Abstract
The ILM web server provides a web interface to two algorithms, iterated loop matching and maximum weighted matching, for efficiently predicting RNA secondary structures with pseudoknots. The algorithms can utilize either thermodynamic or comparative information or both, and thus can work on both aligned and individual sequences. Predicted secondary structures are presented in several formats compatible with a variety of existing visualization tools. The service can be accessed at http://cic.cs.wustl.edu/RNA/.
INTRODUCTION
RNA molecules play many important regulatory, catalytic and structural roles in the cell. A complete understanding of the functions of RNA molecules requires knowledge of their three-dimensional (3D) structures. Since it is often difficult to obtain X-ray diffraction or nuclear magnetic resonance (NMR) data for large RNA molecules to inspect their structures, reliable prediction of RNA structures from their primary sequences is highly desirable.
Many computational methods have been developed for predicting RNA secondary structures. Thermodynamic approaches (1,2) compute for a single RNA sequence an optimal secondary structure with globally minimal free energy, and have been successful for relatively short RNAs. When a number of aligned homologous sequences are available, comparative approaches (3–6) are more reliable than thermodynamic approaches and have been used to establish the structures of most known RNA families. In addition, several methods (7–11) have combined the advantages of thermodynamic and comparative methods. By taking both thermodynamic stability and sequence covariance into consideration, these methods are able to achieve much higher prediction accuracies.
On the other hand, relatively little work has been done on predicting pseudoknotted RNA secondary structures. Pseudoknots are important RNA structures and often have important functional roles (12). However, optimally predicting pseudoknots in RNA secondary structures is difficult and computationally expensive (13–16). A graph algorithm, maximum weighted matching (MWM), has been used as a practical solution for pseudoknot prediction (17–19). Although efficient, the prediction accuracy of MWM is low when the number of homologous sequences is small.
We recently developed an algorithm, iterated loop matching (ILM), to predict pseudoknotted RNA secondary structures (20). This method can utilize either thermodynamic or comparative information or both, and thus can be applied to both aligned and individual sequences. Our experiments have shown that the method is accurate and efficient. However, the software can only be executed with Unix commands and requires users to set up various parameters, making it difficult for biologists to use.
To meet the high demand of a service for predicting RNA secondary structures with pseudoknots, we have developed an easy-to-use web interface to provide most features of ILM online. We have also provided an option for the user to choose the MWM algorithm and compare their results.
Several web servers for RNA secondary structure prediction were introduced in last year's web server special issue, including MFold (21), Pfold (22), the Vienna RNA package (23) and GPRM (24). ILM differs from the first three in that it supports pseudoknots. GPRM is designed to find common secondary structure elements in a set of homologous RNA sequences and cannot be applied to a single RNA sequence or a small dataset, e.g. a family of fewer than 10 sequences.
OVERVIEW OF THE SERVICE
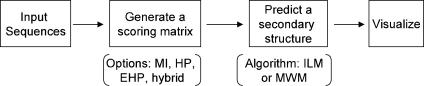
The technical details of the MWM and ILM algorithms can be found in their original publications (19,20). Here, we highlight only the basic steps of the web service (Figure 1) and the parameters that need user intervention. The first step after reading in the RNA sequences is to generate a scoring matrix, which describes the likelihood of every base pairing. The methods to generate scoring matrices include mutual information, helix plot, extended helix plot and their combinations (20). In the second step, the ILM or MWM algorithm is applied to predict a secondary structure with pseudoknots allowed. Finally, the output of predicted structures can be viewed with certain external visualization tools.
Figure 1.
Overview of the ILM web service. Abbreviations used: MI, mutual information; HP, helix plot; EHP, extended helix plot; ILM, iterated loop matching; MWM, maximum weighted matching.
INPUT
The service can be accessed through a web interface (http://cic.cs.wustl.edu/RNA/). The web interface takes a single RNA sequence or a set of aligned RNA sequences as input in FASTA format. The user may choose to upload a sequence file or ‘cut and paste’ sequences into the web interface directly. Currently, the maximum length of each individual sequence is 2000 bases and the maximum size of a sequence file is 10 kb.
The web server provides optimized default values for all parameters, which may also be adjusted for different needs. The user may choose to calculate a scoring matrix with only mutual information or (extended) helix plot, or choose a combined scoring matrix and vary the relative weights of these two scoring methods. When the number of sequences is small (<10), mutual information scores are usually not reliable and should receive a lower weight. If only one sequence is provided, mutual information scores cannot be calculated; therefore the provided weights are ignored, and only helix plot or extended helix plot can be used. In other cases, a default weight ratio of 1:1 is suggested by the server. The user may select to use either the ILM or the MWM algorithm for the prediction and may also customize parameters such as the minimum helix length and minimum loop length. (See text on the website for information on the meaning and suggested value of each parameter.)
OUTPUT
For small tasks, i.e. sequences up to 300 bases, the results will be presented immediately, while for large tasks, the user will be notified by email on how to access to the results when the tasks are completed.
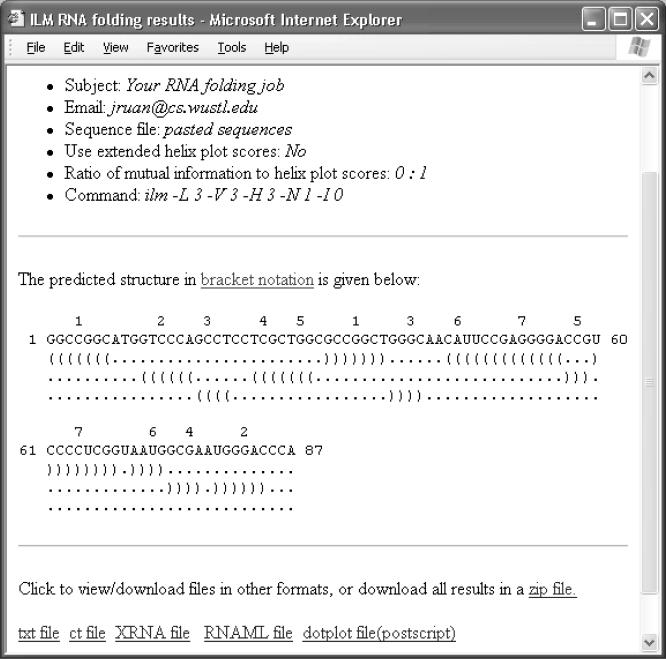
The output is self-documented (Figure 2). The page headings include user information, a dataset name and parameters specified by the user. As shown in the middle of Figure 2, the predicted secondary structure is given in a modified dot–bracket notation, where a base pair is represented by a pair of opening and closing brackets. When pseudoknots are present, we break a secondary structure into several levels. Base pairs on the same level do not cross each other, while base pairs from different levels can cross each other, forming pseudoknots. This notation is able to represent pseudoknots of any complexity. In Figure 2, for example, helix 3 (helix indices are shown on top of the primary sequence) intersects with helix 1 and helix 5, while helix 1 also intersects with helix 5.
Figure 2.
Example output generated by the ILM web server.
In addition, the output provides links to several files in different formats compatible with existing visualization tools. An automatic drawing of RNA secondary structures with pseudoknots is notoriously difficult and many existing visualization tools handle this in a user-interactive way. We thus do not attempt to provide a graphic presentation of the secondary structures on our web site. Instead, we generate the output in several formats that can be directly imported into a variety of existing visualization tools for RNA secondary structures.
Currently supported formats include ct, xrna and rnaml files. The format of ct files is supported by several programs, including the RNAviz program (25), which we recommend for drawing pseudoknotted RNA secondary structures. An xrna file contains a primary sequence and helix descriptions that can be separately copy–pasted into the XRNA program (http://rna.ucsc.edu/rnacenter/xrna/xrna.html). The rnaml format has been proposed as a standard for exchanging RNA sequence and structure information between programs (26). Our rnaml syntax is compatible with the DTD (Document Type Definition) version 1.1 (http://www-lbit.iro.umontreal.ca/rnaml/current/rnaml.dtd).
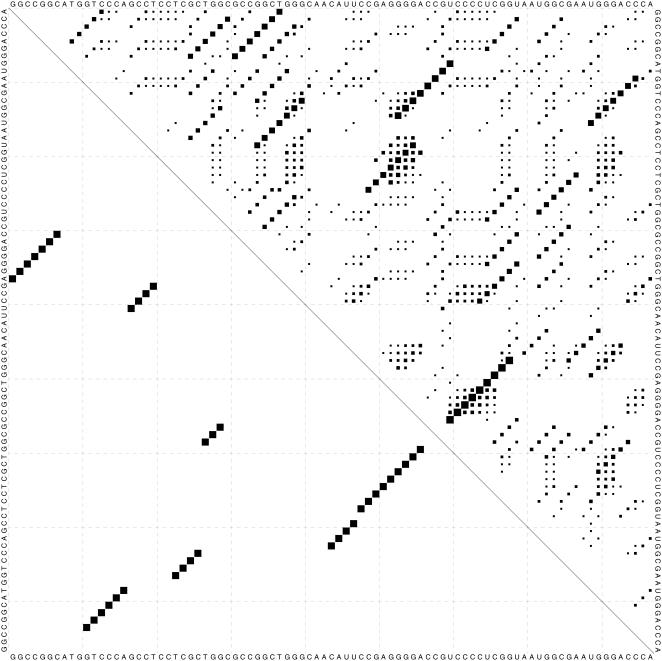
Finally, together with structure results, a dot plot (Figure 3) is provided in postscript format, which allows the user to view the scoring matrix and predicted structure at the same time. The actual scores are embedded in the postscript file and can be parsed using computer programs.
Figure 3.
A dot plot. In the upper triangle, the area of a dot in row i and column j represents the relative score for a base pair between the i-th and j-th bases. A dot in the lower triangle means that the corresponding base pair is predicted by the algorithm.
IMPLEMENTATION
The web service is implemented in static HTML pages and dynamic CGI scripts in PERL. The ILM and MWM programs are implemented in ANSI C. The server is currently running on a machine with dual AMD Athlon 1.6 GHz CPUs and 2 GB of RAM, running Redhat Linux version 2.4.18 and Apache web server. In the future, we plan to use a batch queuing system to distribute large tasks to other machines.
Acknowledgments
ACKNOWLEDGEMENTS
J.R. thanks Ivo Hofacker for providing the dotplot routine and Mark Bober for setting up the web server. We also thank two anonymous reviewers for their very useful comments. This research was supported in part by NSF grants IIS-0196057 and EIA-0113618 under the ITR program. G.D.S. was supported by NIH grant HG00249.
REFERENCES
- 1.Hofacker I., Fontana,W., Stadler,P., Bonhoeffer,L., Tacker,M. and Schuster,P. (1994) Fast folding and comparison of RNA secondary structures. Monatsh. Chem., 125, 167–188. [Google Scholar]
- 2.Zuker M. and Stiegler,P. (1981) Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res., 9, 133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akmaev V., Kelley,S. and Stormo,G. (1999) A phylogenetic approach to RNA structure prediction. Proc. Int. Conf. Intell. Syst. Mol. Biol., 7, 10–17. [PubMed] [Google Scholar]
- 4.Chiu D. and Kolodziejczak,T. (1991) Inferring consensus structure from nucleic acid sequences. Comput. Appl. Biosci., 7, 347–352. [DOI] [PubMed] [Google Scholar]
- 5.Gulko B. and Haussler,D. (1996) Using multiple alignments and phylogenetic trees to detect RNA secondary structure. Proc. Pac. Symp. Biocomput., 1, 350–367. [PubMed] [Google Scholar]
- 6.Gutell R., Power,A., Hertz,G., Putz,E. and Stormo,G. (1992) Identifying constraints on the higher-order structure of RNA: continued development and application of comparative sequence analysis methods. Nucleic Acids Res., 20, 5785–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J., Le,S. and Maizel,J. (2000) Prediction of common secondary structures of RNAs: a genetic algorithm approach. Nucleic Acids Res., 28, 991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofacker I., Fekete,M. and Stadler,P. (2002) Secondary structure prediction for aligned RNA sequences. J. Mol. Biol., 319, 1059–1066. [DOI] [PubMed] [Google Scholar]
- 9.Juan V. and Wilson,C. (1999) RNA secondary structure prediction based on free energy and phylogenetic analysis. J. Mol. Biol., 289, 935–947. [DOI] [PubMed] [Google Scholar]
- 10.Luck R., Graf,S. and Steger,G. (1999) ConStruct: a tool for thermodynamic controlled prediction of conserved secondary structure. Nucleic Acids Res., 27, 4208–4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathews D. and Turner,D. (2002) Dynalign: an algorithm for finding the secondary structure common to two RNA sequences. J. Mol. Biol., 317, 191–203. [DOI] [PubMed] [Google Scholar]
- 12.Dam E., Pleij,K. and Draper,D. (1992) Structural and functional aspects of RNA pseudoknots. Biochemistry, 31, 11665–11676. [DOI] [PubMed] [Google Scholar]
- 13.Akutsu T. (2000) Dynamic programming algorithms for RNA secondary structure prediction with pseudoknots. Disc. Appl. Math., 104, 45–62. [Google Scholar]
- 14.Lyngsø R. and Pedersen,C. (2000) RNA pseudoknot prediction in energy-based models. J. Comput. Biol., 7, 409–427. [DOI] [PubMed] [Google Scholar]
- 15.Rivas E. and Eddy,S. (1999) A dynamic programming algorithm for RNA structure prediction including pseudoknots. J. Mol. Biol., 285, 2053–2068. [DOI] [PubMed] [Google Scholar]
- 16.Uemura Y., Hasegawa,A., Kobayashi,S. and Yokomori,T. (1999) Tree adjoining grammars for RNA structure prediction. Theor. Comp. Sci., 210, 277–303. [Google Scholar]
- 17.Cary R. and Stormo,G. (1995) Graph-theoretic approach to RNA modeling using comparative data. Proc. Int. Conf. Intell. Syst. Mol. Biol., 3, 75–80. [PubMed] [Google Scholar]
- 18.Page R. (2000) Comparative analysis of secondary structure of insect mitochondrial small subunit ribosomal RNA using maximum weighted matching. Nucleic Acids Res., 28, 3839–3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tabaska J., Cary,R., Gabow,H., and Stormo,G. (1998) An RNA folding method capable of identifying pseudoknots and base triples. Bioinformatics, 14, 691–699. [DOI] [PubMed] [Google Scholar]
- 20.Ruan J., Stormo,G. and Zhang,W. (2004) An iterated loop matching approach to the prediction of RNA secondary structures with pseudoknots. Bioinformatics, 20, 58–66. [DOI] [PubMed] [Google Scholar]
- 21.Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res., 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knudsen B. and Hein,J. (2003) Pfold: RNA secondary structure prediction using stochastic context-free grammars. Nucleic Acids Res., 31, 3423–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofacker I. (2003) Vienna RNA secondary structure server. Nucleic Acids Res., 31, 3429–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Y. (2003) GPRM: a genetic programming approach to finding common RNA secondary structure elements. Nucleic Acids Res., 31, 3446–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rijk P.D., Wuyts,J. and Wachter,R.D. (2003) Rnaviz 2: an improved representation of RNA secondary structure. Bioinformatics, 19, 299–300. [DOI] [PubMed] [Google Scholar]
- 26.Waugh A., Gendron,P., Altman,R., Brown,J., Case,D., Gautheret,D., Harvey,S., Leontis,N., Westbrook,J., Westhof,E., Zuker,M. and Major,F. (2002) RNAML: a standard syntax for exchanging RNA information. RNA, 8, 707–717. [DOI] [PMC free article] [PubMed] [Google Scholar]