Abstract
The number of cotton (Gossypium sp.) ovule epidermal cells differentiating into fiber initials is an important factor affecting cotton yield and fiber quality. Despite extensive efforts in determining the molecular mechanisms regulating fiber initial differentiation, only a few genes responsible for fiber initial differentiation have been discovered. To identify putative genes directly involved in the fiber initiation process, we used a cotton ovule culture technique that controls the timing of fiber initial differentiation by exogenous phytohormone application in combination with comparative expression analyses between wild type and three fiberless mutants. The addition of exogenous auxin and gibberellins to pre-anthesis wild type ovules that did not have visible fiber initials increased the expression of genes affecting auxin, ethylene, ABA and jasmonic acid signaling pathways within 1 h after treatment. Most transcripts expressed differentially by the phytohormone treatment in vitro were also differentially expressed in the ovules of wild type and fiberless mutants that were grown in planta. In addition to MYB25-like, a gene that was previously shown to be associated with the differentiation of fiber initials, several other differentially expressed genes, including auxin/indole-3-acetic acid (AUX/IAA) involved in auxin signaling, ACC oxidase involved in ethylene biosynthesis, and abscisic acid (ABA) 8'-hydroxylase an enzyme that controls the rate of ABA catabolism, were co-regulated in the pre-anthesis ovules of both wild type and fiberless mutants. These results support the hypothesis that phytohormonal signaling networks regulate the temporal expression of genes responsible for differentiation of cotton fiber initials in vitro and in planta.
Introduction
Cotton (Gossypium sp.) is the most commercially important natural fiber in the world [1]. Cotton fibers are single-celled trichomes that differentiate from epidermis of cotton ovules [2]. On the day of anthesis (0 days post-anthesis, DPA) 15–30% of the epidermal cells on cotton (Gossypium hirsutum L.) ovules differentiate into fiber initials [3,4,5]. Fiber initials are observed first at the chalazal end of the ovules on 0 DPA [3,6]. Although fiber initials at the chalazal end immediately enter into the elongation phase, new fiber initials bulge from the epidermis in a progressive wave toward the micropylar end until 4 DPA [3,7]. Cotton fiber initiation is considered to be quasi-synchronized in each developing ovule and among ovules within each ovary, and cotton ovules between 0 and 4 DPA contain a mixture of fiber initials and elongating fibers [8].
The density of fiber initials is generally considered an indicator of cotton yield [9] and a potential target for improving yield by biotechnological manipulations [5], although one claim to the contrary has been made [10]. More recent work demonstrated that a higher density of fiber initials presumably caused by higher auxin levels in the ovule epidermis at the fiber initiation stage resulted in higher fiber yield and finer fibers [11]. Thus, the ratio of fiber initials to non-fiber cells on the epidermis of cotton ovules may affect yield and cotton fiber quality.
Due to the potential for improving cotton yield and quality by increasing the number and/or density of fiber initials, several investigations have attempted to identify cotton genes that are mainly responsible for the differentiation of fiber initials from the epidermis of cotton ovules. The first transcriptome profiles compared wild type (WT) ovules at a late fiber initiation stage (3 to 5 DPA) with isogenic ovules from fuzzless and lintless (fl) or a naked (N1) mutant [12,13]. Since large portions of the differentially expressed genes (DEGs) identified in these studies overlapped with those known to be involved in fiber elongation, the second transcriptome profiles used transcripts extracted from fiber initials at an early fiber initiation stage (0 to 1 DPA) using advanced extraction techniques such as laser-capture microdissection [14] or by vortexing a mixture of frozen cotton ovules and glass beads [15]. Expression analyses of fiber initials compared with other tissues from WTs or mutants identified candidate genes involved in the fiber initiation process [14,15,16].
Among the DEGs identified by genome-wide transcriptome expression analyses were GhMYB1-6, GhMyb109, dehydration-induced RD22-like protein (GhRDL), acetyltransferase (GhACY), fiddlehead-like protein (GhFDH), serine carboxylpeptidase protein (GhSCP), tubulins (GhTUA6 and GhTUB1), cellulose synthase catalytic subunit (GhCesA5), actin (GhACT), sterol-C-methyltransferase (GhSMT), sucrose synthase (GhSuS), homeobox 1, calmodulin, and ER lumen protein-retaining receptor, all of which are involved in both fiber initiation and elongation [12,13,15,17,18,19,20,21]. The other DEGs including GhMYB25, GhMYB25-like, homeodomain-leucine zipper transcription factor (GhHD-1), GaMYB2, dehydration-induced RD22-like protein 1 (GARDL1), protodermal factor (GhPDF1), a putative cell wall protein (AY464064), fatty acid elongase (AY464065), lipid transfer protein (AY464062), and a small glycine rich protein were proposed to be involved in fiber initiation [13,14,16,22].
Subsequent functional analyses of individual candidate genes, however, has shown that many genes identified by transcriptome profiles appear to be involved in fiber elongation rather than in the fiber initiation process. When GbPDF1, GaHOX1, GhMYB109, GaRDL1, or GaMYb2 was individually suppressed in transgenic cotton plants, fiber length was reduced [17,22,23,24,25]. The short fiber length implies that these candidate genes are not involved in the differentiation of fiber initials from ovular epidermal cells but may be involved in early fiber development. None of these candidate genes were highly expressed at 0 DPA when the fiber initials were first detected in the ovules. Transcript abundance increased from 0 to 3–5 DPA and then decreased. RNAi-mediated suppression of GhMYB25 slightly reduces the number of fiber initials but markedly reduces fiber length [26]. Ectopic overexpression of GhMYB25 increases the number of fiber initials and leaf trichome number. The phenotypes of transgenic cotton plants with suppressed levels of MYB25, MYB109, or PDF1 indicate that these candidate genes may play roles in fiber elongation although they were originally indentified as genes involved in fiber initiation.
Among numerous candidate genes identified by extensive transcriptome profiles, two transcription factors, GhMYB25-like [27] and GhHD-1 [28], have been verified to be involved in differentiation of fiber initials from the cotton ovule epidermis. Suppression of GhMYB25-like expression resulted in no differentiation of fiber initials on cotton ovules, yet overexpression did not increase the number of fiber initials, suggesting that other factors interacting with GhMYB25-like may be required for fiber initial differentiation. Suppression of GhHD-1, a homeodomain-leucine zipper transcription factor that is structurally similar to the Arabidopsis GLABRA2 transcription factor (GL2) involved in leaf trichome differentiation, delayed the timing of fiber initiation. Overexpression of GhHD-1 increased the number of fiber initials on the seed without affecting leaf trichomes [28]. In addition, functional analyses with transgenic cottons have shown that two enzymes, sucrose synthase (GhSus) [18,29] and a vacuolar invertase (GhVIN 1) [20,21], are involved in both fiber initiation and elongation.
Despite the recent progress in identifying two transcription factors, GhMYB25-like and GhHD-1, responsible for the differentiation of fiber initials from epidermis, the molecular mechanisms regulating the cotton fiber initiation process are mostly unknown. In the model plant Arabidopsis, the processes regulating the differentiation of leaf trichomes and root hairs from epidermal cells have been well characterized, and the number and the types of regulatory interactions between multiple transcription factors are complex [30,31]. Some common transcription factors such as GL2, GL3/EGL3, and TTG1 regulate the differentiation of both trichomes and root hair cells from epidermal cells; however, these processes are regulated by different phytohormones [30]. The number and density of trichome cells in Arabidopsis can be increased by treatments with gibberellin acid (GA), cytokinins, and jasmonic acid, and decreased by salicylic acid treatment. In contrast, root hair formation in Arabidopsis is promoted by auxin and ethylene.
Cotton fiber differentiation from ovule epidermal cells is likely regulated by multiple transcription factors that are regulated by phytohormones as the case in Arabidopsis. Indeed, unknown cotton proteins were reported to interact with GhMYB25-like transcription factor [27]. Cotton expressed sequence tag (EST) analyses also showed that MYB and WRKY transcription factors predicted to be involved in auxin, GA, ethylene, brassinosteroid (BR), and abscisic acid (ABA) signaling pathways were more enriched in cotton ovules at the fiber initiation stage (-3, 0, and +3 DPA) than in other non-fiber tissues [32]. Auxin and GA are essential for differentiating fiber initials in vitro [33]. Although there is a general agreement that multiple networks of phytohormonal regulators and transcription factors are required for differentiating fiber initials, only very limited numbers of the candidate genes have been verified [12,13,14,15,16,34].
Based on the temporal and spatial expression patterns of GhMYB25-like and GhHD-1, their indispensable roles in the differentiation of fiber initials and their up-regulation in pre-anthesis ovules (-2 to -1 DPA) containing no fiber initials [27,28], we hypothesize that pre-anthesis cotton ovules are enriched with the transcripts required for fiber differentiation.
In this paper, we used comparative transcript profiling to identify genes whose expression changed after the exogenous application of auxin and GA to pre-anthesis cotton ovules grown in vitro. Candidate DEGs identified from in cultured cotton ovules were verified as important for fiber initiation by measuring the transcript abundance of candidate genes in the ovules of three fiberless (fl) mutants compared with a WT grown in planta. Our results support the hypothesis that phytohormonal signaling networks and crosstalk regulate the temporal expression of genes responsible for the differentiation of fiber initials in vitro and in planta.
Materials and Methods
Plant material
G. hirsutum WT lines (TM-1 and Xu-142) and three fiberless mutant lines (Xu-142 fl, MD17, and SL-1-7-1) were grown in a field at the USDA-ARS, Southern Regional Research Center in New Orleans, LA. Standard growing practices were applied during the growing season. The soil type in New Orleans was aquent dredged-over alluvium in an elevated location to provide adequate drainage. Two biological replications (6 bolls per replicate) of unfertilized ovules from cotton squares (TM-1, -1 DPA) were collected for cotton ovule culture. For DEG validation, three biological replications (4 bolls per replicate) of cotton ovules (-3, -1, 0, 1, and 3 DPA) from a WT (Xu-142) and the three fiberless mutant lines were harvested. All ovules were collected in the morning between 0800 and 0900. The collected ovules were either frozen with liquid nitrogen for RNA extraction and qPCR analyses or fixed for microscopic analyses.
Cotton ovule culture
Cotton ovaries harvested from the cotton squares (-1 DPA) were surface-sterilized and dissected under sterile conditions. The unfertilized ovules were transferred to a liquid basal medium in the presence or absence of 5μM indole-3-acetic acid (IAA) and 1μM GA and incubated at 32°C in a 5% CO2 atmosphere [33,35]. Cotton ovules incubated for 1, 3, 6, 12, and 24 h were harvested for RNA extraction, microarray and image analyses.
Scanning electron microscopy
Cotton ovules were fixed with 3% (v/v) glutaraldehyde in 0.1 M sodium phosphate, pH 7.0, dehydrated in a graded ethanol series starting from 20% (v/v) up to 100% (v/v) ethanol and mounted on standard Cambridge SEM stubs. SEM images were taken with a XL30 Environmental Scanning Electron Microscope (FEI Company, Hillsboro, OR) at an accelerating voltage from 10–15 kV under high vacuum conditions.
RNA extraction from cotton fibers
Total RNA was extracted from the frozen ovules using a Sigma Spectrum Plant Total RNA Kit (Sigma-Aldrich, St. Louis, MO) with DNase1 digestion according to the manufacturer’s protocol. The quality and quantity of total RNAs were determined using a NanoDrop 2000 spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE) and an Agilent Bioanalyzer 2100 (Agilent Technologies Inc., Santa Clara, CA).
Microarray hybridizations and data analysis
Cotton oligonucleotide arrays version 2 [36] representing 22,787 cotton probes designed from Gossypium expressed sequence tag (EST) sequences were used to compare the expression of genes in cultured ovules (-1 DPA) treated with or without phytohormones (5μM IAA and 1μM GA). The phytohormone incubation periods for -1 DPA unfertilized ovules were 0, 1, 3, 6, and 12 h with two biological replicates at each time-point. Procedures for RNA amplification, labeling with Cy5 or Cy3, hybridization, scanning, data normalization and assessment of statistically and biologically significant genes were performed as described previously [37]. Statistical analyses were performed using JMP Genomics version 3.2 (SAS Institute Inc., Cary, NC, USA) to identify significant DEGs (fold change > 2) in fibers of each cotton NIL at each time point with p-value ≤ 0.05. Primary annotation was performed using Blast2GO [58], and blastx in The Arabidopsis Information Resource (http:/www.arabidopsis.org/Blast/) and Gossypium raimondii genome sequence (http:/www.phytozome.net/cotton.php). Gene Ontology (GO) enrichment and visualization of microarray results were performed according to the described method [38]. For the singular enrichment analysis, the identified DEGs were compared with the Gossypium raimondii genome [39]. For the sequence analyses, Arabidopsis orthologues of the identified DEGs from the cotton ovules were compared with Arabidopsis genome sequences (http:/www.arabidopsis.org/index.jsp). For statistical analyses and GO enrichment, the p-value cutoff for significance was 0.05. The GEO accession number was assigned as GSE66251.
Reverse transcription quantitative PCR (RT-qPCR)
The experimental procedures and data analysis related to RT-qPCR were performed according to the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines [61]. The cDNA synthesis reactions were performed using the iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer’s instructions with 1 μg of total RNA per reaction used as template. Control cDNA synthesis reactions to check for genomic DNA contamination during RT-qPCR consisted of the same template and components as the experimental reactions without the reverse transcriptase enzyme. Thirty-seven pairs of specific primers were designed from twenty-nine DEGs for validation of the microarray results. The RT-qPCR reactions used iTaq SYBR Green Supermix (Bio-Rad Laboratories) in a Bio-Rad CFX96 real time PCR detection system. Thermal cycler parameters for RT-qPCR were as follows: 95°C 3 minutes, 50 cycles of 95°C 15 seconds, 60°C 30 seconds. A dissociation curve was generated and used to validate that a single amplicon was present for each RT-qPCR reaction. The calculations for amplification efficiencies of the target and reference genes and the relative quantifications of the different target gene transcript abundance were performed using the comparative Cq method as described [61] with the following modification: the average of three reference gene Cq values was determined by taking the geometric mean which was used to calculate the ΔCq values for the individual target genes. The endogenous reference genes used in the RT-qPCR reactions were the 18S rRNA (U42827), ubiquitin-conjugating protein (AI730710), and α-tubulin 4 (AF106570). The reference and target gene primer sequences are shown in S1 Table. Three biological replications and five technical replications were used for each time-point sample.
Results
Differentiation of fiber initials on pre-anthesis cotton ovules grown in planta
To determine when and how fiber initials are differentiated on cotton ovules (G. hirsutum, TM-1) grown in our field conditions, we obtained scanning electron microscopic (SEM) images from unfertilized Upland cotton TM-1 ovules harvested from field grown plants at two different time points (-1 and 0 DPA). There were no visible fiber initials on unfertilized ovules harvested at -1DPA (0900) (Fig 1A and 1D), whereas visible fiber initials were evident at very top of the chalazal end by 1700 on the same day (Fig 1B and 1E). Unfertilized ovules harvested at 0800 on 0 DPA had fiber initials covering most of the ovular surface except the micropylar ends (Fig 1C and 1F).
Fig 1. Fiber initiation of G. hirsutum grown in planta.
Scanning microscopic images of G. hirsutum TM-1 unfertilized ovules (top panel) and epidermal tissue (bottom panel) were photographed from field-grown ovules that were harvested at 0900 on -1 DPA (A & D) and 1700 on -1 DPA (B & E), and 0800 on 0 DPA (C & F). Scale bars equal 50μm.
Differentiation of fiber initials on pre-anthesis ovules grown in vitro with exogenous phytohormones
Since no fiber initials had differentiated before 0900 on -1 DPA, we tested whether the exogenous application of auxin and GA could promote fiber initiation in vitro. When the -1 DPA ovules were cultured with 5μM IAA and 1μM GA for 8 days, the 7 DPA ovules were covered with developing fibers (Fig 2A). In contrast, no visible fibers were detected on the 7 DPA ovules cultured in the absence of phytohormones, although ovule size substantially increased as compared with those at -1 DPA.
Fig 2. Fiber initiation of G. hirsutum grown in vitro.
A. Phytohormonal effects on fiber development in cultured ovules. One day before anthesis (-1 DPA) ovules (G. hirsutum, TM-1) containing no fiber initials were treated with exogenous phytohormones (+P) consisting of 5μM IAA and 1μM GA until 7 DPA. No phytohormone (-P) was used as a negative control. B. Differentiation of fiber initials cultured with exogenous phytohormones. Scanning microscopic images of G. hirsutum TM-1 unfertilized ovules (top panel) and epidermal tissue (bottom panel) were photographed before (a & d) and after culture for 24 h on basal medium either in the absence (b & e) or presence (c & f) of phytohormones (5μM IAA and 1μM GA). The bars represent lengths of 50μm.
Fig 2B showed that the phytohormone treatment differentiated fiber initials from the -1 DPA ovules within 24 hours. The unfertilized -1 DPA ovules used for the culture contained no fiber initials on the surface (Fig 2B, a & d). Cotton ovules cultured for 24h without the phytohormones maintained the same ovule surface containing no fiber initials (Fig 2B, b & e), whereas the ovules cultured for 24h with the phytohormones differentiated fiber initials on the surface of the ovule (Fig 2B, c & f).
Transcriptome profiles of pre-anthesis ovules grown in vitro with exogenous phytohormones
Transcriptome profiles were obtained from ovules harvested before 0900 on -1 DPA and treated with phytohormones (5μM IAA and 1μM GA) for 1, 3, 6 and 12 hours in ovule cultures. Ovule cultures with no phytohormone treatment served as the control. Among the 22,787 probes contained on the cotton oligonucleotide array chip, 1,043 unique transcripts were differentially expressed with more than a 2-fold difference (S1 File). As the incubation period with the phytohormones increased from 1, 3, 6, to 12 h, the numbers of DEGs increased from 181, to 507, to 631, to 764, respectively (Fig 3A). Comparisons of the DEGs at the four different time points showed three distinctive patterns of temporal regulation (Fig 3B). Of the DEGs, 54, 250, and 396 DEGs specifically identified at 1, 3h and both 6 and 12h, respectively appear to be involved in immediate, intermediate, and late responses to the exogenous phytohormones.
Fig 3. Summary of microarray analysis of cotton ovules cultured with phytohormones.
A. Comparison of up- or down-regulated DEGs in cultured cotton ovules (-1 DPA) that were treated for 1, 3, 6, and 12 hours with IAA + GA. B. Venn diagrams representing the common and specific DEGs among the four time points in cultured cotton ovules (-1 DPA). C. GO enrichment analysis. Singular enrichment analysis was used to identify GO categories potentially governing differential expression of genes involved in fiber initiation caused by exogenous phytohormone treatment. The color and numbers adjacent to the GO identifier represent p-values.
Gene ontology analyses of differentially expressed genes in cultured ovules
To identify the potential processes governing the 1,043 DEGs that may be involved in the differentiation of fiber initials by phytohormones, singular enrichment analysis was used from agriGO [38]. Significantly enriched (p-value ≤ 0.05) genes included those encoding proteins with transcription regulator activity, protein-binding activity, and catalytic activities (Fig 3C). The catalytic activities found among the DEGs included oxidoreductases, UDP-glucosyltransferases, kinases, hydrolases, and phosphatases. Transcription factors such as IAA29, IAA11, IAA7, IAA16, IAA4, and ARF16 that are involved in auxin signaling pathways in Arabidopsis were mainly up-regulated within 1h (Table 1). Among them, IAA29 was the most up-regulated (1,673 fold at 3 h) by the exogenous phytohormone treatment (Table 1). A WRKY23 is a transcription factor that responds to auxin and nematodes in Arabidopsis. In addition, MYB and TCP transcription factors [40,41] known to be involved in fiber initiation and elongation in cotton were also up-regulated within a 1 h treatment (Table 1). The GhHD-1 transcription factor (Gorai.013G167800) proposed to be involved in fiber initiation was up-regulated after 3 or 6 h of phytohormone treatment (S1 File).
Table 1. Transcription factors regulated in pre-anthesis cotton ovules (-1 DPA) by auxin and GA.
| No | Transcriptional factor names | G. raimondii ID | Cotton EST ID | TAIR ID | Array transcript levels | ||||
|---|---|---|---|---|---|---|---|---|---|
| 0h | 1h | 3h | 6h | 12h | |||||
| 1 | indole-3-acetic acid inducible 29 (IAA 29) | Gorai.001G043900 | CGI8_TC65241; | At4g32280 | 1.0 | 100.3 | 1673.0 | 147.9 | 36.1 |
| Cotton12_11151_01 | |||||||||
| 2 | bHLH DNA-binding protein | Gorai.001G103300 | Cotton12_18505_01 | At3g07340 | 1.0 | 9.8 | - | - | 1.3 |
| 3 | GATA transcription factor 26 (GATA26) | Gorai.006G214400 | Cotton12_17078_01 | At4g17570 | 1.0 | 7.5 | 5.1 | 3.1 | 0.8 |
| 4 | High mobility group B2 | Gorai.002G115100 | AI054798 | At1g20693 | 1.0 | 6.5 | 2.1 | 1.3 | 1.5 |
| 5 | MYB | Gorai.013G196800 | CGI8_TC75953 | At1g08810 | 1.0 | 5.3 | 8.2 | 3.0 | 3.6 |
| 6 | C2H2 type zinc finger protein | Gorai.013G155400 | Cotton12_05478_01 | At4g27240 | 1.0 | 5.0 | 53.3 | 1.3 | 1.1 |
| 7 | indole-3-acetic acid inducible 11 (IAA 11) | Gorai.007G150000 | Cotton12_21593_01; | At4g28640 | 1.0 | 3.8 | 2.7 | 2.4 | 2.7 |
| Cotton12_15680_01 | |||||||||
| 8 | auxin-responsive protein iaa7 (IAA 7) | Gorai.009G132200 | Cotton12_AJ458442; | At3g04730 | 1.0 | 3.1 | 3.7 | 2.8 | 2.4 |
| Cotton12_16710_01 | |||||||||
| 9 | B-box type zinc finger protein | Gorai.004G113000 | Cotton12_15879_01 | At1g25440 | 1.0 | 2.7 | 1.2 | 12.0 | 2.2 |
| 10 | Indole acetic acid-induced protein 16 (IAA 16) | Gorai.007G276900 | CGI8_TC72612; | At3g04730 | 1.0 | 2.4 | 3.1 | 3.2 | 1.0 |
| Cotton12_00037_06 | |||||||||
| 11 | TCP transcription factor | Gorai.012G166500 | Cotton12_03262_01 | At1g58100 | 1.0 | 2.2 | 3.1 | 2.2 | 2.2 |
| 12 | NF-YB8, nuclear factor Y, subunit B8 | Gorai.011G241600 | Cotton12_06501_01 | At2g37060 | 1.0 | 2.2 | 11.5 | 3.4 | 1.2 |
| 13 | AUX2-11 (IAA4) | Gorai.007G277000 | Cotton12_08080_01 | At5g43700 | 1.0 | 2.0 | 2.1 | 2.5 | 1.3 |
| 14 | WRKY23 DNA-binding protein | Gorai.006G265200 | Cotton12_10616_01 | At2g47260 | 1.0 | 1.7 | 2.3 | 0.6 | 1.8 |
| 15 | LHW transcription factor | Gorai.009G082800 | Cotton12_21049_01 | At2g27230 | 1.0 | 1.1 | 0.2 | 0.2 | 0.0 |
| 16 | C2H2-like zinc finger | Gorai.007G002600 | CGI8_TC79812 | At2g01940 | 1.0 | 1.0 | 6.8 | 0.8 | 0.1 |
In contrast, few genes associated with GA signaling pathways were identified among the DEGs. The genes for gibberellin 20-oxidase and gibberellin 3 β-hydroxylase, two GA biosynthetic enzymes, were down-regulated when the ovules were incubated with auxin and GA for 12 h (Table 2). Interestingly, the exogenous application of auxin and GA affected the expression levels of genes involved in other phytohormone signaling pathways such as ethylene, abscisic acid (ABA), and jasmonic acid (JA). 1-Aminocyclopropane-1-carboxylate (ACC) oxidase, a key enzyme for ethylene biosynthesis [42], was up-regulated at 1h, and then transcript abundance gradually decreased. ABA 8'-hydroxylase, involved in ABA degradation [43], was up-regulated at 1h and continued to be highly expressed up to 12 h (Table 2).
Table 2. Phytohormonal pathways regulated in pre-anthesis cotton ovules (-1 DPA) by auxin and GA.
| No | Phytohormone signaling pathway | G. raimondii ID | Cotton EST ID | TAIR ID | Array transcript levels | ||||
|---|---|---|---|---|---|---|---|---|---|
| 0h | 1h | 3h | 6h | 12h | |||||
| GA signaling pathway | |||||||||
| 1 | gibberellin 20-oxidase | Gorai.006G005500 | CGI8_TC64916 | At5g51810 | 0.99 | 1.17 | 1.06 | 1.40 | 0.33 |
| 2 | gibberellin 3 β-hydroxylase | Gorai.010G227700 | CGI8_TC73128 | At1g15550 | 1.00 | 1.03 | 1.01 | 0.43 | 0.35 |
| Ethylene signaling pathway | |||||||||
| 3 | ACC oxidase | Gorai.001G011100 | Cotton12_11109_01 | At2g19590 | 1.00 | 5.41 | 2.06 | 0.17 | 0.71 |
| Cotton12_34165_01 | At2g19590 | 1.00 | 3.62 | 0.71 | 0.07 | 0.27 | |||
| 4 | ACC oxidase | Gorai.009G182300 | Cotton12_04011_01 | At1g05010 | 1.00 | 1.42 | 0.97 | 0.46 | 0.15 |
| ABA signaling pathway | |||||||||
| 5 | ABA 8'-hydroxylase | Gorai.004G177200 | CGI8_TC80236 | At4g19230 | 1.00 | 2.11 | 2.26 | - | 1.83 |
| Jasmonic acid/ H 2 O 2 common pathway | |||||||||
| 6 | proline dehydrogenase | Gorai.007G186600 | Cotton12_CD485620 | At5g38710 | 1.00 | 1.05 | 3.36 | 1.95 | 0.96 |
| 7 | WRKY33 | Gorai.012G119600 | Cotton12_AJ458442 | At2g38470 | 1.00 | 3.14 | 3.70 | 2.78 | 2.36 |
| 8 | jasmonate resistant 1 (JAR 1) | Gorai.001G156600 | CGI8_TC78256 | At2g46370 | 1.00 | 1.34 | 0.30 | 0.53 | 0.10 |
Validation of differentially expressed genes in cultured ovules
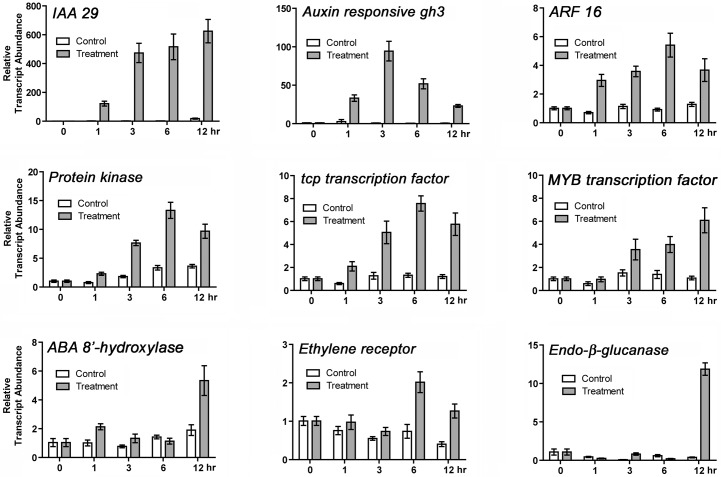
The DEGs identified by the arrays were validated with reverse transcription quantitative PCR analysis (RT-qPCR). Consistent with the array results (Table 1 and S1 File), the IAA 29 transcription factor belonging to an auxin/indole-3-acetic acid-responsive (AUX/IAA) gene family was induced by the exogenous phytohormone treatment within 1 hr and continued to be highly expressed for 12 h (Fig 4). The expression patterns of other auxin signaling pathway-related transcripts such as auxin responsive gh3, ARF 16 and protein kinase were similar to that of IAA 29. The expression of tcp transcription factor and MYB transcription factor involved in fiber development [35, 36] was also up-regulated by the phytohormones. ABA 8´-hydroxylase, a key enzyme for ABA catabolism, was slightly induced after a 1h treatment with auxin and GA and up-regulated more significantly at 12 h. An ethylene receptor was up-regulated after 6 h. Endo-β-glucanase whose transcripts were highly expressed in actively elongating fibers [44] was induced after 12 h of phytohormone treatment.
Fig 4. Validation of array data by RT-qPCR analysis.
Genes differentially expressed due to exogenous auxin and GA treatment of unfertilized cotton ovules (-1 DPA) for 0, 1, 3, 6, and 12 h included: IAA 29 (Gorai.001G043900)), auxin responsive gh3 (Gorai.005G234200), ARF 16 (Gorai.011G238900), protein kinase (Gorai.008G294700), tcp transcription factor (Gorai.012G166500), MYB transcription factor (Gorai.013G196800), ABA 8'-hydroylase (Gorai.004G177200), ethylene receptor (Gorai.002G038300), endo-β -glucanase (Gorai.007G126900). All RT-qPCR analyses were performed with three biological replications for each time point with five technical replications. The error bars represent the SD.
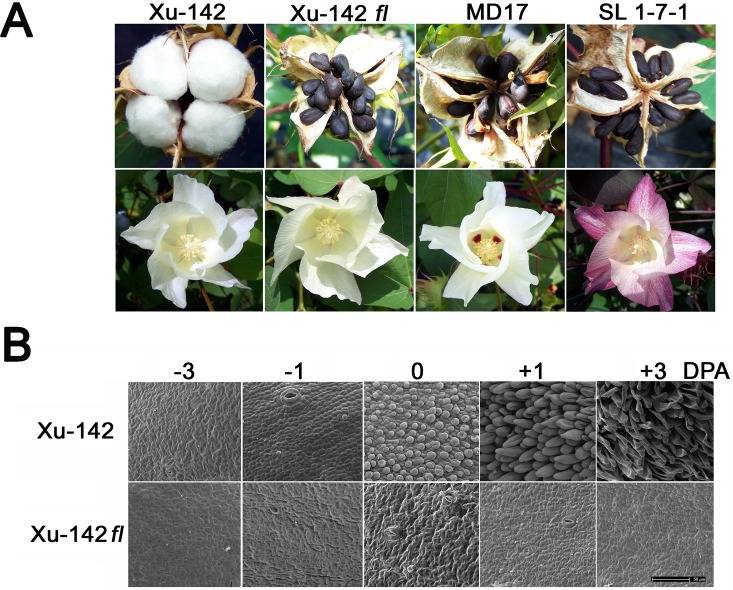
Phenotypes of wild type and three fiberless cotton mutants in planta
To determine if the DEGs identified from the cultured ovules differentiating fiber initials by exogenous phytohormones are really involved in fiber differentiation in planta, we used a fuzzless and lintless (fl) Xu-142 fl mutant and its isogenic WT Xu-142 that have been extensively used to study fiber initiation [45]. In addition, two other fiberless mutants, MD17 and SL1-7-1 that have noticeable variances in flower phenotypes from Xu-142 fl were also used [46,47] (Fig 5A). The SEM images of the cotton ovule surface showed that fiber initials had differentiated between -1 and 0 DPA on the WT cotton ovules and the initials had elongated from 0 to 3 DPA, whereas no fiber initials had differentiated on ovules (-3 to 3 DPA) of the Xu-142 fl mutant (Fig 5B). Like the Xu-142 fl mutant, the two other fiberless mutants had no fiber initials on the 0 DPA ovules when they were photographed by our group (S1 Fig 1) as well as others [16,48].
Fig 5. Phenotypic comparisons of a wild type and three different fiberless cotton mutants.
A. Cotton bolls (top panel) and flowers (bottom panel) are from WT (Xu-142) and three fiberless mutants (Xu-142 fl, MD 17, and SL 1-7-1). B. SEM images of epidermal tissue from G. hirsutum WT Xu-142 ovules (top panel) and fiberless Xu-142 fl mutant ovules (bottom panel). The ovules were harvested around 0900 on -3, -1, 0, +1, and +3 DPA. The bar represents the length of 50μm.
Temporal expression of the DEGs between wild type and three fiberless mutants in planta
The comparative transcriptome analyses of the cultured cotton ovules in which fiber initial differentiation is triggered by exogenous phytohormones was enriched with putative genes involved in fiber initiation. Among the DEGs identified by the transcriptome analyses, some genes may be involved in ovule development alone, whereas other genes may be regulated by phytohormones but are not involved in fiber and ovule development. To identify authentic DEGs involved in the differentiation of fiber initials in planta as well as in vitro, we compared the temporal expression of the DEGs between WT and the three fiberless mutants grown in planta by RT-qPCR. Based on their temporal expression in cotton ovules during the initiation stage of fiber development (-3 to +3 DPA) between the WT and three fiberless mutants grown in planta, the expression patterns of the DEGs were classified into three different classes.
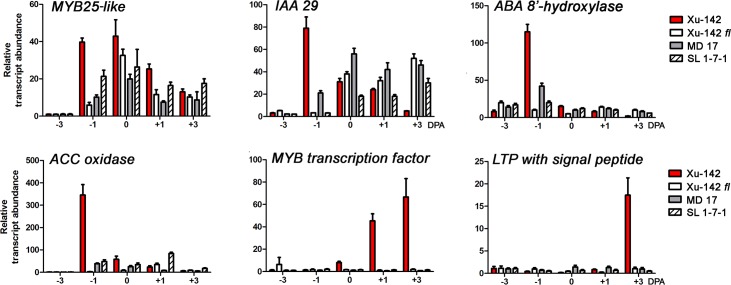
Class 1 DEGs, “DEGs more highly expressed in the WT than in the fiberless mutants”, were temporarily and specifically expressed in initiating and/or elongating fibers grown in planta as well as in vitro. Among them, three DEGs IAA 29, ACC oxidase, and ABA 8'-hydroxylase that were up-regulated in cultured ovules by 1h phytohormone treatment were identically co-regulated with MYB25-like known to be involved in the differentiation of fiber initials [27]. They were all up-regulated specifically at -1 DPA WT ovules over the three fiberless mutants (Fig 6). The temporal expression pattern for IAA29, ACC oxidase, and ABA 8'-hydroxylase, namely higher expression at -1DPA in WT ovules compared with the three fiberless mutants, suggests that these genes may be involved in the differentiation of fiber initials from cotton ovules as was documented for MYB25-like. Up-regulated expression of a MYB transcription factor in WT ovules occurred on 0 DPA and increased until 3 DPA. As a lipid transfer protein (LTP) was up-regulated in cultured ovules by the 12 h exogenous phytohormone treatment, the expression of both an LTP containing a signal peptide (TC275175) and another LTP missing a signal peptide (TC275775) began to be up-regulated at 3 DPA in WT ovules in planta (Fig 6).
Fig 6. Expression patterns of Class 1 DEGs, genes more highly expressed in wild type ovules than the fiberless mutant ovules.
Transcript levels were measured by RT-qPCR from a WT (Xu-142) and three fiberless mutants (Xu-142 fl, MD 17, and SL 1-7-1) that were harvested on -3, -1, 0, 1, and 3 DPA. MYB25-like (Gorai.008G179600), IAA 29 (Gorai.001G043900)), ABA 8'-hydroylase (Gorai.004G177200), ACC oxidase (Gorai.001G011100), MYB transcription factor (Gorai.013G196800), LTP with a signal peptide (Gorai.008G057100). All RT-qPCR analyses were performed with three biological replications at each time point with five technical replications. The error bars represent the SD.
Class 2 DEGs, “DEGs more highly expressed in fiberless mutant ovules than in WT ovules at 3 DPA”, were equivalently expressed in the ovules of WT and the three fiberless mutants from -3 to 1 DPA, but their expression levels were noticeably different at 3 DPA. The expression levels of IAA7, ARF16, ethylene receptor, MYB transcription factor, tcp transcription factor, and endo-β-glucanase were all higher in the fiberless mutant ovules at 3 DPA than in the WT ovules containing elongating fibers (Fig 7). In addition to the six genes presented in Fig 7, many other genes including programmed cell death protein (Gorai.007G244700), auxin-response gh3-like protein (Gorai.005G234200), zinc finger transcription factor (Gorai.011G072500), serine carboxylase peptidase 2 (Gorai.006G103600), heat shock Hsp 101 chaperone (Gorai.012G003700), and heat shock protein-associated protein (Gorai.007G021300) belong to Class 2 DEGs.
Fig 7. Expression patterns of Class 2 DEGs, genes more highly expressed in fiberless mutant ovules than wild type ovules at 3DPA.
Transcript levels were measured by RT-qPCR from a WT (Xu-142) and three fiberless mutants (Xu-142 fl, MD 17, and SL 1-7-1) that were harvested on -3, -1, 0, 1, and 3 DPA. IAA7 (Gorai.009G132200), ARF16 (Gorai.011G238900), ethylene receptor (Gorai.002G038300), MYB transcription factor (Gorai.013G196800), tcp transcription factor (Gorai.012G166500), and Endo-β-glucanase (Gorai.007G126900). All RT-qPCR analyses were performed with three biological replications at each time point with five technical replications. The error bars represent SD.
Class 3 DEGs, “DEGs constitutively expressed in WT and mutants”, were not differentially expressed in WT ovules and fiberless mutant ovules although these genes were differentially regulated in cultured ovules by exogenous phytohormone treatment that triggered fiber initial differentiation. MEK kinase and protein kinase that are involved in the mitogenic activated protein kinase (MAPK) signaling pathway belong to Class 3 DEGs (Fig 8).
Fig 8. Expression patterns of Class 3 DEGs, genes constitutively expressed in wild type and the fiberless mutants.
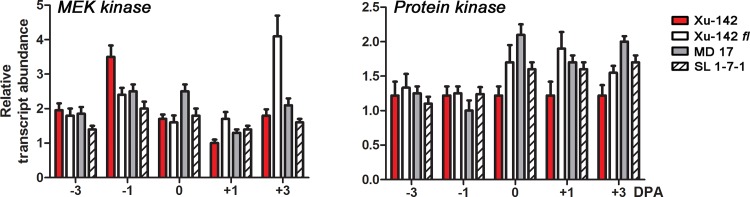
Transcript levels were measured by RT-qPCR from a WT (Xu-142) and three fiberless mutants (Xu-142 fl, MD 17, and SL 1-7-1) that were harvested on -3, -1, 0, 1, and 3 DPA. MEK kinase (Gorai.012G034500), and protein kinase (Gorai.008G294700). All RT-qPCR analyses were performed with three biological replications at each time point with five technical replications. The error bars represent the SD.
Discussion
Cotton fiber differentiation in pre-anthesis ovules is regulated by an exogenous application of phytohormones
To understand the molecular mechanisms regulating cotton fiber differentiation from ovular epidermal cells, we took advantage of the cotton ovule culture technique. Cotton ovule culture methods have been optimized, and many groups have found that fiber development and cell wall composition in vitro closely mimic fiber formation in planta [33,49]. Despite either gibberellic acid (GA) or auxin can differentiate fiber initials from unfertilized ovules, abnormal fiber development [50] or inefficient differentiation of fiber initials [51] were observed when unfertilized ovules were cultured with either phytohormone. Thus, the optimized concentrations of two phytohormones, indole-3-acetic acid (IAA) and GA producing normal fiber development were used for differentiating fiber initials from unfertilized cotton ovules [33,49]. Due to the biochemical and developmental similarities between the fibers cultured with IAA and GA in vitro and the fibers grown in planta, cotton ovule culture has been used to identify regulatory factors affecting fiber elongation by brassinosteroids [52,53], ethylene [42], phytosulfokine peptide hormone, extracellular ATP levels [54], and reactive oxygen species [55]. Although the same exogenous phytohormone treatments promote the differentiation of fiber initials on pre-anthesis (-3 to -1 DPA) cultured ovules, the culture technique has not been used for determining transcriptome profiles during the fiber initiation stage of fiber development.
To study cotton fiber initial differentiation, we started all ovule cultures with -1 DPA cotton ovules harvested before 0900 with no fiber initials on the surface (Fig 1). Exogenous application of auxin and GA enabled the differentiation of fiber initials from the -1 DPA ovular epidermal cells within 24 hours, whereas the control cultures without the phytohormones did not differentiate fiber initials (Fig 2). Thus, -1 DPA ovules may be more enriched in transcripts involved in the differentiation of fiber initials than the 0 and 1 DPA ovules that were previously used for other transcriptome analyses [14,15,16]. Our conclusion is also supported by biochemical analyses revealing that fiber cell fate determination occurs prior to the formation of morphologically visible fiber initials [5,13,56].
IAA29, the most up-regulated gene in pre-anthesis cotton ovules by exogenous phytohormones, is co-regulated in planta with MYB25-like, a gene involved in cotton fiber differentiation
Using the cultured ovules, transcriptome profiles were monitored frequently (1, 3, 6 and 12 h) after the addition of auxin and GA, phytohormones that together triggered the differentiation of fiber initials. Previously reported transcriptome analyses [12,13,14,15,16,34] were mostly conducted at one time point or multiple time points over several days, making it difficult to determine the cascade of phytohormone signaling and networks that affect the temporal regulation of genes involved in the differentiation of fiber initials (Figs 1 and 2).
The transcript abundance of both MYB25-like and HD-1, reportedly involved in the differentiation of fiber initials, was up-regulated around one or two days pre-anthesis [27,28]. In our study, striking differences in MYB25-like transcript abundance between WT and three fiberless mutants were detected in -1 DPA ovules (Fig 7). Thus, the comparison of transcript levels in cotton ovules at -1 DPA between WT and three genetically different fiberless mutants provided us with a means to identify genes that are co-regulated with MYB25-like and HD-1. Among the transcription factors identified by GO enrichment analysis (Table 1), genes involved in auxin signaling were temporally up-regulated during fiber initial differentiation in vitro. A pattern of expression similar to MYB25-like [26] was evident for INDOLE-3-ACETIC ACID INDUCIBLE 29 (IAA29), a Class 1 DEG and the most highly up-regulated phytohormone-induced gene after 1 h phytohormone treatment in the cultured ovules. Like MYB25-like, IAA29 was substantially up-regulated in planta in WT ovules at -1 DPA over ovules of the same age from the fiberless mutants (Figs 6 and 7). In contrast to MYB25- like whose expression levels decreased in WT ovules at 1 DPA, IAA29 decreased in WT ovules at 0 DPA. Walford et al [27] previously showed that transcript abundance of an upstream MYB25- like gene diminished on earlier than that of a downstream MYB109 gene involved in fiber development. IAA29 (AT4G32280) belongs to the auxin/indole-3-acetic acid-responsive (AUX/IAA) protein family of genes that play crucial roles in auxin regulation of Arabidopsis development, such uni-dimensional cell growth, cell wall loosening, cell proliferation, cell expansion, nucleosome organization, and DNA-protein complex formation [57,58]. The Aux/IAA proteins interact with ARFs to regulate auxin-responsive genes by controlling the activity of ARFs [59,60]. Among twenty-nine Aux/IAA genes identified in Arabidopsis, auxin-inducible IAA29 is involved in clock-controlled plant developmental events such as photoperiodic control of flowering time and hypocotyl elongation [61]. Transgenic cotton lines containing higher auxin levels in the ovule epidermis at anthesis from the overexpression of iaaM, an IAA biosynthetic gene, had higher densities of fiber initials [11]. Thus, it would be interesting to test if IAA29 may be an upstream gene of MYB25-like that controls fiber initial differentiation. The expression patterns of nine Aux/IAA transcripts (GhAux1 to GhAux9) from G. hirsutum were previously compared between WT and fiberless mutants [62]. In ovules at the initiation phase of fiber development (-3 to 1 DPA), most GhAux genes were down-regulated in WT as compared with the fiberless mutants unlike IAA29 that was up-regulated in WT as compared with the fiberless mutants (Fig 6). Our results showing that the auxin signaling pathway is mainly involved in fiber differentiation are consistent with previous reports that auxin is required for fiber development from unfertilized cotton ovules but not from fertilized ovules that already have fiber initials [33,35]. Numerous AUX/IAA transcriptional regulators of the auxin signal transduction pathway are known to be involved in the differentiation of root hair cells in Arabidopsis [30], and GhTCP14 functions in auxin-mediated epidermal cell differentiation and elongation during cotton fiber development [41].
Other Class 2 DEGs involved in auxin signaling pathways such as AUX/IAA genes (IAA 4, 7, 11, and 16), ARF16, auxin-response gh3-like protein, and a TCP transcription factor were up-regulated in vitro by the exogenous auxin and GA within 1 h (Table 1 and Fig 4). During fiber initiation (-3 to 1 DPA) in planta, there was little difference in the expression levels of these Class 2 DEGs between WT and three fiberless mutants, whereas they decreased as fiber elongated in 3 DPA WT ovules unlike the corresponding fiberless mutant ovules in which their expressions were maintained (Fig 7). As results, they may be involved in ovule development rather than fiber initial differentiation.
Several Class 3 DEGs, such as MEK kinase and protein kinase, that are involved in the auxin-induced MAP kinase signaling pathway responding to wounding and root development in Arabidopsis [63,64] were also up-regulated in vitro by the exogenously applied phytohormones. Since these Class 3 DEGs were equivalently expressed in planta between WT and fiberless mutants, they are unlikely to be involved directly in fiber initial differentiation; however, we do not rule out a potential involvement of the MAP kinase pathway in early fiber and/or ovule development since multiple genes involved in jasmonic acid signaling pathways for defense responses were also identified by our transcriptome analysis (Table 2). Exogenous application of hydrogen peroxide is known to be involved in signaling pathways responding to defense and wound-facilitated fiber initiation in another fiberless mutant, XinFLM [65]. Thus, further research is required to conclude if and how the auxin-induced MAP kinase signaling pathway and jasmonic acid signaling pathways affect differentiation of cotton fiber initials.
Crosstalk among the auxin, ethylene, and ABA signaling pathways contributes to cotton fiber differentiation from epidermal cells
In addition to IAA29 in the auxin signaling pathway, the expression patterns of ACC oxidase involved in ethylene production and ABA 8'-hydroxylase involved in ABA catabolism were co-regulated both in vitro and in planta with MYB25-like and GhHD-1 that are indispensible for cotton fiber differentiation (Table 2, Figs 4 and 6). Unlike MYB25-like and IAA29 that were differentially expressed in the ovules from both WT and fiberless mutants, ACC oxidase and ABA 8’-hydroxylase were mostly found in WT ovules but little in the ovules from the fiberless mutants (Fig 6). In addition to the tissue specific expression, their temporal expression patterns at -1 DPA just before differentiating fiber initials in WT ovules also suggest potential involvement in fiber initiation process.
ACC oxidase (EC 1.14.17.4) catalyses the final step in the biosynthesis of the phytohormone ethylene from 1-aminocyclopropane-1-carboxylate (ACC) as a substrate. Synergistic interactions between auxin and ethylene-dependent phenomena have been reported from Arabidopsis [42,66,67]. In Arabidopsis root development, auxin is a positive regulator for ethylene-mediated response [66], and normal auxin signaling is required for a proper ethylene response in Arabidopsis hypocotyls [67]. Identical up-regulation of both IAA29 for auxin signaling and ACC oxidase for ethylene signaling during the fiber initiation phase (Fig 6) suggests that synergistic interactions between auxin and ethylene may be necessary for the differentiation of fiber initials.
Abscisic acid (ABA) 8'-hydroxylase is also known as a cytochrome P450 (CYP707A) that catalyzes the first step in the oxidative inactivation of ABA [43,68]. The ABA 8'-hydroxylase converts ABA to 8'-hydroxy-ABA that is unstable and isomerizes to form phaseic acid. Therefore, ABA 8'-hydroxylase can regulate physiologically active ABA levels during plant developmental process. ABA is known to inhibit fiber initiation in cultured ovules [69]. The elevated levels of ABA 8'-hydroxylase in WT ovules (-1 DPA) compared with the fiberless mutant ovules should reduce the level of ABA that could inhibit fiber development, thereby stimulating the differentiation of cotton fiber initials (Figs 4 & 6).
Conclusions
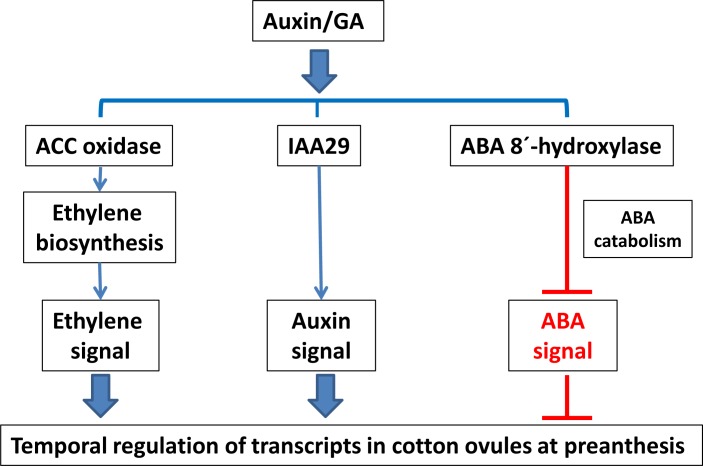
Technical difficulties in monitoring the rapid process of fiber initial differentiation in ovaries and extracting RNAs from very short and small fiber initials without contamination with epidermal tissues has hindered the progress of cotton fiber initiation research. Pioneering investigations using the fiberless cotton mutants [16] and fiber initials extracted by a laser capture microdissection technique [14] produced transcriptome profiles from fiber initials differentiating on 0 DPA cotton ovules. Despite these efforts, recent functional analyses of the candidate genes identified by these earlier transcriptome studies showed that the candidate genes are more likely to be involved in fiber elongation than in fiber initial differentiation. To identify putative genes involved in fiber initial differentiation and understand the molecular mechanisms of differentiating fiber initials, we improved the conditions for the comparative transcriptome expression analyses to enrich for transcripts involved in cotton fiber initial differentiation in vitro and in planta. Our study identified genes that are co-regulated with MYB25-like and HD-1, two genes previously identified as key transcriptional factors responsible for fiber initial differentiation (Fig 9). We have identified candidate genes for fiber initiation whose contribution must be more completely analyzed to discover how phytohormonal signaling pathways and crosstalk regulate the temporal expression of genes responsible for the differentiation of fiber initials in vitro and in planta.
Fig 9. Genes co-regulated in vitro and in planta with MYB25-like.
Supporting Information
(TIF)
(XLSX)
(DOCX)
Acknowledgments
The authors thank Bruce F. Ingber for photographing SEM images and Tracy Condon for managing cotton plants. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA that is an equal opportunity employer.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by the USDA-ARS CRIS Project #6435-21000-016-00D, NSF DBI0624077 and ISO1025947, and Cotton Incorporated-sponsored project #07-161. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Wakelyn PJ, Bertoniere NR, French AD, Thibodeaux DP, Triplett BA, et al. (2010) Cotton fiber chemistry and technology: CRC Press. [Google Scholar]
- 2. Kim HJ, Triplett BA (2001) Cotton fiber growth in planta and in vitro. Models for plant cell elongation and cell wall biogenesis. Plant Physiology 127: 1361–1366. [PMC free article] [PubMed] [Google Scholar]
- 3.Stewart JM (1975) Fiber initiation on the cotton ovule (Gossypium hirsutum). American Journal of Botany: 723–730.
- 4. Applequist WL, Cronn R, Wendel JF (2001) Comparative development of fiber in wild and cultivated cotton. Evolution & development 3: 3–17. [DOI] [PubMed] [Google Scholar]
- 5. Lee JJ, Woodward AW, Chen ZJ (2007) Gene expression changes and early events in cotton fibre development. Annals of botany 100: 1391–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ryser U (1999) Cotton fiber initiation and histodifferentiation Cotton Fibers: Developmental Biology, Quality Improvement, and Textile Processing, The Haworth Press, New York: 1–46. [Google Scholar]
- 7. Lang AG (1938) The origin of lint and fuzz hairs of cotton. Journal of Agricultural Research 56: 507–521. [Google Scholar]
- 8. Basra AS, Malik C (1984) Development of the cotton fiber. International Review of Cytology 89: 65–113. [Google Scholar]
- 9. Li C, Guo W, Zhang T (2009) Fiber initiation development in Upland cotton (Gossypium hirsutum L.) cultivars varying in lint percentage. Euphytica 165: 223–230. [Google Scholar]
- 10. Romano GB, Taliercio EW, Turley RB, A SJ (2011) Fiber Initiation in 18 cultivars and experimental lines of three Gossypium species. Journal of Cotton Science 15: 61–72. [Google Scholar]
- 11. Zhang M, Zheng X, Song S, Zeng Q, Hou L, et al. (2011) Spatiotemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fiber yield and quality. Nature Biotechnology 29: 453–458. 10.1038/nbt.1843 [DOI] [PubMed] [Google Scholar]
- 12. Li C-H, Zhu Y-Q, Meng Y-L, Wang J-W, Xu K-X, et al. (2002) Isolation of genes preferentially expressed in cotton fibers by cDNA filter arrays and RT-PCR. Plant Science 163: 1113–1120. [Google Scholar]
- 13. Lee JJ, Hassan OS, Gao W, Wei NE, Kohel RJ, et al. (2006) Developmental and gene expression analyses of a cotton naked seed mutant. Planta 223: 418–432. [DOI] [PubMed] [Google Scholar]
- 14. Wu Y, Llewellyn DJ, White R, Ruggiero K, Al-Ghazi Y, et al. (2007) Laser capture microdissection and cDNA microarrays used to generate gene expression profiles of the rapidly expanding fibre initial cells on the surface of cotton ovules. Planta 226: 1475–1490. [DOI] [PubMed] [Google Scholar]
- 15. Taliercio EW, Boykin D (2007) Analysis of gene expression in cotton fiber initials. BMC Plant Biology 7: 22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu Y, Machado AC, White RG, Llewellyn DJ, Dennis ES (2006) Expression profiling identifies genes expressed early during lint fibre initiation in cotton. Plant and Cell Physiology 47: 107–127. [DOI] [PubMed] [Google Scholar]
- 17. Suo J, Liang X, Pu L, Zhang Y, Xue Y (2003) Identification ofGhMYB109 encoding a R2R3 MYB transcription factor that expressed specifically in fiber initials and elongating fibers of cotton Gossypium hirsutum L. Biochimica et Biophysica Acta (BBA)-Gene Structure and Expression 1630: 25–34. [DOI] [PubMed] [Google Scholar]
- 18. Ruan Y-L, Chourey PS (1998) A fiberless seed mutation in cotton is associated with lack of fiber cell initiation in ovule epidermis and alterations in sucrose synthase expression and carbon partitioning in developing seeds. Plant Physiology 118: 399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Loguercio L, Zhang J-Q, Wilkins T (1999) Differential regulation of six novel MYB-domain genes defines two distinct expression patterns in allotetraploid cotton (Gossypium hirsutum L.). Molecular and General Genetics 261: 660–671. [DOI] [PubMed] [Google Scholar]
- 20. Wang L, Cook A, Patrick JW, Chen X-Y, Ruan Y-L (2014) Silencing the vacuolar invertase geneGhVIN1blocks cotton fiber initiation from the ovule epidermis, probably by suppressing a cohort of regulatory genes via sugar signaling. The Plant Journal 78: 686–696. 10.1111/tpj.12512 [DOI] [PubMed] [Google Scholar]
- 21. Wang L, Li X-R, Lian H, Ni D-A, He Y-k, et al. (2010) Evidence that high activity of vacuolar invertase is required for cotton fiber and Arabidopsis root elongation through osmotic dependent and independent pathways, respectively. Plant Physiology 154: 744–756. 10.1104/pp.110.162487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang S, Wang J-W, Yu N, Li C-H, Luo B, et al. (2004) Control of plant trichome development by a cotton fiber MYB gene. The Plant Cell 16: 2323–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Deng F, Tu L, Tan J, Li Y, Nie Y, et al. (2011) GbPDF1 Is Involved in Cotton Fiber Initiation via the Core cis-Element HDZIP2ATATHB2. Plant Physiology 158: 890–904. 10.1104/pp.111.186742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guan XY, Li QJ, Shan CM, Wang S, Mao YB, et al. (2008) The HD‐Zip IV gene GaHOX1 from cotton is a functional homologue of the Arabidopsis GLABRA2. Physiologia Plantarum 134: 174–182. 10.1111/j.1399-3054.2008.01115.x [DOI] [PubMed] [Google Scholar]
- 25. Pu L, Li Q, Fan X, Yang W, Xue Y (2008) The R2R3 MYB Transcription Factor GhMYB109 Is Required for Cotton Fiber Development. Genetics 180: 811–820. 10.1534/genetics.108.093070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Machado A, Wu Y, Yang Y, Llewellyn DJ, Dennis ES (2009) The MYB transcription factor GhMYB25 regulates early fibre and trichome development. The Plant Journal 59: 52–62. 10.1111/j.1365-313X.2009.03847.x [DOI] [PubMed] [Google Scholar]
- 27. Walford S-A, Wu Y, Llewellyn DJ, Dennis ES (2011) GhMYB25-like: a key factor in early cotton fibre development. The Plant Journal 65: 785–797. 10.1111/j.1365-313X.2010.04464.x [DOI] [PubMed] [Google Scholar]
- 28.Walford S-A, Wu Y, Llewellyn DJ, Dennis ES (2012) Epidermal cell differentiation in cotton mediated by the homeodomain leucine zipper gene, GhHD-1. The Plant Journal: no-no. [DOI] [PubMed]
- 29. Ruan Y-L, Llewellyn DJ, Furbank RT (2003) Suppression of sucrose synthase gene expression represses cotton fiber cell initiation, elongation, and seed development. The Plant Cell 15: 952–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ishida T, Kurata T, Okada K, Wada T (2008) A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol 59: 365–386. 10.1146/annurev.arplant.59.032607.092949 [DOI] [PubMed] [Google Scholar]
- 31. Schiefelbein J, Huang L, Zheng X (2014) Regulation of epidermal cell fate in Arabidopsis roots: the importance of multiple feedback loops. Frontiers in Plant Science 5: 47 10.3389/fpls.2014.00047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang SS, Cheung F, Lee JJ, Ha M, Wei NE, et al. (2006) Accumulation of genome‐specific transcripts, transcription factors and phytohormonal regulators during early stages of fiber cell development in allotetraploid cotton. The Plant Journal 47: 761–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Beasley C, Ting IP (1974) Effects of plant growth substances on in vitro fiber development from unfertilized cotton ovules. American Journal of Botany 61: 188–194. [Google Scholar]
- 34. Wang QQ, Liu F, Chen XS, Ma XJ, Zeng HQ, et al. (2010) Transcriptome profiling of early developing cotton fiber by deep-sequencing reveals significantly differential expression of genes in a fuzzless/lintless mutant. Genomics 96: 369–376. 10.1016/j.ygeno.2010.08.009 [DOI] [PubMed] [Google Scholar]
- 35. Beasley C, Ting IP (1973) The effects of plant growth substances on in vitro fiber development from fertilized cotton ovules. American Journal of Botany 60: 130–139. [Google Scholar]
- 36. Udall JA, Flagel LE, Cheung F, Woodward AW, Hovav R, et al. (2007) Spotted cotton oligonucleotide microarrays for gene expression analysis. BMC Genomics 8: 81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hinchliffe DJ, Meredith WR, Yeater KM, Kim HJ, Woodward AW, et al. (2010) Near-isogenic cotton germplasm lines that differ in fiber-bundle strength have temporal differences in fiber gene expression patterns as revealed by comparative high-throughput profiling. Theoretical and Applied Genetics 120: 1347–1366. 10.1007/s00122-010-1260-6 [DOI] [PubMed] [Google Scholar]
- 38. Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Research 38: W64–W70. 10.1093/nar/gkq310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, et al. (2012) Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492: 423–427. 10.1038/nature11798 [DOI] [PubMed] [Google Scholar]
- 40. Hao J, Tu L, Hu H, Tan J, Deng F, et al. (2012) GbTCP, a cotton TCP transcription factor, confers fibre elongation and root hair development by a complex regulating system. Journal of Experimental Botany 63: 6267–6281. 10.1093/jxb/ers278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang MY, Zhao PM, Cheng HQ, Han LB, Wu XM, et al. (2013) The Cotton Transcription Factor TCP14 Functions in Auxin-Mediated Epidermal Cell Differentiation and Elongation. Plant Physiology 162: 1669–1680. 10.1104/pp.113.215673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shi Y-H, Zhu S-W, Mao X-Z, Feng J-X, Qin Y-M, et al. (2006) Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. The Plant Cell 18: 651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Saito S, Hirai N, Matsumoto C, Ohigashi H, Ohta D, et al. (2004) Arabidopsis CYP707As encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiology 134: 1439–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shimizu Y, Aotsuka S, Hasegawa O, Kawada T, Sakuno T, et al. (1997) Changes in levels of mRNAs for cell wall-related enzymes in growing cotton fiber cells. Plant and Cell Physiology 38: 375–378. [DOI] [PubMed] [Google Scholar]
- 45. Zhang T, Pan J (1991) Genetic analysis of a fuzzless-lintless mutant in Gossypium hirsutum L. Jiangsu J Agric Sci 7: 13–16. [Google Scholar]
- 46. Turley R, Kloth R (2002) Identification of a third fuzzless seed locus in upland cotton (Gossypium hirsutum L.). Journal of Heredity 93: 359–364. [DOI] [PubMed] [Google Scholar]
- 47. Turley RB, Kloth RH (2008) The inheritance model for the fiberless trait in upland cotton (Gossypium hirsutum L.) line SL1-7-1: variation on a theme. Euphytica 164: 123–132. [Google Scholar]
- 48. Zhang DY, Zhang TZ, Sang ZQ, Guo WZ (2007) Comparative Development of Lint and Fuzz Using Different Cotton Fiber‐specific Developmental Mutants in Gossypium hirsutum. Journal of Integrative Plant Biology 49: 1038–1046. [Google Scholar]
- 49. Meinert MC, Delmer DP (1977) Changes in biochemical composition of the cell wall of the cotton fiber during development. Plant Physiology 59: 1088–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kosmidou-Dimitropoulou K (1986) Hormonal influences on fiber development In: Mauney JR, Stewart JM, editors. Cotton Physiology. Memphis: The Cotton Foundation; pp. 361–373. [Google Scholar]
- 51. Gialvalis S, Seagull RW (2001) Plant hormones alter fiber initiation in unfertilized, cultured ovules of Gossypium hirsutum. Journal of Cotton Science 5: 252–258. [Google Scholar]
- 52. Sun Y, Veerabomma S, Abdel-Mageed HA, Fokar M, Asami T, et al. (2005) Brassinosteroid regulates fiber development on cultured cotton ovules. Plant and Cell Physiology 46: 1384–1391. [DOI] [PubMed] [Google Scholar]
- 53. Yang Z, Zhang C, Yang X, Liu K, Wu Z, et al. (2014) PAG1, a cotton brassinosteroid catabolism gene, modulates fiber elongation. New Phytologist 203: 437–448. 10.1111/nph.12824 [DOI] [PubMed] [Google Scholar]
- 54. Clark G, Torres J, Finlayson S, Guan X, Handley C, et al. (2010) Apyrase (nucleoside triphosphate-diphosphohydrolase) and extracellular nucleotides regulate cotton fiber elongation in cultured ovules. Plant Physiology 152: 1073–1083. 10.1104/pp.109.147637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Potikha TS, Collins CC, Johnson DI, Delmer DP, Levine A (1999) The involvement of hydrogen peroxide in the differentiation of secondary walls in cotton fibers. Plant Physiology 119: 849–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Graves D, Stewart J (1988) Chronology of the differentiation of cotton (Gossypium hirsutum L.) fiber cells. Planta 175: 254–258. 10.1007/BF00392435 [DOI] [PubMed] [Google Scholar]
- 57. Guilfoyle T, Hagen G, Ulmasov T, Murfett J (1998) How does auxin turn on genes? Plant Physiology 118: 341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Leyser O (2002) Molecular genetics of auxin signaling. Annual Review of Plant Biology 53: 377–398. [DOI] [PubMed] [Google Scholar]
- 59. Tiwari SB, Hagen G, Guilfoyle T (2003) The roles of auxin response factor domains in auxin-responsive transcription. The Plant Cell 15: 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. The Plant Cell 9: 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kunihiro A, Yamashino T, Nakamichi N, Niwa Y, Nakanishi H, et al. (2011) Phytochrome-interacting factor 4 and 5 (PIF4 and PIF5) activate the homeobox ATHB2 and auxin-inducible IAA29 genes in the coincidence mechanism underlying photoperiodic control of plant growth of Arabidopsis thaliana. Plant and Cell Physiology 52: 1315–1329. 10.1093/pcp/pcr076 [DOI] [PubMed] [Google Scholar]
- 62. Han X, Xu X, Fang DD, Zhang T, Guo W (2012) Cloning and expression analysis of novel Aux/IAA family genes in Gossypium hirsutum. Gene 503: 83–91. 10.1016/j.gene.2012.03.069 [DOI] [PubMed] [Google Scholar]
- 63. Hadiarto T, Nanmori T, Matsuoka D, Iwasaki T, Sato K-i, et al. (2006) Activation of Arabidopsis MAPK kinase kinase (AtMEKK1) and induction of AtMEKK1–AtMEK1 pathway by wounding. Planta 223: 708–713. [DOI] [PubMed] [Google Scholar]
- 64. Mockaitis K, Howell SH (2000) Auxin induces mitogenic activated protein kinase (MAPK) activation in roots of Arabidopsis seedlings. The Plant Journal 24: 785–796. [DOI] [PubMed] [Google Scholar]
- 65. Zhang D, Zhang T, Guo W (2010) Effect of H2O2 on fiber initiation using fiber retardation initiation mutants in cotton (Gossypium hirsutum). Journal of Plant Physiology 167: 393–399. 10.1016/j.jplph.2009.10.005 [DOI] [PubMed] [Google Scholar]
- 66. Rahman A, Amakawa T, Goto N, Tsurumi S (2001) Auxin is a positive regulator for ethylene-mediated response in the growth of Arabidopsis roots. Plant and Cell Physiology 42: 301–307. [DOI] [PubMed] [Google Scholar]
- 67.Robles LM, Deslauriers SD, Alvarez AA, Larsen PB (2012) A loss-of-function mutation in the nucleoporin AtNUP160 indicates that normal auxin signalling is required for a proper ethylene response in Arabidopsis. Journal of Experimental Botany: err424. [DOI] [PMC free article] [PubMed]
- 68. Ji X, Dong B, Shiran B, Talbot MJ, Edlington JE, et al. (2011) Control of abscisic acid catabolism and abscisic acid homeostasis is important for reproductive stage stress tolerance in cereals. Plant Physiology 156: 647–662. 10.1104/pp.111.176164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang H, Shao M, Qiao Z, Yuan S, Wang X, et al. (2009) Effect of phytohormones on fiber initiation of cotton ovule. Acta physiologiae plantarum 31: 979–986. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(XLSX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.