Abstract
Human monoclonal antibodies (HMAbs) with neutralizing capabilities constitute potential immune-based treatments or prophylaxis against hepatitis C virus (HCV). However, lack of cell culture-derived HCV (HCVcc) harboring authentic envelope proteins (E1/E2) has hindered neutralization investigations across genotypes, subtypes, and isolates. We investigated the breadth of neutralization of 10 HMAbs with therapeutic potential against a panel of 16 JFH1-based HCVcc expressing patient-derived Core-NS2 from genotypes 1a (strains H77, TN, and DH6), 1b (J4, DH1, and DH5), 2a (J6, JFH1, and T9), 2b (J8, DH8, and DH10), 2c (S83), and 3a (S52, DBN, and DH11). Virus stocks used for in vitro neutralization analysis contained authentic E1/E2, with the exception of full-length JFH1 that acquired the N417S substitution in E2. The 50% inhibition concentration (IC50) for each HMAb against the HCVcc panel was determined by dose-response neutralization assays in Huh7.5 cells with antibody concentrations ranging from 0.0012 to 100 μg/ml. Interestingly, IC50-values against the different HCVcc’s exhibited large variations among the HMAbs, and only three HMAbs (HC-1AM, HC84.24, and AR4A) neutralized all 16 HCVcc recombinants. Furthermore, the IC50-values for a given HMAb varied greatly with the HCVcc strain, which supports the use of a diverse virus panel. In cooperation analyses, HMAbs HC84.24, AR3A, and, especially HC84.26, demonstrated synergistic effects towards the majority of the HCVcc’s when combined individually with AR4A. Conclusion: Through a neutralization analysis of 10 clinically relevant HMAbs against 16 JFH1-based Core-NS2 recombinants from genotypes 1a, 1b, 2a, 2b, 2c, and 3a, we identified at least 3 HMAbs with potent and broad neutralization potential. The neutralization synergism obtained when pooling the most potent HMAbs could have significant implications for developing novel strategies to treat and control HCV.
Keywords: HCVcc, cell culture, neutralizing antibodies, therapeutic antibodies, vaccine
Hepatitis C virus (HCV) remains a major health burden, with over 130 million infected individuals at increased risk of developing liver cirrhosis and hepatocellular carcinoma1. Treatment regimens with interferon-α/ribavirin and direct-acting antivirals show variable effectiveness against different viral genotypes, induce viral escape, and cause severe side effects2. Neutralizing antibodies (NAbs) could improve treatment efficacy or be a prophylactic measure following liver transplantation in HCV patients, as in hepatitis B patients3. Recently, interest in humoral immunity has increased following studies showing that HCV-specific NAbs play a protective role against this genetically diverse virus4-8. HCV is divided into six clinically important genotypes and numerous subtypes, which differ on the nucleotide level by ~30% and 15-20%, respectively9. Genotypes 1–3 account for an estimated 80% of HCV infections worldwide10.
The HCV envelope proteins, E1 and E2, form a heterodimer on the virion surface with key roles in the host-cell infection, being the main target for NAbs. The virus has a high amino acid (aa) substitution rate in infected patients, providing evasion against host-derived immune responses11. Combined with extensive global HCV heterogeneity, this is a major challenge for the development of treatment and vaccine strategies11. The utilization of human monoclonal antibodies (HMAbs) with broadly neutralizing capabilities targeting conserved viral epitopes could constitute novel regimens in HCV prophylaxis and therapy. In a recent study, we found that HCV isolates that were resistant to polyclonal antibodies derived from patients with chronic HCV were sensitive to neutralization by HMAbs12. Several HCV-specific HMAbs with clinical potential have been developed13-18. These HMAbs demonstrate neutralizing capabilities in vitro and in vivo, with the most efficient candidates targeting epitopes on E2 or the E1/E2 complex19. Although a number of neutralizing HMAbs have been tested for their cross-neutralizing capabilities against single HCVcc isolates from genotypes 1-613,14,16, a thorough investigation of HMAb efficacy within and among HCV genotypes has not been performed20.
Since the discovery of HCV, the lack of cell culture-derived HCV (HCVcc) panels comprising strains reflecting global HCV diversity has hampered analyses of emerging antiviral drugs and HMAbs for efficacy on a broad range of viral isolates. With the discovery of the 2a JFH1 strain21, and the subsequent generation of JFH1-based Core/E1/E2/p7/NS2 (Core-NS2) recombinants, HCVcc systems for all major genotypes and important subtypes have been developed12,22-28. With the aim of identifying therapeutically relevant HMAbs with broad HCV neutralization capability across genotypes, subtypes, and isolates, we tested the efficacy of 10 HMAbs targeting various sites on HCV E1/E213-18,29 against a panel of 16 JFH1-based recombinants comprising Core-NS2 of genotypes 1-312,22,24,26,27,30. Moreover, HMAbs with complementary neutralization profiles were selected for cooperation analyses in which the degree of neutralization synergy was evaluated.
Materials and Methods
Cell culture assays
Culturing of Huh7.5 cells, in vitro transcription and transfection of HCV RNA genomes, and infection were conducted as described26. The percent infected cells was estimated every 2–3 days by immunostaining using anti-NS5A primary antibody (9E1024) and Alexa Flour 594 goat anti-mouse IgG (H+L) secondary antibody (Invitrogen). Cell supernatants were collected when HCV infection was >80%, and infectivity titers expressed as Focus Forming Units per milliliter (FFU/mL) were determined as described26,27.
JFH1-based recombinants
Previously developed genotypes 1–3 Core-NS2 JFH1-based recombinants were used, including adapted 1a (H77/JFH1V787A,Q1247L, TN/JFH1R1408W, DH6/JFH1V157A,V787A,S905C,Q1247L)26,27, 1b (J4/JFH1F886L,Q1496L, DH1/JFH1F886L,Q1496L, DH5/JFH1F886L,R1369Q,Q1496L)22,27, and 3a (DBN/JFH1W838R,K1398Q, S52/JFH1I793S,K1404Q)22,27 (aa numbering according to H77 reference, GenBank accession number AF009606), as well as 2a (J6/JFH1, T9/JFH1)12,24, 2b (DH8/JFH1, DH10/JFH1, J8/JFH1)12,22, and 2c (S83/JFH1)12 without adaptive mutations. In addition, we used JFH130. Furthermore, we constructed 3a recombinant DH11/JFH112,27. In short, we developed a 3078 nucleotide Core-NS2 consensus clone based on five clones derived from RT-PCR of extracted HCV RNA. The final DH11 Core-NS2 sequence was identical to the consensus nucleotide sequence. DH11/JFH1 was generated through ligation of DH11 Core-NS2 consensus into pJFH1 following AgeI (5′UTR) and SpeI (NS3) digests. T1089A, identified in another 3a recombinant27, was inserted by site-directed mutagenesis. In passaging DH11/JFH1T1089A, V783D was identified and introduced, thus generating DH11/JFH1V783D,T1089A. In the remainder of the text, the HCVcc name relates to the isolate-specific Core-NS2.
For each Core-NS2 recombinant and JFH1, stocks were prepared by inoculating Huh7.5 cells with a multiplicity of infection (MOI) of ~0.003. Virus stocks originated from 2nd or 3rd passage cell culture supernatant. The consensus E1/E2 sequence of virus recovered from final stocks was determined by direct sequencing of amplicons as described26,27. For the E1/E2 alignment, we used Molecular Evolutionary Genetics Analysis (MEGA5).
HCV-specific human monoclonal antibodies
The HMAbs selected for our study were: CBH-5 and CBH-7, which were derived from a HCV genotype 1b-infected patient15; HC-11 and the affinity maturated HC-1 (HC-1AM)18, which are from a 1a-infected individual29; HC33.4.10, HC84.24, and HC84.26, which are from a 2b-infected blood donor;14,16 and AR3A, AR4A, and AR5A, which are also from a 1a-infected patient13,17. The R04 and b6 monoclonal antibodies, which target cytomegalovirus15 and human immunodeficiency virus17 proteins, were used as isotype-matched controls. HMAb stocks were obtained from The Scripps Research Institute and Stanford University School of Medicine. In order to directly compare the antibody concentrations of individual HMAbs, human IgG content was quantified in-house at Hvidovre Hospital using Cobas c-systems (Roche/Hitachi).
HMAbs dose-response neutralization analysis
The neutralization activity of the HMAbs was quantified in a dose-response assay using FFUs as a read-out, as described12. In brief, 6×103 Huh7.5 cells/well were plated in a poly-D-lysine-coated 96-well plate. The following day, a volume of virus stock corresponding to a read-out of 15–300 FFU/well was mixed with a given HMAb in 5-fold dilutions ranging from 0.0012 to 100 μg/ml, incubated 1h at 37°C, and used to infect plated Huh7.5 cells 3h at 37°C. Depending on the HMAb, the isotype-matched antibodies R04 or b6 were included as controls15,17. Cells were washed and incubated for 45h, before HCV-specific staining and neutralization quantification by counting FFUs on an ImmunoSpot 5 UV analyzer (CTL Europe GmbH)27. Prior to the determination of percent neutralization, background FFUs, which were defined as the mean number of FFUs in six uninfected wells, were subtracted from all wells. Percent neutralization was then determined by comparing four replicate wells infected with virus/HMAb mixture relative to six replicate wells infected with virus alone.
The inhibitory concentration for 50% virus neutralization (IC50-value) was computed in GraphPad Prism 6 using a Sigmoidal dose-response curve (variable slope) with bottom and top constraints of 0 and 100. The IC50-value was only calculated when the 100 μg/ml concentration resulted in >50% neutralization. The statistical significance of differences between IC50-values for a given HMAb was determined using the Chi-squared test and Fisher’s exact test.
HMAb cooperation analysis
We used the median effect analysis method of Chou and Talalay and the well-established CompuSyn software (ComboSyn)14. Neutralization assays were performed as described above, but, for each antibody, a 2-fold dilution series was used, that ranged from 2−4 to 23-fold the IC50 value determined for the given antibodies. Dose-response neutralizations were done for the selected HMAbs alone and in combination against the HCVcc’s. The calculated percentage of virus neutralization was fed into CompuSyn as fractional effects (Fa) ranging from 0.01-0.99. Using Fa-values, dose-response curves for HMAbs, alone and in combination, were generated in CompuSyn, and the combination index (CI) was calculated using the Fa-values and curve shape/slope from the dose-response neutralization graph.
Results
Development of a panel of genetically heterogeneous HCVcc genotype 1-3 viruses for HMAb neutralization analysis
To obtain a panel of genotype 1-3 HCVcc’s encompassing the structural proteins of at least three patient-derived strains from each major genotype, we used previously developed JFH130 and JFH1-based Core-NS2 recombinants12,22,24,26,27, and a novel 3a (DH11) Core-NS2 recombinant. Thus, 16 HCVcc virus stocks of 1a (H77, TN, DH6), 1b (J4, DH1, DH5), 2a (J6, T9, JFH1), 2b (DH8, DH10, J8), 2c (S83), and 3a (DH11, DBN, S52) were generated. They had peak infectivity titers ranging from 103.3 to 105.3 FFU/ml and, except for JFH1, did not harbor E1/E2 aa substitutions. JFH1 had E2 aa change N417S, earlier identified as an adaptive mutation31.
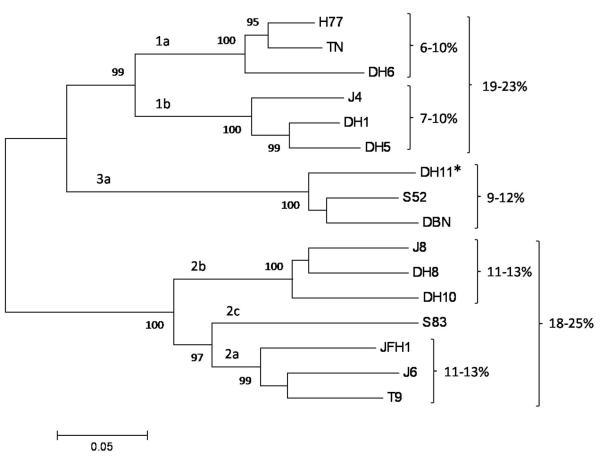
A phylogenetic analysis of the E1/E2 glycoproteins of the genotype 1-3 HCVcc’s is shown in Fig. 1. The percent aa difference between the E1/E2 of isolates from genotype 1 compared to isolates of genotypes 2 and 3 ranged from 30-35% and 28-30%, respectively, and the difference between isolates of genotypes 2 and 3 was 35-39%. Inter-subtype E1/E2 variation within genotypes 1 and 2 was 18-25%, and differences among isolates within subtypes were >6% (Fig. 1). Thus, the genotypes 1-3 HCVcc panel exhibited significant genetic diversity at the isolate, subtype, and genotype level, hence constituting a valuable tool to study the breadth of HMAb neutralization.
Fig. 1.
Phylogenetic analysis of the E1/E2 amino acid sequences of the genotype 1-3 HCVcc panel used for in vitro neutralization analysis. Multiple sequence alignment was computed with ClustalW (MEGA5 software). The phylogenetic tree was generated using a neighbor-joining algorithm. The HCV subtype and isolate names are indicated. A novel genotype 3a JFH1-based Core-NS2 recombinant (DH11/JFH1) is indicated with a star. Numbers at phylogenetic branches represent the percentage (≥75%) at which the included sequences cluster together in 1000 replicate multiple alignments through bootstrapping. The scale bar represents the evolutionary distance expressed as amino acid substitutions per site. Percentage ranges indicated to the right represent the amino acid differences among isolates within specific subtypes and genotypes, respectively.
Breadth of neutralization of HMAbs against the genotype 1-3 HCV panel
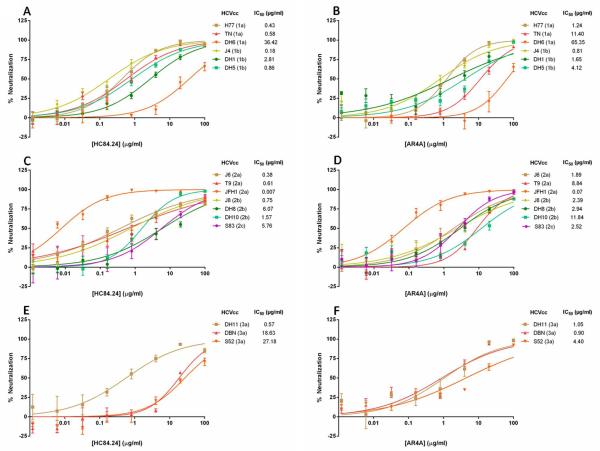
Ten previously identified HMAbs with therapeutic potential were tested for their neutralization capability by FFU reduction using the genetically diverse genotype 1-3 HCVcc panel13-18,29. We tested each HMAb in a dose-response analysis against 16 HCVcc isolates (Supplemental Fig. 1) and determined IC50 values (Table 1). Fig. 2 represents dose-response neutralization using HMAbs HC84.24 (A, C, E) and AR4A (B, D, F), which have distinct envelope targets.
Table 1.
Neutralization analysis using HMAb against HCVcc genotypes 1-3 panel
| Calculated IC50-values (μg/ml) for HMAbs | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| HCV isolate | Genotype | CBH-5 | CBH-7 | HC-11 | HC-1AM | HC33.4.10 | HC84.24 | HC84.26 | AR3A | AR4A | AR5A |
|
| |||||||||||
| H77 | 1a | 55 | 15 | 1.6 | 4.7 | 0.45 | 0.43 | 0.023 | 3.5 | 1.2 | 0.76 |
|
|
|||||||||||
| TN | 1a | 25 | >100 | 0.28 | 0.89 | 0.99 | 0.58 | 0.059 | 3.8 | 11 | 5.4 |
|
|
|||||||||||
| DH6 | 1a | >100 | >100 | >100 | 98 | >100 | 36 | >100 | 80 | 65 | 91 |
|
|
|||||||||||
| J4 | 1b | 0.25 | 5.9 | 0.70 | 0.13 | 0.56 | 0.18 | 0.022 | 75 | 0.81 | 2.8 |
|
|
|||||||||||
| DH1 | 1b | 4.0 | >100 | 3.6 | 2.7 | 15 | 2.8 | 0.43 | 6.8 | 1.6 | 33 |
|
|
|||||||||||
| DH5 | 1b | 0.95 | >100 | 1.6 | 0.28 | 0.78 | 0.86 | 0.24 | 2.1 | 4.1 | 3.3 |
|
|
|||||||||||
| J6 | 2a | 0.14 | 30 | 0.17 | 0.28 | 1.3 | 0.38 | 0.075 | 0.13 | 1.9 | 5.3 |
|
|
|||||||||||
| T9 | 2a | 4.4 | >100 | 23 | 2.9 | 24 | 0.61 | 1.4 | 5.4 | 8.8 | 25 |
|
|
|||||||||||
| JFH1 | 2a | 0.02 | 1.8 | 0.030 | 0.03 | 11 | 0.0078 | 0.014 | 0.0056 | 0.071 | 0.38 |
|
|
|||||||||||
| J8 | 2b | 1.2 | 59 | 0.82 | 1.7 | 6.6 | 0.75 | 0.14 | 1.1 | 2.4 | 6.9 |
|
|
|||||||||||
| DH8 | 2b | 0.27 | 19 | 0.18 | 0.64 | 82 | 6.1 | 0.91 | 0.93 | 2.9 | 30 |
|
|
|||||||||||
| DH10 | 2b | 2.8 | >100 | 23 | 19 | 46 | 1.6 | 0.040 | 3.4 | 12 | 23 |
|
|
|||||||||||
| S83 | 2c | 18 | >100 | 6.6 | 13 | >100 | 5.8 | 2.1 | 12 | 2.5 | >100 |
|
|
|||||||||||
| DH11 | 3a | 1.5 | >100 | 0.54 | 0.81 | 1.3 | 0.57 | 0.26 | 2.6 | 1.1 | 29 |
|
|
|||||||||||
| DBN | 3a | 23 | >100 | 66 | 15 | 6.1 | 19 | >100 | 24 | 0.90 | >100 |
|
|
|||||||||||
| S52 | 3a | 70 | >100 | >100 | 30 | 16 | 27 | 0.82 | >100 | 4.4 | >100 |
HMAbs unable to neutralize 50% of HCV virus input at 100 μg/ml are stated as >100 IC50-values are shown with 2 determining digits.
Fig. 2.
Dose-response neutralization of the genotype 1-3 HCVcc panel by HMAbs HC84.24 and AR4A. (A-B) Neutralization of genotype 1 Core-NS2 JFH1-based recombinants including H77 (1a), TN (1a), DH6 (1a), J4 (1b), DH1 (1b), and DH5 (1b); (C-D) Genotype 2: J6 (2a), T9 (2a), JFH1 (2a), J8 (2b), DH8 (2b), DH10 (2b), and S83 (2c); (E-F) Genotype 3: DH11 (3a), DBN (3a), and S52 (3a). HMAb concentrations ranged from 0.0012 to 100 μg/ml and were incubated in a 5-fold dilution series with the various virus stocks in a 96 well format. Inhibitory concentrations for 50% virus neutralization (IC50) values for each HCVcc are indicated. Controls for this experiment were isotype-matched antibodies R04 (for HC84.24) and b6 (for AR4A) (see Supplemental Fig. 1). All measurements were performed in 4 replicates and error bars show the standard error of the mean (SEM). For data on all HMAbs see Supplemental Fig. 1.
The neutralization effectiveness against the different HCVcc’s varied greatly among the HMAbs. Only HC84.24, AR4A, and HC-1AM neutralized at least 50% of the virus for all genotype 1-3 HCVcc strains at ≤100 μg/ml, thus conferring full breadth of protection (Table 1). The range of IC50-values across virus strains for HC84.24, AR4A, and HC-1AM was 0.01-36 μg/ml, 0.07-65 μg/ml, and 0.03-98 μg/ml, respectively. In addition, AR3A had low IC50-values against most strains (Table 1). Although HC84.26 exhibited poor neutralization against DH6 and DBN (IC50-values >100 μg/ml), this HMAb had the most efficient neutralization (Fig. 3) with an IC50-value range from 0.01-2.08 μg/ml against the 14 remaining strains (Table 1).
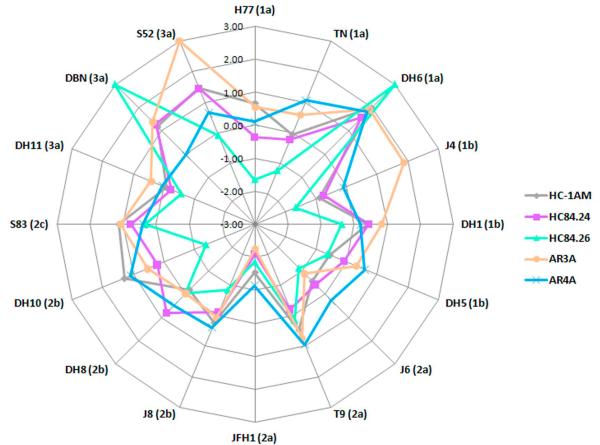
Fig. 3.
Radar chart of HMAb neutralization patterns against the 16 HCVcc JFH1-based Core-NS2 recombinants of the genotype 1-3 panel. Only HMAbs exhibiting the most efficient neutralization ability, i.e. HC84.24, HC84.26, AR3A, AR4A, and HC-1AM are shown. Values in the radar chart display log10(IC50) values, with the maximum measured log(IC50) = 2. HCVcc with IC50 >100 μg/ml were defined as 3. Each data point represents an IC50-value against a specific virus isolate. The color code in the connecting lines represents the various included HMAbs. The selection of antibodies for cooperation analysis was based on minimizing the area towards the center of the graphs and the differences in epitope targets, thus suggesting that two given HMAbs would complement their respective IC50-values.
To identify HMAbs with the highest efficacy, we compared the IC50-value of each HMAb against a given HCVcc to the IC50 values of each of the other HMAbs against the given HCVcc. We then compiled the data for all the viruses in the panel and determined the statistical significance using Fisher’s exact test (Table 2). Thus, neutralization using HC84.26 against specific HCVcc’s was significantly better than 8 of the 9 remaining HMAbs, HC84.24 was significantly better than 5 of the 9, HC-1AM was significantly better than 3 of the 9, and AR4A neutralization was significantly better than 2 of the 9 remaining HMAbs (Table 2).
Table 2.
IC50 values-based neutralization comparison between pairs of HMAb
| HMAb | CBH-5 | CBH-7 | HC-11 | HC-1AM | HC33.4.10 | HC84.24 | HC84.26 | AR3A | AR4A | AR5A |
|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||
| CBH5 | 14 vs 1 | 7 vs 8 | 3 vs 13 | 9 vs 6 | 1 vs 15 | 1 vs 14 | 9 vs 7 | 8 vs 8 | 13 vs 3 | |
|
|
||||||||||
| CBH7 | 0 vs14 | 0 vs 16 | 2 vs 12 | 0 vs 16 | 0 vs 14 | 1 vs 15 | 0 vs 16 | 1 vs 15 | ||
|
|
||||||||||
| HC-11 | 7 vs 9 | 10 vs 5 | 4 vs 12 | 2 vs 13 | 9 vs 6 | 8 vs 8 | 13 vs 2 | |||
|
|
||||||||||
| HC-1AM | 13 vs 3 | 5 vs 11 | 3 vs 13 | 7 vs 9 | 9 vs 7 | 13 vs 3 | ||||
|
|
||||||||||
| HC33.4.10 | 3 vs 13 | 1 vs 14 | 7 vs 9 | 5 vs 11 | 11 vs 4 | |||||
|
|
||||||||||
| HC84.24 | 4 vs 12 | 13 vs 3 | 11 vs 5 | 16 vs 0 | ||||||
|
|
||||||||||
| HC84.26 | 13 vs 3 | 14 vs 2 | 14 vs 1 | |||||||
|
|
||||||||||
| AR3A | 8 vs 8 | 13 vs 2 | ||||||||
|
|
||||||||||
| AR4A | 13 vs 3 | |||||||||
|
|
||||||||||
| AR5A | ||||||||||
The efficacy difference between HMAbs neutralization was analyzed by comparing each HCVcc IC50-values (<100 μg/ml) of one HMAb with IC50-values from the corresponding HCVcc of another HMAb followed by calculation of statistical significance using Fisher’s exact test.
Numbers in the matrix to the left of vs relates to HMAbs in the left column and number to the rigth related to HMAbs in top row Statistical significant p-values <0.05 is displayed in blue and <0.01 in purple. Non-significant values are shown in black.
The HMAb with the least cross-neutralization ability was CBH-7, in which IC50-values for only 6 of 16 HCVcc (H77, J4, J6, JFH1, J8, and DH8) could be calculated. Moreover, CBH-5, HC-11, HC33.4.10, and AR5A had a lower degree of cross-neutralization potential compared to the other HMAbs.
The IC50-values identified for each HMAb against the various HCVcc strains were very diverse. Using Chi-square analysis, the differences in IC50-values against the 16 HCVcc strains for each HMAb were found to be highly statistically significant (p<10−6) supporting the use of a large panel of HCV isolates (Table 3). The DH6 isolate exhibited the highest level of neutralization resistance with the lowest IC50 being 36 μg/ml. In contrast, JFH1 had the lowest IC50-value against all HMAbs except HC33.4.10.
Table 3.
IC50-value heterogenity analysis against HCVcc genotypes 1-3 panel
| CBH5 | CBH7 | HC.11 | HC.1AM | HC33.4.10 | HC84.24 | HC84.26 | AR3A | AR4A | AR5A | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||||
|
Df
p.value |
14 | 5 | 13 | 15 | 13 | 15 | 13 | 14 | 15 | 12 | |
|
|
|||||||||||
| <10−6 | <10−6 | <10−6 | <10−6 | <10−6 | <10−6 | <10−6 | <10−6 | <10−6 | <10−6 | ||
The statistical significant difference between IC50-values within a HMAb was tested with Chi-squared test.
Degrees of freedom was defined as the number of HCVcc with IC50-value <100 μg/ml minus 1.
A p-value <0.01 was considered statistically significant
1:1 pooling of AR4A with HC84.24, HC84.26 or AR3A induced synergy in neutralization of genotype 1-3 HCVcc’s
We investigated the synergistic potential of the HMAbs with the highest neutralization capabilities using cooperation analysis14. HC84.24, HC84.26, and AR4A were selected for this study based on the observed low IC50-values and reported differences in epitope targets (Supplemental Fig. 2). AR4A was used as the main candidate, since it complemented HC84.24 and HC84.26 in HCVcc neutralization efficacy across isolates (Fig. 3) and has a different epitope target13,16,29. In addition, selection of these HMAbs for additional investigation was supported by recent studies, which demonstrated their neutralization ability across genotypes13,16. Although, overall, HC84.26 exhibited the lowest IC50-values, it was unable to neutralize DBN and DH6. In addition, due to insufficient amounts of HC84.24, the cooperation analysis against these recombinants could not be performed. Hence, the AR3A was included to investigate possible synergy against the neutralization resistant DH6 and DBN. Thus, we performed the cooperation analysis, using AR4A in combination with HC84.24, HC84.26 or AR3A, respectively, against the genotype 1-3 HCVcc panel.
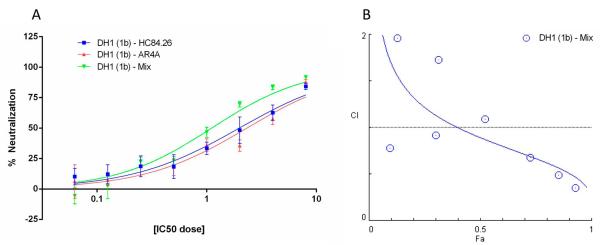
In general, HCV isolates for which the IC50-value was ≤100 μg/ml in the initial neutralization (Table 1) were included for investigation. Figure 4A shows a representative dose-neutralization curve for the cooperation analysis of HC84.26 and AR4A in the neutralization of DH1. Percent neutralizations at a given antibody concentration were input into the CompuSyn software as a fractional effect (Fa), and the Fa values were used to calculate the Combination Index (CI), which represents a quantitative measurement of the degree of cooperation at a given Fa endpoint. Fig. 4B illustrates a sample CI-Fa plot of HC84.26 and AR4A. CI-values at essential Fa endpoints (50%, 75%, and 90% effective dose (ED)) are shown in Table 4 for HMAb combinations against the HCVcc panel. Interestingly, the majority of the HMAbs tested had CI-values defined as synergistic in HCVcc neutralization. The synergistic effect ranged from slight to strong synergism, although the majority had moderate synergism (Table 4).
Fig. 4.
Cooperativity in virus neutralization between HMAbs HC84.26 and AR4A against the DH1(1b) recombinant. (A) Dose-response neutralization of HC84.26/AR4A alone and in combination in a constant ratio 2-fold dilution series using concentrations ranging from 2−4- to 23-fold of the previously determined IC50-values. (B) Neutralization percentages, represented as a fractional effect (Fa) from 0.01-0.99, were input into the CompuSyn software in relation to the corresponding HMAb dose. The combination index (CI) plot, displaying the calculated CI for the combined HC84.26/AR4A in relation to Fa, was subsequently computed as described previously13,14,38. The CI value was calculated based on the Fa values from the HC84.26/AR4A combination relative to the Fa values obtained for the HMAbs when tested individually. Blue symbols represent combined HC84.26/AR4A. The various CI-values for HC84.26/AR4A against the HCVcc panel are shown in Table 4. Following the CompuSyn recommendations, a CI-value of 0.1-0.3 was defined as strong synergism; 0.3-0.7 as synergism; 0.7-0.85 as moderate synergism; 0.85-0.9 as slight synergism; 0.9-1.1 as nearly additive; 1.1-1.2 as slight antagonism; 1.2-1.45 as moderate antagonism; 1.45-3.3 as antagonism; 3.3-10 as strong antagonism.
Table 4.
Corporation analysis using HMAb HC84.24/AR4A, HC84.26/AR4A and AR3A/AR4A
| HCV isolate | Genotype | HC84.24 + AR4A | Curve r-values | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| ED50 | ED70 | ED90 | HC84.24 | AR4A | Mix | ||
|
|
|||||||
| H77 | 1a | 0.92 | 0.91 | 0.90 | 0.98 | 0.98 | 0.97 |
|
|
|||||||
| TN | 1a | 1.38 | 0.88 | 0.57 | 0.92 | 0.98 | 0.94 |
|
|
|||||||
| DH6 | 1a | N/A | N/A | N/A | N/A | N/A | N/A |
|
|
|||||||
| J4 | 1b | 0.88 | 0.72 | 0.66 | 0.97 | 0.98 | 0.96 |
|
|
|||||||
| DH1 | 1b | 1.75 | 1.45 | 1.20 | 0.99 | 0.98 | 0.98 |
|
|
|||||||
| DH5 | 1b | 0.21 | 0.32 | 0.47 | 0.97 | 0.98 | 0.98 |
|
|
|||||||
| J6 | 2a | 0.44 | 0.38 | 0.52 | 0.97 | 0.95 | 0.98 |
|
|
|||||||
| T9 | 2a | 1.06 | 0.87 | 0.72 | 0.99 | 0.99 | 0.99 |
|
|
|||||||
| JFH1 | 2a | 1.18 | 0.97 | 0.81 | 0.93 | 0.96 | 1.00 |
|
|
|||||||
| J8 | 2b | 0.81 | 0.44 | 0.36 | 0.99 | 0.99 | 0.98 |
|
|
|||||||
| DH8 | 2b | 0.46 | 0.30 | 0.20 | 0.99 | 0.98 | 0.97 |
|
|
|||||||
| DH10 | 2b | 0.88 | 0.70 | 0.56 | 0.95 | 1.00 | 0.98 |
|
|
|||||||
| S83 | 2c | 0.55 | 0.51 | 0.48 | 0.98 | 0.94 | 0.98 |
|
|
|||||||
| DH11 | 3a | 0.34 | 0.38 | 0.42 | 0.87 | 0.76 | 0.91 |
|
|
|||||||
| DBN | 3a | N/A | N/A | N/A | N/A | N/A | N/A |
|
|
|||||||
| S52 | 3a | 0.90 | 0.99 | 1.11 | 0.82 | 0.96 | 0.89 |
| HCV isolate | Genotype | HC84.26 + AR4A | Curve r-values | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| ED50 | ED75 | ED90 | HC84.26 | AR4A | Mix | ||
|
|
|||||||
| H77 | 1a | 1.03 | 0.74 | 0.54 | 0.95 | 0.99 | 0.99 |
|
|
|||||||
| TN | 1a | 0.65 | 0.55 | 0.50 | 0.88 | 0.96 | 0.94 |
|
|
|||||||
| DH6 | 1a | N/A | N/A | N/A | N/A | N/A | N/A |
|
|
|||||||
| J4 | 1b | 0.40 | 0.51 | 0.67 | 0.95 | 0.98 | 0.99 |
|
|
|||||||
| DH1 | 1b | 0.84 | 0.71 | 0.61 | 0.98 | 0.94 | 0.98 |
|
|
|||||||
| DH5 | 1b | 0.75 | 0.60 | 0.49 | 0.97 | 0.94 | 0.96 |
|
|
|||||||
| J6 | 2a | 0.62 | 0.69 | 0.78 | 0.97 | 0.98 | 0.95 |
|
|
|||||||
| T9 | 2a | 0.61 | 0.54 | 0.49 | 0.98 | 0.99 | 0.97 |
|
|
|||||||
| JFH1 | 2a | 1.22 | 1.06 | 0.93 | 0.99 | 0.99 | 0.99 |
|
|
|||||||
| J8 | 2b | 1.05 | 0.89 | 0.83 | 0.99 | 0.98 | 0.99 |
|
|
|||||||
| DH8 | 2b | 0.19 | 0.37 | 0.74 | 0.98 | 0.94 | 0.94 |
|
|
|||||||
| DH10 | 2b | 0.63 | 0.63 | 0.66 | 0.96 | 0.97 | 0.99 |
|
|
|||||||
| S83 | 2c | 0.80 | 0.82 | 0.85 | 0.98 | 0.98 | 0.99 |
|
|
|||||||
| DH11 | 3a | 1.27 | 1.20 | 1.16 | 0.98 | 0.95 | 0.97 |
|
|
|||||||
| DBN | 3a | N/A | N/A | N/A | N/A | N/A | N/A |
|
|
|||||||
| S52 | 3a | 0.50 | 0.45 | 0.43 | 0.98 | 0.99 | 0.97 |
| HCV isolate | Genotype | AR3A + AR4A | Curve r-values | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| ED50 | ED75 | ED90 | AR3A | AR4A | Mix | ||
|
|
|||||||
| H77 | 1a | 1.54 | 0.85 | 0.51 | 0.94 | 0.99 | 0.93 |
|
|
|||||||
| TN | 1a | 1.07 | 0.74 | 0.54 | 0.98 | 0.95 | 0.97 |
|
|
|||||||
| DH6 | 1a | 0.52 | 0.57 | 0.72 | 0.95 | 0.99 | 0.95 |
|
|
|||||||
| J4 | 1b | 0.19 | 0.12 | 0.08 | 0.98 | 0.99 | 0.98 |
|
|
|||||||
| DH1 | 1b | 0.81 | 0.75 | 0.71 | 0.99 | 0.98 | 0.99 |
|
|
|||||||
| DH5 | 1b | 0.49 | 0.37 | 0.32 | 0.90 | 0.97 | 0.97 |
|
|
|||||||
| J6 | 2a | 0.97 | 0.82 | 0.74 | 0.97 | 0.99 | 0.98 |
|
|
|||||||
| T9 | 2a | 0.54 | 0.49 | 0.48 | 0.93 | 0.97 | 0.98 |
|
|
|||||||
| JFH1 | 2a | 1.23 | 0.98 | 0.80 | 0.97 | 0.97 | 0.99 |
|
|
|||||||
| J8 | 2b | 0.59 | 0.95 | 1.84 | 0.97 | 0.98 | 0.98 |
|
|
|||||||
| DH8 | 2b | 1.80 | 1.86 | 2.44 | 0.96 | 0.97 | 0.94 |
|
|
|||||||
| DH10 | 2b | 0.58 | 0.51 | 0.54 | 0.95 | 0.94 | 0.97 |
|
|
|||||||
| S83 | 2c | 0.70 | 0.49 | 0.40 | 0.97 | 0.97 | 0.98 |
|
|
|||||||
| DH11 | 3a | 5.10 | 2.80 | 1.55 | 0.93 | 0.95 | 0.95 |
|
|
|||||||
| DBN | 3a | 0.93 | 0.92 | 0.92 | 0.97 | 0.95 | 0.94 |
|
|
|||||||
| S52 | 3a | N/A | N/A | N/A | N/A | N/A | N/A |
CI-values of HC84.24, HC84.26 or AR3A combined with AR4A at Fa-values of ED50, ED75, and ED90 are shown.
CI-value of 0.1-0.3 was defined as strong synergism; 0.3-0.7 as synergism; 0.7-0.85 as moderate synergism; 0.85-0.9 as slight synergism; 0.9-1.1 as nearly additive; 1.1-1.2 as slight antagonism; 1.2-1.45 as moderate antagonism; 1.45-3.3 as antagonism; 3.3-10 as strong antagonism.
Based on the CI-values at ED75 and ED90, synergism was identified in 10 of 14 HCVcc’s when combining HC84.24/AR4A, with CI-values ranging from 0.20-0.88; in 12 of 14 strains for HC84.26/AR4A, with CI-values ranging from 0.37-0.89; and in 10 of 15 HCVcc for AR3A/AR4A, with CI-values ranging from 0.08-0.85 (Table 4). Only H77, DH1, JFH1, and S52 (HC84.24/AR4A), JFH1 and DH11 (HC84.26/AR4A), and JFH1, J8, DH8, DH11, and DBN (AR3A/AR4A) did not exhibit direct synergy at ED75 and ED90, showing CI-values from 0.90-2.80, which define a nearly additive-to-antagonistic effect. At ED50 9 of 14 (HC84.24/AR4A), 10 of 14 (HC84.26/AR4A), and 8 of 15 (AR3A/AR4A) HCVcc isolates exhibited synergism with CI-values ranging from 0.19-0.90. At ED50, only a minority of the HMAbs’ cooperative effect against the HCVcc recombinants converted from synergistic to nearly additive or antagonistic compared to the ED75 and ED90 (Table 4). Even though synergistic effects of HMAbs were not seen against some viruses in the HCVcc panel, pooling of these HMAbs still efficiently neutralized these viruses in a dose-response manner. Interestingly, analysis of AR3A/AR4A cooperation against the neutralization resistant DH6 showed a synergistic effect ranging from 0.52-0.72 at ED50, ED75, and ED90 (Table 4).
Overall, these results indicate synergistic effects of combining AR4A with HC84.24, HC84.26 or AR3A against genotypes 1-3 HCVcc strains. Thus, the approach for selection of NAbs for possible synergistic effect, i.e. complementing neutralization efficacy and distinct epitope targets between HMAbs, seems feasible. Synergism could be identified in the majority of cases, albeit with a wide range of CI-values. This further supports the use of a diverse HCVcc panel.
Discussion
We used a panel of 16 JFH1-based Core-NS2 HCVcc virus stocks representing clinically important genotypes 1 (subtypes 1a and 1b), 2 (subtypes 2a, 2b, and 2c), and 3 (subtype 3a) to investigate the neutralization potential of 10 HMAbs that target various epitopes on the HCV envelope glycoproteins. The HCVcc panel represented the expected genetic heterogeneity (Fig. 1), thus, enabling identification of differences between genotypes, subtypes, and isolates. We observed a high level of variation in the neutralization capability between HMAbs in dose-response analyses. Importantly, this diversity extended to the individual HMAbs, which demonstrated significant differences between IC50-values. Combining highly potent HMAbs, we identified synergistic effects for HC84.24/AR4A, HC84.26/AR4A, and AR3A/AR4A towards most HCV strains, which may be relevant for future HCV prophylaxis and therapy.
Identification of conserved immunogenic envelope epitopes has been a challenge in the development of broadly neutralizing antibodies against HCV. Most epitopes are located in highly variable domains, such as hypervariable region 1, where NAb efficacy is influenced by single-isolate specificity and viral escape mutations32. In contrast, antigenic domains B and D (nomenclature as defined in15,16) within E2 show a higher level of conservation with verified immunogenicity16. In addition, these domains overlap with the CD81 binding site and feature overlapping conformational epitopes of which one or more amino acids serve as targets for many of the HMAbs included in this study, which show significant neutralization ability against multiple HCVcc strains. CBH-5, HC-1AM, and HC-11 target residues in domain B and HC84.24 and HC84.26 target residues in domain D15,16,18,29, and this study further confirms the broad immunogenicity of these E2 glycoprotein epitopes (Table 1 and 2). These two adjacent antigenic domains are located within the well-defined antigenic region 3 (AR3) of the E2 crystal structure33. In addition, we found that CBH-7, which targets antigenic domain C in E215, exhibits the lowest breadth of neutralization (Table 1). Finally, there are efficient HMAbs, such as AR4A and AR5A, with targets outside of antigenic domains B, C, and D. These antibodies target conserved discontinuous epitopes in the E1/E2 quaternary structure, which involve residue D698 for AR4A and R639 for AR5A13.
It would be natural to ascribe variation in neutralization among the HMAbs to genetic heterogeneity resulting in epitope differences, particularly between subtypes and genotypes. However, our data does not support any obvious correlation between the HCV genotype infecting the patient from whom the antibody was derived and their neutralization capability against the genotypes of the HCVcc panel (Table 1). Similar findings were previously reported using polyclonal antibodies20. In general, genotypes 1b and 2a appeared to have the highest neutralization susceptibility. However, it should be noted that genotype 3a HCVcc isolates exhibited the overall highest IC50 values and that none of the included HMAbs originated from genotype 3-infected patients (Table 1). Moreover, our results did not suggest that differences in HCVcc neutralization sensitivity could be explained by specific differences in mapped determinants of the given epitope (Supplemental Fig. 2). S83 represents one such example in which lack of neutralization using HC33.4.10 and AR5A cannot be explained by differences in mapped determinant residues for these antibodies (HC33.4.10: L413, G418, W420, and N423; AR5A: R639)13-15. This phenomenon was most pronounced for DH6, as this isolate was highly resistant to neutralization despite having intact target epitopes in the consensus envelope protein sequences. Hence, only five HMAbs were able to neutralize the DH6 recombinant (Table 1). Interestingly, a recent study showed that introduction of the I414T and Y444H adaptive mutations into DH6 increased the HCVcc neutralization sensitivity by several orders of magnitude when tested against patient-derived polyclonal NAbs34. An HCV genotype 1a isolate containing Y444 in concert with N501 and A506 has been reported to resist neutralization by all antigenic domain B antibodies, as well as an antibody to the highly conserved E2 segment spanning amino acids 412-42335. Neutralization sensitivity was restored with Y444H or Y444Q substitutions, in combination with N501S and A506V, in this 1a isolate. One explanation for this might involve the unshielding of target epitopes with importance for HMAb neutralization. Furthermore, secondary and tertiary structures in the HCV envelope proteins, including variation in co-factor interactions, could influence neutralization.
Our results support the use of a diverse panel of cell culture virus systems expressing authentic envelope proteins in assessing the neutralization efficacy of MAbs. Except for genotype 2 recombinants, all HCVcc required adaptive mutations for efficient spread in cell culture12,22,24,26,27. Nevertheless, the envelope proteins in the HCVcc panel used in this study did not contain adaptive mutations. The JFH1 virus exhibited, in all cases but one (HC33.4.10), the lowest IC50-value. However, the N417S E2 substitution, previously demonstrated to be relevant for virus in vitro viability31, was identified. Since several of the tested HMAbs target epitopes that encompass or are adjacent to position 417, this aa could influence neutralization potential. This assumption is supported by the significant increase in neutralization in a DH6/JFH1 recombinant with I414T and Y444H34. Positions 414, 417, and 444 are all located in neutralization epitopes13,14,16. More importantly, N417 represents a potential glycosylation site involved in the reported glycan shield of HCV36. As JFH1 is one of the most frequently used isolates in HCV neutralization analyses, these findings could have implications on the conclusions derived from these studies.
Based on the pattern of IC50-values and the previously mapped epitope targets of the HMAbs13,16, we pooled HC84.24/AR4A and HC84.26/AR4A for cooperation analysis (Table 4). In addition, we tested AR3A/AR4A in an attempt at attaining synergism against the neutralization resistant DH6. Our finding that AR4A demonstrates broad synergism corroborates the study by Giang et al.13, in which AR4A, in combination with AR3A and/or AR5A, showed synergism against H77(1a) HCV pseudo-particles. HC84.26 has been shown to have slight synergism against JFH1 HCVcc, but only at high antibody concentrations and in combination with HMAbs targeting epitopes in close proximity14. Contact residues for HC84.24 include C429, F442 and Y443, while contact residues for HC84.26 include L441 and F442, and AR3A primarily targets S424, G523, P525, G530, D535, V538, and N540 in the CD81 binding site. In contrast, AR4A requires the presence of D698 on a properly folded E1E2 complex. Hence, differences in HCV epitope targets could facilitate the broad synergism observed, perhaps by decreasing competition in antibody binding, thus inducing a cumulative effect on particle entry inhibition. This could be mediated through direct inhibition by prevention of host cell receptor binding, or indirectly, by interference with co-receptors and/or entry factors5.
This study presents, to our knowledge, the most potent synergistic effect demonstrated on HCV virus particles. Synergism between HC84.24/AR4A, HC84.26/AR4A, and AR3A/AR4A markedly increases their therapeutic relevance, since few HMAbs have previously shown cooperative potential13,14. The diversity of the HCVcc panel used for these analyses offers hope that a truly cross-genotype/subtype effective HMAb mixture against HCV can be found. A previous study demonstrated that the AR4A antibody inhibited HCV genotypes 1b and 2a infection in a mouse model13, thus demonstrating the in vivo relevance of this HMAb. It would be of great interest to further compare the efficiency of the HMAb mixtures tested in this study in vivo. Finally, the ability of HCV to escape the neutralizing effects of these antibodies by the development of specific escape mutations should be investigated and compared. Clearly, combining escape-resistant HMAbs with a broad and often synergistic effect against HCV isolates of any genotype/subtype would constitute an ideal choice in cases where immunotherapy is relevant.
Passive immunity in patients using NAbs has been under much investigation. Although this strategy against HCV is not yet applicable in humans, previous studies have shown the ability of neutralizing antibodies to prevent HCV infection in animal models6,8,13,37. In light of this, the results using HC84.24, HC84.26, AR3A, and AR4A could potentially constitute the basis for future antibody-based HCV treatments, and prevention of recurrent infection following liver transplantation. The genotype 1-3 HCVcc panel described herein represents a valuable tool for further HMAb efficacy analysis as well as for future studies on HCV envelope proteins, and their use in HCV vaccine development.
Supplementary Material
Supplemental Fig. 1. Dose-response neutralization of 16 genotype 1-3 HCVcc’s by the HMAbs CBH-5, CBH-7, HC-11, HC-AM1, HC33.4.10, HC84.24, HC84.26, AR3A, AR4A, and AR5A. The JFH1-based Core-NS2 HCVcc recombinants were: Genotype 1: H77 (1a), TN (1a), DH6 (1a), J4 (1b), DH1(1b), and DH5 (1b); Genotype 2: J6 (2a), T9 (2a), JFH1 (2a), J8 (2b), DH8 (2b), DH10 (2b), and S83 (2c); Genotype 3: DH11 (3a), DBN (3a), and S52 (3a). HMAb concentrations ranged from 0.0012-100 μg/ml, and were incubated in 5-fold dilutions with appropriate FFU input readout titers of the various virus stocks in a 96 well format. Inhibitory concentrations for 50% virus neutralization (IC50) values for each HCVcc are indicated. Control for this experiment was isotype-matched antibodies R04 (CBH-5, CBH-7, HC-11, HC-AM1, HC33.4.10, HC84.24, and HC84.26) and b6 (AR3A, AR4A, and AR5A) (open symbols). All measurements were performed in 4 replicates and error bars show the standard error of the mean (SEM) of the replicates.
Supplemental Fig. 2. Multiple sequence alignment of E1/E2 amino acid sequences of the 16 JFH1-based genotype 1-3 Core-NS2 recombinants constituting the HCVcc panel. The HCVcc strains were: Genotype 1: H77 (1a), TN (1a), DH6 (1a), J4 (1b), DH1 (1b), and DH5 (1b); Genotype 2: J6 (2a), T9 (2a), JFH1 (2a), J8 (2b), DH8 (2b), DH10 (2b), and S83 (2c); Genotype 3: DH11 (3a), DBN (3a), and S52 (3a). Conserved amino acids among the HCVcc strains are labeled in white and non-conserved are labeled in gray. HMAb target residues or domains are presented in color-coding as indicated. Potential glycosylation sites are indicated in purple. Residues with dual coloring represent residues involved in binding of either more than one HMAb or a shared glycosylation/antibody target. Multiple sequence alignment was computed with ClustalW (MEGA5 software).
Acknowledgements
We thank Troels Scheel, Nanna Hansen and Judith Gottwein for their contribution to the development of the DH11/JFH1 recombinant, Judith Gottwein for her input on analysis of synergy, Daryl Humes for proofreading and editorial input, Anna-Louise Sørensen for sample handling, and Steen Ladelund for statistical advice (all from Hvidovre Hospital).
In addition we are grateful to the Department of Clinical Biochemistry, Hvidovre Hospital, for in-house quantification of IgG. Also we thank Jens Ole Nielsen, Bjarne Ørskov Lindhart and Ove Andersen (Hvidovre Hospital) for supporting this study, and Charles Rice (Rockefeller University, New York) and Takaji Wakita (National Institute of Infectious Diseases, Tokyo) for providing reagents.
This study was supported by the Lundbeck Foundation (JCP, JB), the Danish Cancer Society (JB), the Novo Nordisk Foundation (JB), the RegionH Research Fund (JB), the Danish Council for Independent Research (JB), a Ph.D. stipend from the Faculty of Health and Medical Sciences, University of Copenhagen (THRC), and an Individual Postdoctoral Stipend from the Danish Council for Independent Research (JCP). ML is supported by the United States National Institutes of Health grant number AI79031. SF is supported by the United States National Institutes of Health grant number AI108024.
Abbreviations
- HCV
hepatitis C virus
- HCVcc
cell culture derived hepatitis C virus
- NAb
neutralizing antibody
- HMAb
human monoclonal antibody
- MOI
multiplicity of infection
- FFU
Focus Forming Units
- aa
amino acid
- SEM
standard error of the mean
Footnotes
Potential conflict of interest: Nothing to report
Reference List
- 1.Mohd HK, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57(4):1333–1342. doi: 10.1002/hep.26141. [DOI] [PubMed] [Google Scholar]
- 2.Scheel TK, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med. 2013;19(7):837–849. doi: 10.1038/nm.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guillouche P, Feray C. Systematic review: anti-viral therapy of recurrent hepatitis C after liver transplantation. Aliment Pharmacol Ther. 2011;33(2):163–174. doi: 10.1111/j.1365-2036.2010.04505.x. [DOI] [PubMed] [Google Scholar]
- 4.Bartosch B, Bukh J, Meunier JC, Granier C, Engle RE, Blackwelder WC, et al. In vitro assay for neutralizing antibody to hepatitis C virus: evidence for broadly conserved neutralization epitopes. Proc Natl Acad Sci U S A. 2003;100(24):14199–14204. doi: 10.1073/pnas.2335981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorner M, Horwitz JA, Robbins JB, Barry WT, Feng Q, Mu K, et al. A genetically humanized mouse model for hepatitis C virus infection. Nature. 2011;474(7350):208–211. doi: 10.1038/nature10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meuleman P, Bukh J, Verhoye L, Farhoudi A, Vanwolleghem T, Wang RY, et al. In vivo evaluation of the cross-genotype neutralizing activity of polyclonal antibodies against hepatitis C virus. Hepatology. 2011;53(3):755–762. doi: 10.1002/hep.24171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pestka JM, Zeisel MB, Blaser E, Schurmann P, Bartosch B, Cosset FL, et al. Rapid induction of virus-neutralizing antibodies and viral clearance in a single-source outbreak of hepatitis C. Proc Natl Acad Sci U S A. 2007;104(14):6025–6030. doi: 10.1073/pnas.0607026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vanwolleghem T, Bukh J, Meuleman P, Desombere I, Meunier JC, Alter H, et al. Polyclonal immunoglobulins from a chronic hepatitis C virus patient protect human liver-chimeric mice from infection with a homologous hepatitis C virus strain. Hepatology. 2008;47(6):1846–1855. doi: 10.1002/hep.22244. [DOI] [PubMed] [Google Scholar]
- 9.Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, et al. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment Web resource. Hepatology. 2014;59(1):318–327. doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li YP, Ramirez S, Humes D, Jensen SB, Gottwein JM, Bukh J. Differential sensitivity of 5′UTR-NS5A recombinants of hepatitis C virus genotypes 1-6 to protease and NS5A inhibitors. Gastroenterology. 2014;146(3):812–821. doi: 10.1053/j.gastro.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 11.Dustin LB, Rice CM. Flying under the radar: the immunobiology of hepatitis C. Annu Rev Immunol. 2007;25:71–99. doi: 10.1146/annurev.immunol.25.022106.141602. [DOI] [PubMed] [Google Scholar]
- 12.Pedersen J, Carlsen TH, Prentoe J, Ramirez S, Jensen TB, Forns X, et al. Neutralization resistance of hepatitis C virus can be overcome by recombinant human monoclonal antibodies. Hepatology. 2013;58(5):1587–1597. doi: 10.1002/hep.26524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giang E, Dorner M, Prentoe JC, Dreux M, Evans MJ, Bukh J, et al. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc Natl Acad Sci U S A. 2012;109(16):6205–6210. doi: 10.1073/pnas.1114927109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keck Z, Wang W, Wang Y, Lau P, Carlsen TH, Prentoe J, et al. Cooperativity in virus neutralization by human monoclonal antibodies to two adjacent regions located at the amino terminus of hepatitis C virus E2 glycoprotein. J Virol. 2013;87(1):37–51. doi: 10.1128/JVI.01941-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keck ZY, Op De BA, Hadlock KG, Xia J, Li TK, Dubuisson J, et al. Hepatitis C virus E2 has three immunogenic domains containing conformational epitopes with distinct properties and biological functions. J Virol. 2004;78(17):9224–9232. doi: 10.1128/JVI.78.17.9224-9232.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keck ZY, Xia J, Wang Y, Wang W, Krey T, Prentoe J, et al. Human monoclonal antibodies to a novel cluster of conformational epitopes on HCV E2 with resistance to neutralization escape in a genotype 2a isolate. PLoS Pathog. 2012;8(4):e1002653. doi: 10.1371/journal.ppat.1002653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Law M, Maruyama T, Lewis J, Giang E, Tarr AW, Stamataki Z, et al. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat Med. 2008;14(1):25–27. doi: 10.1038/nm1698. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Keck ZY, Saha A, Xia J, Conrad F, Lou J, et al. Affinity maturation to improve human monoclonal antibody neutralization potency and breadth against hepatitis C virus. J Biol Chem. 2011;286(51):44218–44233. doi: 10.1074/jbc.M111.290783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeisel MB, Felmlee DJ, Baumert TF. Hepatitis C virus entry. Curr Top Microbiol Immunol. 2013;369:87–112. doi: 10.1007/978-3-642-27340-7_4. [DOI] [PubMed] [Google Scholar]
- 20.Tarr AW, Urbanowicz RA, Hamed MR, Albecka A, McClure CP, Brown RJ, et al. Hepatitis C patient-derived glycoproteins exhibit marked differences in susceptibility to serum neutralizing antibodies: genetic subtype defines antigenic but not neutralization serotype. J Virol. 2011;85(9):4246–4257. doi: 10.1128/JVI.01332-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11(7):791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gottwein JM, Scheel TK, Jensen TB, Lademann JB, Prentoe JC, Knudsen ML, et al. Development and characterization of hepatitis C virus genotype 1-7 cell culture systems: role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology. 2009;49(2):364–377. doi: 10.1002/hep.22673. [DOI] [PubMed] [Google Scholar]
- 23.Jensen TB, Gottwein JM, Scheel TK, Hoegh AM, Eugen-Olsen J, Bukh J. Highly efficient JFH1-based cell-culture system for hepatitis C virus genotype 5a: failure of homologous neutralizing-antibody treatment to control infection. J Infect Dis. 2008;198(12):1756–1765. doi: 10.1086/593021. [DOI] [PubMed] [Google Scholar]
- 24.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309(5734):623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 25.Pietschmann T, Kaul A, Koutsoudakis G, Shavinskaya A, Kallis S, Steinmann E, et al. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc Natl Acad Sci U S A. 2006;103(19):7408–7413. doi: 10.1073/pnas.0504877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheel TK, Gottwein JM, Jensen TB, Prentoe JC, Hoegh AM, Alter HJ, et al. Development of JFH1-based cell culture systems for hepatitis C virus genotype 4a and evidence for cross-genotype neutralization. Proc Natl Acad Sci U S A. 2008;105(3):997–1002. doi: 10.1073/pnas.0711044105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scheel TK, Gottwein JM, Carlsen TH, Li YP, Jensen TB, Spengler U, et al. Efficient culture adaptation of hepatitis C virus recombinants with genotype-specific core-NS2 by using previously identified mutations. J Virol. 2011;85(6):2891–2906. doi: 10.1128/JVI.01605-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yi M, Ma Y, Yates J, Lemon SM. Compensatory mutations in E1, p7, NS2, and NS3 enhance yields of cell culture-infectious intergenotypic chimeric hepatitis C virus. J Virol. 2007;81(2):629–638. doi: 10.1128/JVI.01890-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keck ZY, Li TK, Xia J, Gal-Tanamy M, Olson O, Li SH, et al. Definition of a conserved immunodominant domain on hepatitis C virus E2 glycoprotein by neutralizing human monoclonal antibodies. J Virol. 2008;82(12):6061–6066. doi: 10.1128/JVI.02475-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li YP, Ramirez S, Gottwein JM, Scheel TK, Mikkelsen L, Purcell RH, et al. Robust full-length hepatitis C virus genotype 2a and 2b infectious cultures using mutations identified by a systematic approach applicable to patient strains. Proc Natl Acad Sci U S A. 2012;109(18):E1101–E1110. doi: 10.1073/pnas.1203829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russell RS, Meunier JC, Takikawa S, Faulk K, Engle RE, Bukh J, et al. Advantages of a single-cycle production assay to study cell culture-adaptive mutations of hepatitis C virus. Proc Natl Acad Sci U S A. 2008;105(11):4370–4375. doi: 10.1073/pnas.0800422105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Hahn T, Yoon JC, Alter H, Rice CM, Rehermann B, Balfe P, et al. Hepatitis C virus continuously escapes from neutralizing antibody and T-cell responses during chronic infection in vivo. Gastroenterology. 2007;132(2):667–678. doi: 10.1053/j.gastro.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 33.Kong L, Giang E, Nieusma T, Kadam RU, Cogburn KE, Hua Y, et al. Hepatitis C virus E2 envelope glycoprotein core structure. Science. 2013;342(6162):1090–1094. doi: 10.1126/science.1243876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pedersen J, Jensen TB, Carlsen TH, Schonning K, Christensen PB, Laursen AL, et al. Neutralizing Antibodies in Patients with Chronic Hepatitis C, Genotype 1, against a Panel of Genotype 1 Culture Viruses: Lack of Correlation to Treatment Outcome. PLoS One. 2013;8(5):e62674. doi: 10.1371/journal.pone.0062674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keck ZY, Li SH, Xia J, von HT, Balfe P, McKeating JA, et al. Mutations in hepatitis C virus E2 located outside the CD81 binding sites lead to escape from broadly neutralizing antibodies but compromise virus infectivity. J Virol. 2009;83(12):6149–6160. doi: 10.1128/JVI.00248-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Helle F, Goffard A, Morel V, Duverlie G, McKeating J, Keck ZY, et al. The neutralizing activity of anti-hepatitis C virus antibodies is modulated by specific glycans on the E2 envelope protein. J Virol. 2007;81(15):8101–8111. doi: 10.1128/JVI.00127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morin TJ, Broering TJ, Leav BA, Blair BM, Rowley KJ, Boucher EN, et al. Human monoclonal antibody HCV1 effectively prevents and treats HCV infection in chimpanzees. PLoS Pathog. 2012;8(8):e1002895. doi: 10.1371/journal.ppat.1002895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70(2):440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1. Dose-response neutralization of 16 genotype 1-3 HCVcc’s by the HMAbs CBH-5, CBH-7, HC-11, HC-AM1, HC33.4.10, HC84.24, HC84.26, AR3A, AR4A, and AR5A. The JFH1-based Core-NS2 HCVcc recombinants were: Genotype 1: H77 (1a), TN (1a), DH6 (1a), J4 (1b), DH1(1b), and DH5 (1b); Genotype 2: J6 (2a), T9 (2a), JFH1 (2a), J8 (2b), DH8 (2b), DH10 (2b), and S83 (2c); Genotype 3: DH11 (3a), DBN (3a), and S52 (3a). HMAb concentrations ranged from 0.0012-100 μg/ml, and were incubated in 5-fold dilutions with appropriate FFU input readout titers of the various virus stocks in a 96 well format. Inhibitory concentrations for 50% virus neutralization (IC50) values for each HCVcc are indicated. Control for this experiment was isotype-matched antibodies R04 (CBH-5, CBH-7, HC-11, HC-AM1, HC33.4.10, HC84.24, and HC84.26) and b6 (AR3A, AR4A, and AR5A) (open symbols). All measurements were performed in 4 replicates and error bars show the standard error of the mean (SEM) of the replicates.
Supplemental Fig. 2. Multiple sequence alignment of E1/E2 amino acid sequences of the 16 JFH1-based genotype 1-3 Core-NS2 recombinants constituting the HCVcc panel. The HCVcc strains were: Genotype 1: H77 (1a), TN (1a), DH6 (1a), J4 (1b), DH1 (1b), and DH5 (1b); Genotype 2: J6 (2a), T9 (2a), JFH1 (2a), J8 (2b), DH8 (2b), DH10 (2b), and S83 (2c); Genotype 3: DH11 (3a), DBN (3a), and S52 (3a). Conserved amino acids among the HCVcc strains are labeled in white and non-conserved are labeled in gray. HMAb target residues or domains are presented in color-coding as indicated. Potential glycosylation sites are indicated in purple. Residues with dual coloring represent residues involved in binding of either more than one HMAb or a shared glycosylation/antibody target. Multiple sequence alignment was computed with ClustalW (MEGA5 software).