,000 BC (N = 41), 'POST_HG' for European hunter-gatherers after 4,000 BC (N = 30), 'PRE_Central_F' for Central European Neolithic farmers anterior to 4,000 BC (N = 147), 'POST_Central_F' for Central European Neolithic farmers after 4,000 BC (N = 28), 'PRE_South_F' for Southern European Neolithic farmers anterior to 4,000 BC (N = 56), and 'POST_South_F' for Southern European Neolithic farmers after 4,000 BC (N = 49). Ancient groups anterior to 4,000 BC, which were sufficiently large for comparison at the population level are also shown (Derenburg, Halberstadt, Karsdorf, Salzmünde, from Germany [11,12,14,15]; Los Cascajos from Spain [19] and Les Treilles form southern France [54]).
The F ST values calculated between Gurgy and (i) the 'PRE_Central_F' and (ii) the 'PRE_South_F' groups are almost perfectly equal (0.03333 and 0.03176, respectively) and much lower than the F ST between Gurgy and the 'PRE_HG' group (Fig 4 and S10 Table). We also computed F ST between Gurgy and ancient groups sufficiently large enough for comparison at the population level (specific archaeological sites providing more than 20 HVS-I sequences) (Fig 4). Interestingly, low F ST values suggest similar genetic proximity between Gurgy and some LBK populations in Germany (Karsdorf, Salzmünde [11,12]) or Los Cascajos in Spain [19]). The MDS constructed using the F ST value reported above places the Gurgy group in an intermediate position between southern and Central farmer groups (Fig 2).
The PCA presented above highlights another substantial degree of variation distinguishing 'PRE_Central_F' and 'PRE_South_F', following the two major migration waves associated with Neolithic diffusion in Europe (Fig 1). In the PCA, the population of Gurgy that shares haplogroups with both Southern and Central farmer groups also revealed an expected intermediate position. Notably, Gurgy showed three individuals carrying mutations characteristic of the N1a haplogroup, which is considered a genetic marker of the Danubian expansion route because it is common among Early Neolithic farmers from Central Europe (14 N1a sequences are currently associated with LBK groups [11,14,15]) and has not been detected in farmers groups associated with the Mediterranean migration route so far. Although the three N1a sequences from the Gurgy group differ from those reported for the LBK groups, these findings might support the implication of LBK descendants in Gurgy group formation.
In summary, all descriptive analyses highlight strong genetic differentiation between Gurgy and European hunter-gatherers and even stronger and equal genetic affinities between Gurgy and ancient farmers from both Central and South Europe. These observations might reflect distinct evolutionary scenarios such as (i) a tree-like population history with three ancestral farmer populations leading to the three farmer groups analyzed (Gurgy/Danubian farmers/Mediterranean farmers) or (ii) admixture, i.e., the Gurgy gene pool was likely shaped by a similar contribution of farmers originating from the Mediterranean region and Central Europe associated with the Danubian and Mediterranean Neolithization routes. Although the descriptive analyses did not favor any evolutionary scenario (patterns might be affected by sample composition and interpretations in terms of admixture events were not straightforward), the available archaeological data for this region support the admixture model.
Concordant archaeological and paleogenetic clues of gene flow between Early/Middle Neolithic Paris Basin and southern France farmers
Gurgy is localized in the southern area of the Paris Basin, which lies at the westernmost part of the LBK cultural influence (S1 Fig and S2 Fig) [2,24]. The Gurgy necropolis shows a potential RRBP cultural tradition (clearly LBK derived) in terms of burial type, position or orientation (S1 File) [64]. However, some archaeologists have observed parallels with western Switzerland [34] and demonstrated cultural exchanges between Paris Basin populations and groups from southern France associated with Cardium and Chasséen cultures [27,28,30,65]. Various artifacts or cultural influences from southern France, corresponding to pottery decorations [27], bone industry [29,66] or personal ornaments (Spondylus shells or white limestone rings [67,68]), have been observed in the RRBP or Villeneuve-Saint-Germain (VSG) burials of the Paris Basin. Two hypotheses have been proposed to explain this southern cultural influence in farmer groups from the Paris Basin, including either an appropriation of techniques and style through genetic contact and admixture between populations or a simple exchange of artifacts and raw materials without gene flow [29]. We propose that the archaeological data and the paleogenetic results obtained on the Gurgy necropolis are consistent with the hypothesis that the Mediterranean cultural influence on the Paris Basin entails some gene flow, implying the northerly migration of Mediterranean farmers in the Paris Basin and admixture with farmer descendants of LBK populations from Central Europe. As proposed by Sidéra [29,69], exogamy between groups might explain the fusion of the material culture of both Central and southern Neolithic spheres in the Paris Basin and the mixture of southern/Central Europe farmer lineages observed in the Gurgy necropolis.
Gurgy and the evolution of farmer groups during the Middle/Late Neolithic period
To unravel the genetic affinities of the Gurgy group with later European groups, the Gurgy maternal gene pool was compared with that of hunter-gatherer or farmer populations dated between 4,000 BC and 2,000 BC: European hunter-gatherers after 4,000 BC ('POST_HG'; N = 30), Central European Neolithic farmers after 4,000 BC ('POST_Central_F'; N = 28), and southern European Neolithic farmers after 4,000 BC ('POST_South_F'; N = 49) (S4 Table and Fig 3). Gurgy mtDNA diversity showed clear genetic affinities (in term of lowest F ST) with 'POST_Central_F' (0.00965), 'POST_South_F' (0,01774), or the French group of Les Treilles (0.01006; southern France, 3,000 BC [54]). Paleogenetic data for the Middle or Late Neolithic periods indicated that migratory/demographic events have largely remodeled the gene pool of farmer groups throughout the Neolithic period. This evidence was recently supported by an analysis of a time transect spanning the >3,500 years of the Central European Neolithic period highlighting substantial gene flow throughout the European continent during the Late Neolithic period [11]. Consequently, farmer groups from the Late Neolithic period were formed from a mosaic of maternal lineages from diverse European origins, and this mixed constitution might explain the genetic similarity of these farmer groups to Gurgy.
The relationship of Gurgy farmers with extant European populations
The Gurgy mitochondrial gene pool wascompared with 20,535 HVS-I sequences compiled from 78 European and Near Eastern modern populations to assess the similarity between the Gurgy and modern European gene pools (S4 Table and S5 Fig). PCA revealed genetic differentiation between modern populations from the Near East and those from Europe (S4 Fig) and high genetic homogeneity of modern European populations. Interestingly, the Gurgy group was isolated from the both blocks, suggesting that global European gene pool remodeling since the Neolithic period contributed to the observed discontinuity between early Neolithic groups (including Gurgy) and modern European mitochondrial gene pools [11,12,14,15,70].
We next assessed haplotypes shared between the Gurgy and modern European populations. Excluding the four common and non-informative haplotypes observed in Gurgy and a large number of European populations (mainly basal haplotypes) and excluding the eleven haplotypes unique to Gurgy, twelve remaining informative haplotypes were shared between Gurgy and specific European and Near East populations (S7 Table). We considered that these informative haplotypes might potentially highlight extant populations with affinities to the Gurgy group, presumably representing a genetic legacy from the Neolithic period [14]. The map specifically reporting these shared informative haplotype frequencies shows interesting hotspots in Turkey and the Balkan Peninsula and along Danube Valley and the Mediterranean shores (S6 Fig). Thus, the geographic distribution of these informative haplotypes in modern European populations might reflect some aspects of alleged Gurgy group history, i.e., the scenario proposing that the group comprised an admixture between the descendants of the farmers from Danubian and Mediterranean expansion routes, who are the descendants of farmers from the Balkans and Anatolia.
Conclusion
We conducted a paleogenetic study on the Gurgy 'Les Noisats' necropolis, presenting the first genetic data on Neolithic settlers (Early/Middle Neolithic transition) in France. Localized in the southern part of the Paris Basin, the necropolis provides a unique opportunity to document the processes implied in the Neolithization of a region where cultural exchanges between culturally distinct farmer groups have been highlighted. Descriptive analyses performed using Gurgy mtDNA diversity highlight a gene pool clearly intermediate between those characterized for the farmers associated with both Danubian and Mediterranean migration routes. Even if these findings were consistent with different evolutionary scenarios, we propose that the scenario in which the Gurgy gene pool resulted from equivalent contributions of maternal lineages from farmer groups associated with the Danubian and Mediterranean expansion routes is the most parsimonious. These arguments notably corroborate archaeological evidence of cultural exchanges between farmers from the Paris Basin and those from southern France, indicating that the observed cultural exchanges reflect genetic admixture between groups. However, we are fully aware that these data do not definitively prove admixture between farmers associated with both Neolithization routes and that the proposed admixture model must be tested under a robust analytical framework, such as the coalescent theory that would account for genetic drift and population demography within a gene genealogy [71].
Supporting Information
(TIF)
(TIF)
(TIF)
Map featuring the frequency distribution of informative haplotypes shared between Gurgy and modern populations (S5 Table).
(TIF)
Performed using the HVS-I sequences (nps 16,056–16,380) available for hunter-gatherers and farmers anterior to 4,000 BC (S4 Table).
(EPS)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We would like to warmly thank P. Gerbault for text editing and instructive discussions. Special thanks to M. Le Roy who has provided the Gurgy necropolis map. We also acknowledge everyone who contributed to the excavation of the Gurgy site.
Data Availability
The sequences were deposited in the GenBank database (http://www.ncbi.nlm.nih.gov/genbank/; accession numbers KP863031-KP863069).
Funding Statement
This study benefitted from excavation grant support from the region of Bourgogne, France, and the Service Régional de l'Archéologie de Bourgogne (89 198 008). This research is funded by a ministerial grant from the Research National Agency as a program of prospects investments ANR-10-LABX-52 (project 'Diversité biologique et culturelle de l'Homme de la fin de la Préhistoire à la Protohistoire'; dir: SR; Université de Bordeaux 1, LaScArBx-ANR; 2012–14) and has also been performed thanks to the PhD research grant from the Ministère de l'Enseignement Supérieur et de la Recherche for MR. Part of the experiments (SNPs analyses) presented were performed at the Genomic and Sequencing Facility of Bordeaux (grants from the Conseil Regional d’Aquitaine n°20030304002FA and 20040305003FA and from the European Union, FEDER n°2003227 and from Investissements d'avenir, Convention attributive d’aide N°ANR-10-EQPX-16-01). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Childe VG. The Dawn of European Civilization. London: Kegan Paul; 1925. [Google Scholar]
- 2. Whittle AW. Europe in the Neolithic: the creation of new worlds: Cambridge University Press; 1996. 443 p. [Google Scholar]
- 3. Price T. Europe's first farmers: Cambridge University Press; 2000. [Google Scholar]
- 4.Zvelebil M. The agricultural transition and the origins of Neolithic society in Europe. Documenta Praehistorica. 2001; XXVIII.
- 5. Ammerman AJ, Cavalli-Sforza LL. The Neolithic transition and the genetics of populations in Europe: Princeton University Press; Princeton; 1984. 176 p. [Google Scholar]
- 6. Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V, et al. Tracing European founder lineages in the Near Eastern mtDNA pool. American Journal of Human Genetics. 2000; 67: 1251–1276. [PMC free article] [PubMed] [Google Scholar]
- 7. Sykes B. The molecular genetics of European ancestry. Philosophical Transactions of the Royal Society of London Series B: Biological Sciences. 1999; 354: 131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chikhi L, Destro-Bisol G, Bertorelle G, Pascali V, Barbujani G. Clines of nuclear DNA markers suggest a largely Neolithic ancestry of the European gene pool. Proceedings of the National Academy of Sciences. 1998; 95: 9053–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rasteiro R, Chikhi L. Female and male perspectives on the neolithic transition in Europe: clues from ancient and modern genetic data. PLoS One. 2013; 8: e60944 10.1371/journal.pone.0060944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bramanti B, Thomas M, Haak W, Unterlaender M, Jores P, Tambets K, et al. Genetic discontinuity between local hunter-gatherers and central Europe’s first farmers. Science. 2009; 326: 137–140. 10.1126/science.1176869 [DOI] [PubMed] [Google Scholar]
- 11. Brandt G, Haak W, Adler CJ, Roth C, Szécsényi-Nagy A, Karimnia S, et al. Ancient DNA Reveals Key Stages in the Formation of Central European Mitochondrial Genetic Diversity. Science. 2013; 342: 257–261. 10.1126/science.1241844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brotherton P, Haak W, Templeton J, Brandt G, Soubrier J, Adler CJ, et al. Neolithic mitochondrial haplogroup H genomes and the genetic origins of Europeans. Nature communications. 2013; 4: 1764 10.1038/ncomms2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Burger J, Kirchner M, Bramanti B, Haak W, Thomas MG. Absence of the lactase-persistence-associated allele in early Neolithic Europeans. Proceedings of the National Academy of Sciences. 2007; 104: 3736–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haak W, Balanovsky O, Sanchez JJ, Koshel S, Zaporozhchenko V, Adler CJ, et al. Ancient DNA from European early neolithic farmers reveals their near eastern affinities. PLoS biology. 2010; 8: e1000536 10.1371/journal.pbio.1000536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haak W, Forster P, Bramanti B, Matsumura S, Brandt G, Tänzer M, et al. Ancient DNA from the First European Farmers in 7500-Year-Old Neolithic Sites. Science. 2005; 310: 1016–1018. [DOI] [PubMed] [Google Scholar]
- 16. Lee EJ, Krause-Kyora B, Rinne C, Schütt R, Harder M, Müller J, et al. Ancient DNA insights from the Middle Neolithic in Germany. Archaeological and Anthropological Sciences. 2013; 6: 199–204. [Google Scholar]
- 17. Lacan M, Keyser C, Ricaut F-X, Brucato N, Tarrús J, Bosch A, et al. Ancient DNA suggests the leading role played by men in the Neolithic dissemination. Proceedings of the National Academy of Sciences. 2011; 108: 18255–18259. 10.1073/pnas.1113061108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gamba C, Fernández E, Tirado M, Deguilloux MF, Pemonge MH, Utrilla P, et al. Ancient DNA from an Early Neolithic Iberian population supports a pioneer colonization by first farmers. Molecular Ecology. 2011; 21: 45–56. 10.1111/j.1365-294X.2011.05361.x [DOI] [PubMed] [Google Scholar]
- 19. Hervella M, Izagirre N, Alonso S, Fregel R, Alonso A, Cabrera VM, et al. Ancient DNA from hunter-gatherer and farmer groups from Northern Spain supports a random dispersion model for the Neolithic expansion into Europe. PLoS One. 2012; 7: e34417 10.1371/journal.pone.0034417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bollongino R, Nehlich O, Richards MP, Orschiedt J, Thomas MG, Sell C, et al. 2000 years of parallel societies in Stone Age Central Europe. Science. 2013; 342: 479–481. 10.1126/science.1245049 [DOI] [PubMed] [Google Scholar]
- 21.Lazaridis I, Patterson N, Mittnik A, Renaud G, Mallick S. Ancient human genomes suggest three ancestral populations for present-day Europeans. bioRxiv. 2014. [DOI] [PMC free article] [PubMed]
- 22. Gamba C, Jones ER, Teasdale MD, McLaughlin RL, Gonzalez-Fortes G, Mattiangeli V, et al. Genome flux and stasis in a five millennium transect of European prehistory. Nature communications. 2014; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vander Linden M. To tame a land: archaeological cultures and the spread of the Neolithic in western Europe Investigating Archaeological Cultures: Springer; 2011. pp. 289–319. [Google Scholar]
- 24. Jeunesse C. Les groupes régionaux occidentaux du Rubané (Rhin et Bassin parisien) à travers les pratiques funéraires. Gallia préhistoire. 1995; 37: 115–154. [Google Scholar]
- 25. Demoule J-P. De l'Europe centrale au Bassin parisien (5200–4400) In: Demoule J-P, editor. La révolution néolithique en France. Paris: La découverte; 2007. pp. 42–59. [Google Scholar]
- 26. Constantin C, Ilett M. Culture de Blicquy-Villeneuve-Saint-Germain, rapports chronologiques avec les cultures rhénanes. Anthropologie et préhistoire. 1998; 109: 207–216. [Google Scholar]
- 27. Lichardus-Itten M. Premières influences méditerranéennes dans le Néolithique du Bassin parisien, contribution au débat In: Demoule J-P, Guilaine J, editors. Le Néolithique de la France Hommage à G Bailloud. Paris: Picard; 1986. pp. 147–160. [Google Scholar]
- 28.Hauzeur A. Affinités et influences dans le Néolithique ancien d'Europe occidentale: le Rubané de la moyenne vallée de la Moselle et la culture de Blicquy-Villeneuve-Saint-Germain. In: Otte M, editor. Préhistoire de la Grande Plaine du Nord de l'Europe. Liège; 2002. pp. 167–182.
- 29. Sidéra I. De mains méridionales en mains septentrionales. Le long transit des objets et des savoir-faire en Europe occidentale, vers 5100 av. J.-C. Mélanges de la Casa de Velázquez Nouvelle série. 2010; 40: 17–32. [Google Scholar]
- 30.Guilaine J, Manen C. Contacts sud-nord au Néolithique ancien: témoignages de la grotte de Gazel en Languedoc; 1997. pp. 301–311.
- 31. Deguilloux M-F, Soler L, Pemonge M-H, Scarre C, Joussaume R, Laporte L. News from the west: Ancient DNA from a French megalithic burial chamber. American Journal of Physical Anthropology. 2010; 144: 108–118. 10.1002/ajpa.21376 [DOI] [PubMed] [Google Scholar]
- 32. Rottier S, Mordant C, Chambon P, Thevenet C. Découverte de plus d'une centaine de sépultures du Néolithique moyen à Gurgy, les Noisats (Yonne). Bulletin de la Société préhistorique française. 2005; 102: 641–645. [Google Scholar]
- 33.Thomas A. Identités funéraires, variants biologiques et facteurs chronologiques: une nouvelle perception de contexte culturel et social du Cerny (Bassin parisien, 4700–4300 avant J.-C.): PhD thesis, Bordeaux 1. 2011. 788 p.
- 34.Chambon P. Des Chamblandes au centre de la France? In: Moinat P, Chambon P, editors. Les cistes de Chamblandes et la place des coffres dans les pratiques funéraires du Néolithique moyen occidental. Actes du colloques de Lausanne, 12 et 13 mai 2006: Cahiers d'archéologie romande 110 et Société préhistorique française XLIII. 2007. pp. 75–89.
- 35. Dubouloz J. Datation absolue du premier Néolithique du Bassin parisien: complément et relecture des données RRBP et VSG. Bulletin de la Société préhistorique française. 2003; 100: 671–689. [Google Scholar]
- 36.Constantin C, Ilett M. Une étape finale dans le Rubané récent du Bassin parisien. In: Jeunesse C, editor. Le Néolithique danubien et ses marge entre Rhin et Seine. Actes du 22e Colloque interrégional sur le Néolithique (27–29 octobre 1995, Strasbourg); 1997. pp. 207–300.
- 37.Demoule J-P. La révolution néolithique en France: Découverte; 2007. 180 p.
- 38. Chambon P, Rottier S, Augereau A, Bonnardin S, Meunier K, Pariat J-G. Evolution, coexistence et confrontation de pratiques funéraires entre 4700 et 4000 av. J.-C. sur un microterritoire dans la vallée de l'Yonne In: Jaubert J, Fourment N, Depaepe P, editors. Transition, ruptures et continuité durant la Préhistoire. Paris: Société Préhistorique Française; 2013. pp. 213–228. [Google Scholar]
- 39. Sénépart I. Premiers bergers et paysans des côtes méditerranéennes (5800–4500) In: Demoule J-P, editor. La révolution néolithique en France. Paris: La découverte; 2007. pp. 26–41. [Google Scholar]
- 40. Demoule J-P, Dubouloz J, Manolakakis L. L'émergence des premières sociétés complexes (4500–3500) In: Demoule J-P, editor. La révolution néolithique en France. Paris: La découverte; 2007. pp. 60–77. [Google Scholar]
- 41. Mendisco F, Keyser C, Hollard C, Seldes V, Nielsen AE, Crubézy E, et al. Application of the iPLEX Gold SNP genotyping method for the analysis of Amerindian ancient DNA samples: Benefits for ancient population studies. Electrophoresis. 2011; 32: 386–393. 10.1002/elps.201000483 [DOI] [PubMed] [Google Scholar]
- 42. Gabriel MN, Huffine EF, Ryan JH, Holland MM, Parsons TJ. Improved MtDNA sequence analysis of forensic remains using a" mini-primer set" amplification strategy. Journal of Forensic Sciences. 2001: 247–253. [PubMed] [Google Scholar]
- 43.Anderson S, Bankier AT, Barrell BG, De Bruijn M, Coulson AR, Drouin J, et al. Sequence and organization of the human mitochondrial genome. 1981. [DOI] [PubMed]
- 44. Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nature genetics. 1999; 23: 147–147. [DOI] [PubMed] [Google Scholar]
- 45. Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005; 1: 47. [PMC free article] [PubMed] [Google Scholar]
- 46. Cooper A, Poinar HN. Ancient DNA: Do It Right or Not at All. Science. 2000; 289: 1139 [DOI] [PubMed] [Google Scholar]
- 47. Krause J, Briggs AW, Kircher M, Maricic T, Zwyns N, Derevianko A, et al. A Complete mtDNA Genome of an Early Modern Human from Kostenki, Russia. Current Biology. 2010; 20: 231–236. 10.1016/j.cub.2009.11.068 [DOI] [PubMed] [Google Scholar]
- 48. Fu Q, Mittnik A, Johnson Philip LF, Bos K, Lari M, Bollongino R, et al. A Revised Timescale for Human Evolution Based on Ancient Mitochondrial Genomes. Current Biology. 2013; 23: 553–559. 10.1016/j.cub.2013.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Caramelli D, Lalueza-Fox C, Vernesi C, Lari M, Casoli A, Mallegni F, et al. Evidence for a genetic discontinuity between Neandertals and 24,000-year-old anatomically modern Europeans. Proceedings of the National Academy of Sciences. 2003; 100: 6593–6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Caramelli D, Milani L, Vai S, Modi A, Pecchioli E, Girardi M, et al. A 28,000 years old Cro-Magnon mtDNA sequence differs from all potentially contaminating modern sequences. PLoS One. 2008; 3: e2700 10.1371/journal.pone.0002700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Di Benedetto G, Nasidze IS, Stenico M, Nigro L, Krings M, Lanzinger M, et al. Mitochondrial DNA sequences in prehistoric human remains from the Alps. European Journal of Human Genetics. 2000; 8: 669–677. [DOI] [PubMed] [Google Scholar]
- 52. Delsate D, Guinet JM, Saverwyns S. De l’ocre sur le crâne mésolithique (haplogroupe U5a) de Reuland-Loschbour (Grand-Duché de Luxembourg). Bulletin de la Société Préhistorique Luxembourgeoise. 2009; 31: 7–30. [Google Scholar]
- 53. Sánchez-Quinto F, Schroeder H, Ramirez O, Ávila-Arcos María C, Pybus M, Olalde I, et al. Genomic Affinities of Two 7,000-Year-Old Iberian Hunter-Gatherers. Current Biology. 2012; 22: 1494–1499. 10.1016/j.cub.2012.06.005 [DOI] [PubMed] [Google Scholar]
- 54. Lacan M, Keyser C, Ricaut F-X, Brucato N, Duranthon F, Guilaine J, et al. Ancient DNA reveals male diffusion through the Neolithic Mediterranean route. Proceedings of the National Academy of Sciences. 2011; 108: 9788–9791. 10.1073/pnas.1100723108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Malmström H, Gilbert MTP, Thomas MG, Brandström M, Storå J, Molnar P, et al. Ancient DNA Reveals Lack of Continuity between Neolithic Hunter-Gatherers and Contemporary Scandinavians. Current Biology. 2009; 19: 1758–1762. 10.1016/j.cub.2009.09.017 [DOI] [PubMed] [Google Scholar]
- 56. Lee EJ, Makarewicz C, Renneberg R, Harder M, Krause‐Kyora B, Müller S, et al. Emerging genetic patterns of the European Neolithic: perspectives from a late Neolithic Bell Beaker burial site in Germany. American Journal of Physical Anthropology. 2012; 148: 571–579. 10.1002/ajpa.22074 [DOI] [PubMed] [Google Scholar]
- 57. Haak W, Brandt G, de Jong HN, Meyer C, Ganslmeier R, Heyd V, et al. Ancient DNA, Strontium isotopes, and osteological analyses shed light on social and kinship organization of the Later Stone Age. Proceedings of the National Academy of Sciences. 2008; 105: 18226–18231. 10.1073/pnas.0807592105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Melchior L, Lynnerup N, Siegismund HR, Kivisild T, Dissing J. Genetic diversity among ancient Nordic populations. PLoS One. 2010; 5: e11898 10.1371/journal.pone.0011898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sampietro M, Lao O, Caramelli D, Lari M, Pou R, Marti M, et al. Palaeogenetic evidence supports a dual model of Neolithic spreading into Europe. Proceedings of the Royal Society B: Biological Sciences. 2007; 274: 2161–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ermini L, Olivieri C, Rizzi E, Corti G, Bonnal R, Soares P, et al. Complete mitochondrial genome sequence of the Tyrolean Iceman. Current Biology. 2008; 18: 1687–1693. 10.1016/j.cub.2008.09.028 [DOI] [PubMed] [Google Scholar]
- 61. Díaz N, Solórzano E, Montiel R, García C, Yañez C, Malgosa A. Determination genétique de l’individu Néolithique de Segudet (Ordino), les restes humains les plus anciens d’Andorre. Anthropo. 2004; 7: 39–44. [Google Scholar]
- 62. Der Sarkissian C, Balanovsky O, Brandt G, Khartanovich V, Buzhilova A, Koshel S, et al. Ancient DNA reveals prehistoric gene-flow from Siberia in the complex human population history of North East Europe. PLoS genetics. 2013; 9: e1003296 10.1371/journal.pgen.1003296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Skoglund P, Malmström H, Omrak A, Raghavan M, Valdiosera C, Günther T, et al. Genomic Diversity and Admixture Differs for Stone-Age Scandinavian Foragers and Farmers. Science. 2014; 344: 747–750. 10.1126/science.1253448 [DOI] [PubMed] [Google Scholar]
- 64.Rottier S. L'architecture funéraire des sépultures du Néolithique moyen des Noisats à Gurgy (Yonne, France). Les cistes de Chamblandes et la place des coffres dans les pratiques funéraires du Néolithique moyen occidental. 2007: 99–107.
- 65. Burnez-Lanotte L, Ilett M, Allard P. Fin des traditions danubiennes dans le Néolithique du Bassin parisien et de la Belgique (5100–4700 BC), Autour des recherches de Claude Constantin: Mémoires de la Société Préhistorique Française 44; 2008. [Google Scholar]
- 66.Sidéra I. Rubané, Villeneuve-Saint-Germain et Cardial: filiations des industries osseuses. In: Burnez Lanotte L, Ilett M, Allard P, editors. Fin des traditions danubiennes dans le néolithique du Bassin parisien et de la Belgique (5100–4700 av J-C) Autour des recherches de C Constantin: Mémoire de la Société Préhistorique Française 44; 2008. pp. 209–219.
- 67. Bonnardin S. La parure funéraire au Néolithique ancien dans les bassins parisien et rhénan: Rubané, Hinkelstein et Villeneuve-Saint-Germain: Société préhitorique française; 2009. 322 p. [Google Scholar]
- 68. Constantin C, Vachard D. Anneaux d'origine méridionale dans le Rubané récent du Bassin parisien. Bulletin de la Société préhistorique française. 2004; 101: 75–83. [Google Scholar]
- 69. Sidéra I. Nouveau regard sur la néolithisation européenne De l'Anatolie au Bassin parisien via Méditerranée: traditions des assemblages osseux. Paris: de Broccard; 2010. [Google Scholar]
- 70. Gamba C, Fernández E, Tirado M, Deguilloux MF, Pemonge MH, Utrilla P, et al. Ancient DNA from an Early Neolithic Iberian population supports a pioneer colonization by first farmers. Molecular Ecology. 2012; 21: 45–56. 10.1111/j.1365-294X.2011.05361.x [DOI] [PubMed] [Google Scholar]
- 71. Wakeley J. Coalescent theory: an introduction: Roberts & Company Publishers; Greenwood Village, Colorado; 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
Map featuring the frequency distribution of informative haplotypes shared between Gurgy and modern populations (S5 Table).
(TIF)
Performed using the HVS-I sequences (nps 16,056–16,380) available for hunter-gatherers and farmers anterior to 4,000 BC (S4 Table).
(EPS)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
The sequences were deposited in the GenBank database (http://www.ncbi.nlm.nih.gov/genbank/; accession numbers KP863031-KP863069).