Abstract
Whole human genome sequencing of individuals is becoming rapid and inexpensive, enabling new strategies for using personal genome information to help diagnose, treat, and even prevent human disorders for which genetic variations are causative or are known to be risk factors. Many of the exploding number of newly discovered genetic variations alter the structure, function, dynamics, stability, and/or interactions of specific proteins and RNA molecules. Accordingly, there are a host of opportunities for biochemists and biophysicists to participate in (1) developing tools to allow accurate and sometimes medically actionable assessment of the potential pathogenicity of individual variations and (2) establishing the mechanistic linkage between pathogenic variations and their physiological consequences, providing a rational basis for treatment or preventive care. In this review, we provide an overview of these opportunities and their associated challenges in light of the current status of genomic science and personalized medicine, the latter often termed precision medicine.
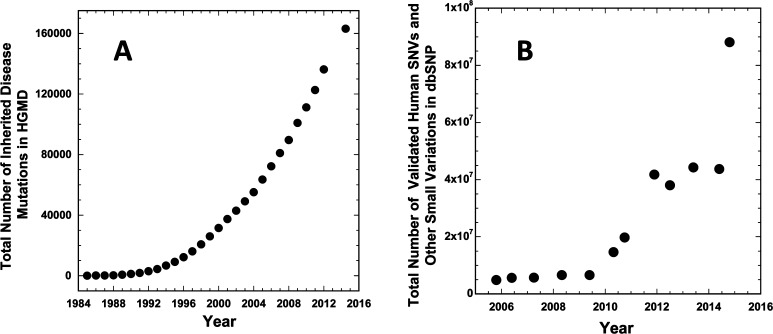
More than 80 million validated rare and common single-nucleotide variations (SNVs) in human genomes have already been catalogued (Figure 1).1,2 Of these, more than 150000 have been documented to predispose individuals to “simple” diseases that exhibit Mendelian (monogenic) inheritance,3 such as cystic fibrosis and familial early onset Alzheimer’s disease. Others impact the responsiveness of patients to certain drug treatments (phamacogenomics).4−6 For example, rare variations in the gene encoding the vitamin K epoxide reductase complex subunit 1 (VKORC1) influence the dosing requirements of the commonly used anticoagulant warfarin, which targets this protein.7 Still other SNVs are risk factors for complex or sporadic disorders. For example, the ε4 variant (encoding Cys112Arg) of apolipoprotein E, which is present in roughly 30% of all human genomes, is a risk factor for the common (sporadic) form of Alzheimer’s disease.8 These advances in medical genetics and human genomics are having transformative effects on medicine and biology.
Figure 1.
(A) Growth with time in the total number of identified human mutations that result in inherited (Mendelian) monogenic disorders. Data from the Human Gene Mutation Database.3 (B) Growth with time in the total number of discovered human genome variations (SNVs and other small scale variations) as logged in the dbSNP Database.2 The small decreases in the number of variations seen in this plot for some time points reflect the consequences of changes in the reference genome and its annotation with time.
In clinical medicine, genetic testing for selected gene variations has become the standard of care in many circumstances, particularly when the information provided is actionable as the basis for prophylactic treatment or improved patient care (cf. refs (9) and (10)). Recently, a new paradigm of human genetic analysis involving complete sequencing of whole exomes or genomes has emerged as being capable of providing comprehensive lists of rare and common variants present in patient genomes that may be disease-predisposing and/or have pharmacogenomic relevance. However, many of these variants will have uncertain clinical significance.
The ever-increasing speed and decreasing cost of exome and genome sequencing are already providing information about patient genetics that was, until very recently, inaccessible to more traditional medical genetic tools.11 As of early 2015, the utilization of such information in the practice of medicine is at a tipping point. A case study documented by Richard Lifton and colleagues at Yale University12 provides a glimpse of both the potential and complexities associated with efforts to bring personal genomic information relating to rare mutations to clinical medicine.
In 2013, a 1-week-old baby was admitted to the Yale-New Haven hospital, presenting with fever and diarrhea. Tests for infections were negative. The child’s condition progressively worsened, during which time whole exome sequencing of the patient and his parents was initiated. Because there was no obvious family history of inherited disease and the parents seemed to be healthy, it was thought that the child’s disorder might be caused by a de novo mutation, a noninherited germline mutation. Unfortunately, 23 days after the onset of symptoms and 1 day before exome sequencing was completed, the infant died of alveolar hemorrhage. Additional tests revealed extensive physiological damage consistent with an aggressively activated immune system. Comparison of the infant’s exome with those of the parents revealed no informative de novo mutations. However, two days after the funeral the father developed a high fever, respiratory distress syndrome, and other symptoms of a highly activated immune system, but without an evident infection. It was only then revealed that the father had suffered from periodic incidents of high fever throughout his life. Similarities in the symptoms and test results between father and son prompted reanalysis of their exomes to search for shared genetic variations. Among 34 previously undocumented shared protein-altering variations was a missense mutation in the NLRC4 gene encoding a valine to alanine substitution in the nucleotide binding domain (NBD) of the NLCR4 protein, an ATPase that promotes inflammasome assembly. On the basis of the crystal structure of NLCR4,13 it was deemed probable that the wild-type valine side chain stabilizes the inactive ADP-bound state of NLCR4, an interaction that is disrupted by mutation to alanine, promoting ADP-for-ATP exchange and constitutive activation of inflammasome assembly. NBD variations that trigger gain of function in a related ATPase, NLPR3, had previously been documented14 to result in dysregulated inflammasome assembly and autoinflammatory syndromes with symptoms similar to those of the father and son of this case study. Subsequent experiments confirmed the pro-inflammatory “gain-of-function” nature of the Val341Ala mutation in transfected macrophages. The condition shared by the father and son in this case study has been named “syndrome of enterocolitis and autoinflammation associated with mutation in NLRC4” (SCAN4). A contemporaneous published study reported an unrelated NLCR4 mutation that resulted in a very similar autoinflammatory syndrome.15 Given that similar syndromes have previously been shown to be relieved by drugs that target interleukin-1β, SCAN4 is probably a treatable disorder, although the father in this case declined this therapy.
One can imagine an imminent future in which the time line for connecting the dots between genetics and diagnosis will be accelerated to the point where there will routinely be little lag time between disease presentation and the availability of actionable genomic insight, even when very rare variations are involved. Indeed, case studies akin to that summarized above but with happier outcomes can already be cited.16,17 The sequencing of genomes in children and their parents may soon become routine, and analyses performed in the perinatal period may allow the compiling of genetic variations in a newborn along with identities of the variant proteins and their predicted function. Moreover, the similarity of the rare variations in genes such as NLRC4 to mutations in homologous proteins previously demonstrated to be disease-associated could be flagged by bioinformatic algorithms. Much effort is underway to develop the infrastructure and tools to address the practical challenges to effective personal genome-informed medicine, sometimes termed “personalized medicine” or “precision medicine”.
In this work, we highlight some of the opportunities and challenges associated with this burgeoning area of genomic medicine that invite the participation of biochemists, biophysicists, and structural biologists. In terms of scope, we generally focus on mutations that occur in germline DNA rather than on somatic mutations that may cause cancer18 or contribute to other disorders.19 However, many of the principles described here also apply to somatic mutations. We also focus on proteins but recognize that many disease-promoting gene variations impact RNA molecules (see Table 1 and ref (20)).
Table 1. Variations in an Individual Human Genome (estimates as of 2015; per person unless otherwise noted)a.
| no. of nucleotide base pairs | 3.2 × 109 |
| no. of protein coding genes | ∼23500 |
| no. of probable essential proteins | 2500–5000 |
| no. of exons | 180000 |
| size of exome (bp) | 30000000 |
| no. of DNA sequence variant (DSVs) | 4000000 |
| no. of de novo variants (noninherited germline) | 30–100 |
| no. of single-nucleotide variants (SNVs) | 3500000 |
| no. of copy number variants (CNVs) and other genome structural variants | 104–105 |
| no. of nonsynonymous SNPs (nsSNVs, encode changes in amino acid) | 10000–13000 |
| no. of synonymous SNPs (sSNVs, alter codon, but not amino acid) | 10000–12000 |
| no. of in-frame insertions/deletions (indels) in exons | 190–210 |
| no. of aberrant stop codons in exons | 25–100 |
| no. of frameshifts in exons | 220–250 |
| no. of splice disruption variants | 40–50 |
| no. of probable protein loss-of-function (LOF)-inducing nsSNVs | 250–500 |
| no. of heritable protein LOF-inducing nsSNVs already logged in the human gene mutation database (HGMD) | 40–100 |
| no. of homozygous LOF-inducing nsSNVs | 40–85 |
| no. of homozygous LOF-inducing nsSNVs already in the HGMD | 3–24 |
| no. of probable disease-causing homozygous LOF-inducing nsSNVs | 1–5 |
| percent of healthy individuals carrying a medically actionable homozygous disease mutation | 1–4 |
| no. of genetic variations logged as being linked to a Mendelian (monogenic) disorder in the HGMD (total to date) | 163000 |
| no. of missense/nonsense variations in HGMD linked to Mendelian disorders (total to date) | 91000 |
| no. of RNA splicing–impacting variations in HGMD linked to Mendelian disorders (total to date) | 15000 |
| no. of small insertions and deletions recorded in HGMD linked to Mendelian disorders (total to date) | 38000 |
Parts of this table were adapted from Table 1 of ref (25). Most other estimates were gleaned from refs (26−28) and (123) or http://www.hgmd.cf.ac.uk/ac/index.php.
Variation in Personal Genomes and Human Disease
Table 1 provides a compilation of genome statistics based on 2015 estimates. In that table, we focus on the fraction of the ∼4 million variations in a typical human genome that impact the exome, which encodes the proteome (Table 1). Most exomic mutations are SNVs, and each individual has 20000–25000 exomic SNVs, approximately half of which are silent at the protein level. Although most silent variants are probably not disease-linked, some are. This seems to be due either to aberrant RNA splicing21 or to alteration of the identity of the codon translated during protein synthesis. The latter can induce protein misfolding stemming from an aberrant change in the translation rate and kinetically linked cotranslational folding due to the switch from a common codon to a synonymous rare codon (or vice versa).22−24
The most common disease-promoting mutations are found among the 10000–13000 nonsynonymous SNVs (nsSNVs) that encode an amino acid change or, more rarely, introduce an aberrant stop codon.25 Of these, it is estimated that for each individual, 100–500 nsSNVs will cause loss of protein function (∼15% involving premature stop codons, and the rest single-amino acid changes), with 20–85 of these nsSNVs being homozygous.26−28 A great many disease-promoting nsSNVs have already been documented for Mendelian disorders3,29 and as risk factors for complex disorders.30 However, for any personal genome, there will be numerous nsSNVs that must initially be classified as “variants of unknown significance” (VUS) because of uncertainty regarding how the variant will impact protein function and whether there will be physiological or pharmacogenomic consequences for the patient. Some VUS will encode previously undocumented mutations in proteins that are known to be subject to disease-associated mutations, making these VUS prime suspects as disease-promoting variations. Of course, numerous VUS—even those in known disease-linked genes—will be harmless neutral variations.
One can imagine the numerous dilemmas in clinical medicine that will arise from the discovery of VUS in patients. Consider the case in which a child is found to have a newly discovered missense mutation in the gene that encodes KCNQ1, a voltage-gated potassium channel that is an essential component of the cardiac action potential. Many of the several hundred known genetic variants that alter the amino acid sequence of KCNQ1 predispose individuals to an arrhythmia known as long-QT syndrome.31 Arrhythmic episodes may go undetected for years but may result in sudden death, often triggered by swimming or exercise.32 Should the child in our imaginary case study be pre-emptively treated with antiarrhythmic drugs33 or a medical device [e.g., internal cardioverter defibrillator (ICD)], which have their own risks? Should the child be permitted to swim? Dilemmas such as these may become commonplace in the very near future. There is clearly an imperative for the continued development of methods that can rapidly determine or predict whether new VUS are harmless or predispose a patient to disease and, if so, how.
Defective Proteins and Disease
When considering how protein-altering nsSNVs result in disease, it should be kept in mind from gene knockout studies of model organisms that only 10–20% of all proteins appear to be essential for birth and survival.34−36 Also, recall that humans are diploid organisms such that there are normally two copies (alleles) of each gene, with the exception of genes on the X and Y chromosomes. Homozygous mutations impact both alleles, whereas heterozygous variations impact only one allele.
Many disease-linked nsSNVs resulting in protein LOF are homozygous, such that loss of that protein’s function is complete. The classic example is provided by inherited mutations in the gene encoding the cystic fibrosis transmembrane regulator (CFTR), a chloride ion channel.37 In contrast, individuals heterozygous for CF mutations do not suffer from the disease because the amount of functional protein originating from the wild-type allele is sufficient to fulfill an essential physiological function of this protein in maintaining proper salt balance in lung alveoli. However, for some proteins, even heterozygous mutations can result in LOF-based disease because of the reduced total functional output of that protein in critical tissues (cf., ref (38)) or because the mutant protein can form oligomers with the wild-type protein, rendering it inactive. This latter possibility applies, for example, to heterozygous mutations in peripheral myelin protein 22 (PMP22) that result in the inherited peripheral neuropathy, Charcot-Marie-Tooth disease. Mutant forms of PMP22 misfold and are targeted for degradation. Because they can form oligomers with the wild-type gene product, some of the wild-type protein is targeted along with the misfolded mutant protein for degradation.39
LOF due to nsSNVs can result from perturbations of global protein structure, active site structure, posttranslational modifications, trafficking, dynamics, and/or protein–protein or protein–ligand interactions.40 However, there is considerable evidence that the most common cause of protein LOF is reduced thermodynamic stability, leading to protein for which the folding equilibrium is shifted toward the nonfunctional unfolded state, possibly coupled to irreversible aggregation (thermal instability) and/or degradation by cellular quality control.41−44
Less common than disease-linked LOF variants are nsSNVs that result in protein that still functions, but in a dysregulated manner, so-called gain-of-function (GOF) mutants. These include the variation in the NLCR4 ATPase described above in the Yale case study and oncogenic mutations in proteins such as the Ras GTPase and the epidermal growth factor receptor family of tyrosine kinases, which often drive cancer tumor cell development and proliferation.45,46 Other examples include a number of G protein-coupled receptors that are subject to disease-causing mutations that induce constitutive activation, resulting in unregulated signaling of normally hormone-regulated pathways.47 These mutations are often heterozygous, reflecting the fact that unregulated protein function triggered by a GOF mutation in a single allele is sometimes sufficient to trigger pathophysiology. Consider the vasopressin V2 receptor (V2R) located in kidney collecting duct epithelia, which controls uptake of water into the bloodstream and formation of urine. GOF mutations in V2R result in constitutive signaling that causes too much water to be absorbed into the blood from the kidney, resulting in a rare disorder characterized by dilute blood and concentrated urine.48 The opposite disorder (diabetes insipidus, which causes hyperosmolar blood and dilute urine) is caused by much more common LOF mutations in V2R that result in misfolding.49
A third general mechanism by which nsSNVs promote disease is promotion of toxic protein aggregation.50−52 The “toxic gain of function” associated with these aggregates is typically unrelated to the native function of the affected protein. This seems to be the case for the amyloid-β polypeptides of Alzheimer’s disease, which form both soluble oligomers that are toxic to neurons and insoluble amyloid aggregates that induce chronic inflammation53,54 in brain tissue. The immediate precursor of the amyloid-β polypeptides is C99, the 99-residue C-terminal domain of the amyloid precursor protein. Some familial Alzheimer’s disease mutations in C99 alter its site specificity for cleavage by γ-secretase, increasing the production ratio between the longer and more toxic forms of the amyloid-β polypeptide relative to shorter and less toxic forms.55 Other inherited mutations in C99, located in its amyloid-β domain, make the released amyloid-β more prone to aggregate.55 Mutations can also promote toxic misfolding through indirect mechanisms. For example, it is generally thought that familial AD mutations in γ-secretase alter its scissile site specificity in C99 so that the production ratio of long to short forms of amyloid-β is increased.56
Finally, it should not be overlooked that other genetic variations besides nsSNVs can result in protein LOF, GOF, or toxic aggregates. Some genes may be completely deleted or inactivated by variations that alter their transcription, while for other genes, protein overdosage may occur when there are more than the usual two copies of the gene or when the gene is subject to aberrantly upregulated transcription.57 While single-site mutations in PMP22 are one cause of Charcot-Marie-Tooth disease, this disorder more commonly results from trisomy (a third copy) of the wild-type gene, resulting in overexpression of wild-type PMP22.58
Mutations that alter either the chaperone or degradative components of cellular protein folding quality control systems can have adverse impacts on the folding of specific client proteins or, more generally, on cellular proteostasis.59,60 The introduction of stop codons into protein open reading frames leads to the production of truncated proteins that may fail to function and possibly misfold, but alternatively may be functional but dysregulated (GOF).61,62 Frameshift mutations lead to proteins with partial wild-type sequence, followed by incorrect amino acids. Chromosomal translocation can lead to the generation of fusion proteins composed of two normally independent proteins (or fragments thereof), sometimes having a highly toxic or aberrantly proliferative function.63
Determining or Predicting the Impact of Genetic Variations on Proteins
Much effort is already being dedicated to developing the infrastructure of databases, methods, protocols, training, counseling approaches, actuarial resources, bioethics, and laws that are needed for personalized genomic information to be widely employed in patient treatment (cf. refs (64−66)). Molecular bioscientists such as biochemists, biophysicists, and structural biologists are already making important contributions to this process67 (e.g., see lists of already available online resources in refs (43) and (68−70)), which may well span decades. It is to be hoped that such investigators will make major contributions to the very important task of assessing exactly how genetic variations impact biomolecular folding, structure, function, and interactions. This information will often be essential to the practice of personalized medicine. For example, there is compelling need to distinguish between true disease-promoting genetic variations and neutral variations to avoid treatment of healthy patients who should therefore not be subjected to expensive, unhelpful, and potentially harmful and expensive pre-emptive treatment. Consider also a disease such as cystic fibrosis for which there are more than 1000 different gene variations known to result in LOF of the CFTR protein.71 It is clear that different mutations can induce LOF through different mechanisms, some (such as the common ΔF508 mutant) by inducing misfolding and others by altering channel properties within correctly folded protein. Given that CF therapeutic drugs are in various states of development that address different classes of potential defects in the CFTR protein,72,73 it may eventually be possible to mechanistically classify each CFTR disease mutation to ensure correct therapeutic matching for each specific CFTR mutation. Given that most drugs have unwanted off-target side effects, there is an added imperative to avoid treatment of unresponsive patients.
The ability to predict with great accuracy and mechanistic detail the possible outcome of nsSNVs in terms of protein LOF, GOF, altered protein–protein interactions, or toxic aggregation would be a great resource. Indeed, there are already numerous algorithms available, often deployed in the form publicly accessible web servers that make such predictions.69,70,74−83 Given that destabilization of proteins leading to LOF may be the most common mechanism by which genetic variations promote disease, it is not surprising that a number of these algorithms are devoted to prediction of mutation-induced protein stability changes. As of late 2014, the authors easily identified several dozen online programs/servers that predict protein stability either as a primary function or as a trait that is factored into overall predictions of whether a particular genetic variation is pathogenic. The very best of the stability predictive algorithms, which can be of formidable sophistication, are able to correctly and quantitatively predict mutation-induced changes in protein stability roughly 85% of the time for relatively small proteins of known three-dimensional structure.(74,84−89) However, this success rate drops by ∼15% when the protein’s three-dimensional structure is not known. In this case, algorithms must rely instead on homology models, sequence conservation patterns, and/or other variables.78 Moreover, the benchmarking used to evaluate the success of these predictions usually lacks data for multidomain proteins, which are especially common in higher organisms. The same deficit holds for membrane proteins, which comprise roughly 20% of all proteins and upward of 50% of the targets of currently used drugs. This problem is fundamental as the rules that govern protein stability may well be altered for interfacial residues in multidomain proteins90−92 and transmembrane segments in membrane proteins.93−96 Therefore, aforementioned methods that are optimized for small soluble proteins would be expected to display decreased accuracy. There remains a compelling need for additional experimental studies of protein stability, especially for membrane proteins and multidomain proteins, for which there may not currently be sufficient data available even to validate and benchmark stability prediction methods.
While most previous effort has been dedicated to targeted studies of particularly common or representative disease variants of a given protein (such as the ΔF508 form of CFTR), experimental studies of modest panels of disease mutant forms of a given protein have been described (cf. refs (97−104)). One might flinch at the thought of conducting experimental studies on hundreds of mutant forms a single protein, such as the more than 500 known retinitis pigmentosa-linked mutant forms of the ABCA4 flippase105 or the more than 15000 cancer-related variants of the p53 tumor suppressor.106 However, the potential power and efficiency of high-throughput experimental methods should not be overlooked when considering how best to test the impact of potential disease-linked sequence variations on protein function, structure, stability, intermolecular interactions, and tendency to aggregate. The high value attached to the availability of a high-quality experimental structure for the protein of interest or a close homologue should also be emphasized, given that our ability to predict mutation-induced perturbations of normal protein traits is often dramatically enhanced by the availability of such a structure.68,107
It has recently been estimated that 35% of all residues in the human proteome have been structurally characterized, through experimental structural determination or being inferred from modeling to homologous protein templates of known experimental structure.108,109 This highlights the tremendous frontier of structural space in the human proteome that remains to be characterized. At the same time, the numerous available experimental structures and homology models for human proteins represent an immediate resource for use in assessing VUS.
As of mid-2015, there are still numerous human proteins for which biochemical and biological function and regulation remain poorly understood, a fact that will confound the degree to which genome variations affecting these proteins can be analyzed for personal medical purposes. The continued need for basic science studies of such proteins reflects the unwavering importance of basic biological research in providing the foundations of molecular medicine.
An additional category of insight needed for personalized medicine to which biochemists and biophysicists can contribute is in assessing how partial or complete GOF or LOF for a variant protein alters the biochemical, cellular, and physiological pathways associated with that protein and how the dysfunction or dysregulation of those pathways impacts cellular and, ultimately, organismal fitness.110,111 Because a great many of these pathways are not isolated but are integrated within complex webs, this realm of analysis will overlap with that of systems biology.
Confounding Factors in Translating Results of Protein Studies into Personalized Medical Decision Making
When considering results for variant proteins, it can be very difficult to predict whether correct assessment of protein function informs on whether a variation actually predisposes the human subject to a related disease.112,113 Indeed, it is now appreciated that many “disease mutations” compiled in databases such as HGMD are probably neutral variations or of low penetrance, meaning they result in disease in only a fraction of the mutation carriers.114 There are a variety of confounding factors, even for mutations associated with “simple” monogenic Mendelian diseases. Among these are data, noted earlier, that show that many proteins seem to be dispensable, at least under some conditions. It may be that life has to some degree evolved so as to be robust by generating molecules with redundant functions and compensatory mechanisms to avoid placing too much importance on single gene products.115 Moreover, in any individual, each genetic variation occurs within the context of the thousands of other genetic variations, some of which may exert a compensatory effect that nullifies the possible physiological consequences of GOF or LOF in another protein. Perhaps this is the case for the dementia-free centenarian who was recently found to be homozygous for the Alzheimer’s-promoting apolipoprotein E ε4 variant.116 The complex genetics of some disorders, where disease occurs only when multiple disease-predisposing variations collude, makes such maladies difficult to predict or mechanistically dissect.117 In complex diseases such as type II diabetes, environmental factors such as diet, exercise, gender, smoking, lifetime exposure to toxins or infectious agents, age, and prior medical history can work, variously, either to reinforce or to offset disease-promoting genetic factors.118 Epigenetics can also complicate the linkage of genome variations with personal medical traits.119 Finally, because protein expression levels can be highly variable from cell to cell, tissue to tissue, and patient to patient, the actual degree of net physiological GOF or LOF associated with an otherwise well-characterized variant protein may be very difficult to predict in an individual.120
The Developing Interface among Biomolecular Science, Bioinformatics, and Personalized Medicine
In light of the contributions that can be made by biochemists and biophysicists to the knowledge infrastructure that will support personalized medicine, we would like to offer a couple suggestions regarding how this interface can be enhanced.
Excellent communication between bioinformaticists and biomolecular scientists will increasingly be required. It will be critical to develop methodologies that combine bioinformatics capabilities in large scale data analysis, machine learning, and biostatistics with data gleaned from biochemical and biophysical experiments and simulations. Bioinformaticists are often well-equipped for tapping into the databases, theory, and information provided by the studies of both geneticists and biomolecular scientists. This is admirably reflected, for example, by the sophistication of some of the available protein stability and pathogenicity prediction methods. However, basic scientists sometimes do not have the required training in statistics and elementary computer programming needed to access and fully understand/assess many bioinformatic analyses, even when the topic (such as protein stability assessment) is familiar. This deficit might well be addressed by injection of a modest dose of formal training in statistics, computer programming, and bioinformatics into graduate programs in the basic biological/biomedical sciences. Along these same lines, a modicum of up-to-date didactic training in the areas of genetics and genomics should perhaps also be regarded as an essential curricular objective.
There also is the continued need to expand the interface between the basic biomedical sciences and the engineering-tinged discipline of systems biology. This is essential to the continued development of methods to assess whether and how a particular variant protein will actually result in disease. Moreover, systems approaches will be required to determine the degree to which multiple genetic variations synergistically promote health problems or alter pharmacogenomic profiles in ways that cannot be predicted on the basis of summing the impacts of the same variants in isolation. The relationship between a single protein and the fitness of the host organism may sometimes be exceedingly complex. At the same time, all diseases arise from problems that are, ultimately, molecular in origin.
Conclusions
Personalized medicine is not a new concept but is being transformed by the advent of widespread exome/genome sequencing. Some of the paradigms for how genome information will be employed are already established, such as the fact that genetics dictates that the optimal treatment for some patients is sometimes different than for others. It is also clear that analysis of genome information will allow rational preemptive treatment of disease prior to the symptoms. Genetic information may, in favorable cases, be used to relieve the anxiety of individuals who fear a particular disorder because of family history or other factors. An interesting emerging area of genetic analysis involves the search for genetic factors that are protective, lowering the risk for various diseases and/or increasing life expectancy.121
Beyond patient care, information from whole genome analysis may well come to be routinely factored into important and highly personal decisions such as those associated with human reproduction. This highlights currently unresolved issues such as the question of how much patients should be told about their genomes and those of family members, who should have access to personal genomic information, and how this information can lawfully and ethically be used in decision making. One wonders whether the impact of the genome on everyday life is currently where personal computers and the computer networks were circa 1980. At that time, it was apparent that the electronic innovations were here to stay, but few could foresee the extraordinary degree to which information technology has come to impact society only 35 years later. It seems probable that widespread genome sequencing will ultimately shape civilization in ways that extend well beyond what we can foresee. There are and will continue to be numerous opportunities for biochemists, biophysicists, structural biologists, and other basic scientists to generate knowledge and approaches that will be critical to the successful ongoing deployment of the genomics revolution. In closing, we suggest that while this revolution will have immediate impact, it is likely to unfold over decades, maybe even centuries. Societal expectations in terms of imminent widespread improvements in healthcare should be tempered. In this regard, it may be helpful to ponder a 2001 observation by Sir David Weatherall, a pioneer in classical genetic studies of hemoglobin disorders such as anemias and thallesemias:122
“The one regret is that the remarkable advances in human genetics, and in the evolution of molecular medicine in general...have done little to help patients with these distressing diseases... The lack of a more definitive approach to the cure of the haemoglobin disorders, despite so much knowledge about their molecular pathology, should not surprise us. The story of the advancement of medical practice over the last century tells us that there is always a long lag period before developments in the basic sciences reach the clinic.”
Acknowledgments
We thank Jonathan Schlebach for helpful discussion.
This work was supported by National Institutes of Health (NIH) Grants RO1 HL122010, RO1 GM106672, and RO1 DC007416. B.M.K. was supported by NIH Training Grant T32 NS007491.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Liu X.; Jian X.; Boerwinkle E. (2013) dbNSFP v2.0: A database of human non-synonymous SNVs and their functional predictions and annotations. Hum. Mutat. 34, E2393–E2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Center for Biotechnology Information, National Library of Medicine (2015) Database of Single Nucleotide Polymorphisms (dbSNP).
- Stenson P. D.; Mort M.; Ball E. V.; Shaw K.; Phillips A.; Cooper D. N. (2014) The Human Gene Mutation Database: Building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet. 133, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr D. F.; Alfirevic A.; Pirmohamed M. (2014) Pharmacogenomics: Current State-of-the-Art. Genes 5, 430–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahti J. L.; Tang G. W.; Capriotti E.; Liu T.; Altman R. B. (2012) Bioinformatics and variability in drug response: A protein structural perspective. J. R. Soc., Interface 9, 1409–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer U. A.; Zanger U. M.; Schwab M. (2013) Omics and drug response. Annu. Rev. Pharmacol. Toxicol. 53, 475–502. [DOI] [PubMed] [Google Scholar]
- Johnson J. A.; Cavallari L. H. (2015) Warfarin pharmacogenetics. Trends Cardiovasc. Med. 25, 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve S.; Brisson D.; Marchant N. L.; Gaudet D. (2014) The potential applications of apolipoprotein E in personalized medicine. Front. Aging Neurosci. 6, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt A. K.; Macrae C. A. (2014) Genetic testing in cardiovascular diseases. Curr. Opin. Cardiol. 29, 235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingles J.; Semsarian C. (2014) The value of cardiac genetic testing. Trends Cardiovasc. Med. 24, 217–224. [DOI] [PubMed] [Google Scholar]
- Newman W. G.; Black G. C. (2014) Delivery of a clinical genomics service. Genes 5, 1001–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romberg N.; Al Moussawi K.; Nelson-Williams C.; Stiegler A. L.; Loring E.; Choi M.; Overton J.; Meffre E.; Khokha M. K.; Huttner A. J.; West B.; Podoltsev N. A.; Boggon T. J.; Kazmierczak B. I.; Lifton R. P. (2014) Mutation of NLRC4 causes a syndrome of enterocolitis and autoinflammation. Nat. Genet. 46, 1135–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z.; Yan C.; Liu P.; Huang Z.; Ma R.; Zhang C.; Wang R.; Zhang Y.; Martinon F.; Miao D.; Deng H.; Wang J.; Chang J.; Chai J. (2013) Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science 341, 172–175. [DOI] [PubMed] [Google Scholar]
- Hoffman H. M.; Mueller J. L.; Broide D. H.; Wanderer A. A.; Kolodner R. D. (2001) Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nat. Genet. 29, 301–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canna S. W.; de Jesus A. A.; Gouni S.; Brooks S. R.; Marrero B.; Liu Y.; DiMattia M. A.; Zaal K. J.; Sanchez G. A.; Kim H.; Chapelle D.; Plass N.; Huang Y.; Villarino A. V.; Biancotto A.; Fleisher T. A.; Duncan J. A.; O’Shea J. J.; Benseler S.; Grom A.; Deng Z.; Laxer R. M.; Goldbach-Mansky R. (2014) An activating NLRC4 inflammasome mutation causes autoinflammation with recurrent macrophage activation syndrome. Nat. Genet. 46, 1140–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinwiddie D. L.; Bracken J. M.; Bass J. A.; Christenson K.; Soden S. E.; Saunders C. J.; Miller N. A.; Singh V.; Zwick D. L.; Roberts C. C.; Dalal J.; Kingsmore S. F. (2013) Molecular diagnosis of infantile onset inflammatory bowel disease by exome sequencing. Genomics 102, 442–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthey E. A.; Mayer A. N.; Syverson G. D.; Helbling D.; Bonacci B. B.; Decker B.; Serpe J. M.; Dasu T.; Tschannen M. R.; Veith R. L.; Basehore M. J.; Broeckel U.; Tomita-Mitchell A.; Arca M. J.; Casper J. T.; Margolis D. A.; Bick D. P.; Hessner M. J.; Routes J. M.; Verbsky J. W.; Jacob H. J.; Dimmock D. P. (2011) Making a definitive diagnosis: Successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet. Med. 13, 255–262. [DOI] [PubMed] [Google Scholar]
- Pon J. R.; Marra M. A.. Driver and Passenger Mutations in Cancer. Annu. Rev. Pathol. 2015, in press [DOI] [PubMed]
- Vijg J. (2014) Somatic mutations, genome mosaicism, cancer and aging. Curr. Opin. Genet. Dev. 26, 141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrdlickova B.; de Almeida R. C.; Borek Z.; Withoff S. (2014) Genetic variation in the non-coding genome: Involvement of micro-RNAs and long non-coding RNAs in disease. Biochim. Biophys. Acta 1842, 1910–1922. [DOI] [PubMed] [Google Scholar]
- Raponi M.; Baralle D. (2010) Alternative splicing: Good and bad effects of translationally silent substitutions. FEBS J. 277, 836–840. [DOI] [PubMed] [Google Scholar]
- Maraia R. J.; Iben J. R. (2014) Different types of secondary information in the genetic code. RNA 20, 977–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer P. S.; Siller E.; Anderson J. F.; Barral J. M. (2012) Silent substitutions predictably alter translation elongation rates and protein folding efficiencies. J. Mol. Biol. 422, 328–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar A. A. (2009) A pause for thought along the co-translational folding pathway. Trends Biochem. Sci. 34, 16–24. [DOI] [PubMed] [Google Scholar]
- Marian A. J. (2012) Challenges in medical applications of whole exome/genome sequencing discoveries. Trends Cardiovasc. Med. 22, 219–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur D. G.; Balasubramanian S.; Frankish A.; Huang N.; Morris J.; Walter K.; Jostins L.; Habegger L.; Pickrell J. K.; Montgomery S. B.; Albers C. A.; Zhang Z. D.; Conrad D. F.; Lunter G.; Zheng H.; Ayub Q.; DePristo M. A.; Banks E.; Hu M.; Handsaker R. E.; Rosenfeld J. A.; Fromer M.; Jin M.; Mu X. J.; Khurana E.; Ye K.; Kay M.; Saunders G. I.; Suner M. M.; Hunt T.; Barnes I. H.; Amid C.; Carvalho-Silva D. R.; Bignell A. H.; Snow C.; Yngvadottir B.; Bumpstead S.; Cooper D. N.; Xue Y.; Romero I. G.; Genomes Project C.; Wang J.; Li Y.; Gibbs R. A.; McCarroll S. A.; Dermitzakis E. T.; Pritchard J. K.; Barrett J. C.; Harrow J.; Hurles M. E.; Gerstein M. B.; Tyler-Smith C. (2012) A systematic survey of loss-of-function variants in human protein-coding genes. Science 335, 823–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y.; Chen Y.; Ayub Q.; Huang N.; Ball E. V.; Mort M.; Phillips A. D.; Shaw K.; Stenson P. D.; Cooper D. N.; Tyler-Smith C.; Genomes Project C. (2012) Deleterious- and disease-allele prevalence in healthy individuals: Insights from current predictions, mutation databases, and population-scale resequencing. Am. J. Hum. Genet. 91, 1022–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium; Abecasis G. R.; Altshuler D.; Auton A.; Brooks L. D.; Durbin R. M.; Gibbs R. A.; Hurles M. E.; McVean G. A. (2010) A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Online Mendelian Inheritance in Man, OMIM (2015) Johns Hopkins University, Baltimore.
- Welter D.; MacArthur J.; Morales J.; Burdett T.; Hall P.; Junkins H.; Klemm A.; Flicek P.; Manolio T.; Hindorff L.; Parkinson H. (2014) The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42, D1001–D1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. A.; Vanoye C. G.; George A. L. Jr.; Meiler J.; Sanders C. R. (2007) Structural models for the KCNQ1 voltage-gated potassium channel. Biochemistry 46, 14141–14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi G.; Kopplin L. J.; Tester D. J.; Will M. L.; Haglund C. M.; Ackerman M. J. (2004) Spectrum and frequency of cardiac channel defects in swimming-triggered arrhythmia syndromes. Circulation 110, 2119–2124. [DOI] [PubMed] [Google Scholar]
- Waddell-Smith K. E.; Earle N.; Skinner J. R. (2015) Must every child with long QT syndrome take a beta blocker?. Arch. Dis. Child. 100, 279–282. [DOI] [PubMed] [Google Scholar]
- Cliften P.; Sudarsanam P.; Desikan A.; Fulton L.; Fulton B.; Majors J.; Waterston R.; Cohen B. A.; Johnston M. (2003) Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301, 71–76. [DOI] [PubMed] [Google Scholar]
- Kamath R. S.; Fraser A. G.; Dong Y.; Poulin G.; Durbin R.; Gotta M.; Kanapin A.; Le Bot N.; Moreno S.; Sohrmann M.; Welchman D. P.; Zipperlen P.; Ahringer J. (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237. [DOI] [PubMed] [Google Scholar]
- Rubin G. M.; Yandell M. D.; Wortman J. R.; Gabor Miklos G. L.; Nelson C. R.; Hariharan I. K.; Fortini M. E.; Li P. W.; Apweiler R.; Fleischmann W.; Cherry J. M.; Henikoff S.; Skupski M. P.; Misra S.; Ashburner M.; Birney E.; Boguski M. S.; Brody T.; Brokstein P.; Celniker S. E.; Chervitz S. A.; Coates D.; Cravchik A.; Gabrielian A.; Galle R. F.; Gelbart W. M.; George R. A.; Goldstein L. S.; Gong F.; Guan P.; Harris N. L.; Hay B. A.; Hoskins R. A.; Li J.; Li Z.; Hynes R. O.; Jones S. J.; Kuehl P. M.; Lemaitre B.; Littleton J. T.; Morrison D. K.; Mungall C.; O’Farrell P. H.; Pickeral O. K.; Shue C.; Vosshall L. B.; Zhang J.; Zhao Q.; Zheng X. H.; Lewis S. (2000) Comparative genomics of the eukaryotes. Science 287, 2204–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan J. R. (2008) CFTR function and prospects for therapy. Annu. Rev. Biochem. 77, 701–726. [DOI] [PubMed] [Google Scholar]
- Faerch M.; Christensen J. H.; Rittig S.; Johansson J. O.; Gregersen N.; de Zegher F.; Corydon T. J. (2009) Diverse vasopressin V2 receptor functionality underlying partial congenital nephrogenic diabetes insipidus. Am. J. Physiol. 297, F1518–F1525. [DOI] [PubMed] [Google Scholar]
- Sanders C. R.; Ismail-Beigi F.; McEnery M. W. (2001) Mutations of peripheral myelin protein 22 result in defective trafficking through mechanisms which may be common to diseases involving tetraspan membrane proteins. Biochemistry 40, 9453–9459. [DOI] [PubMed] [Google Scholar]
- Kucukkal T. G.; Petukh M.; Li L.; Alexov E. (2015) Structural and physico-chemical effects of disease and non-disease nsSNPs on proteins. Curr. Opin. Struct. Biol. 32C, 18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadio R.; Vassura M.; Tiwari S.; Fariselli P.; Luigi Martelli P. (2011) Correlating disease-related mutations to their effect on protein stability: A large-scale analysis of the human proteome. Hum. Mutat. 32, 1161–1170. [DOI] [PubMed] [Google Scholar]
- Shi Z.; Moult J. (2011) Structural and functional impact of cancer-related missense somatic mutations. J. Mol. Biol. 413, 495–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefl S.; Nishi H.; Petukh M.; Panchenko A. R.; Alexov E. (2013) Molecular mechanisms of disease-causing missense mutations. J. Mol. Biol. 425, 3919–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue P.; Li Z.; Moult J. (2005) Loss of protein structure stability as a major causative factor in monogenic disease. J. Mol. Biol. 353, 459–473. [DOI] [PubMed] [Google Scholar]
- Arteaga C. L.; Engelman J. A. (2014) ERBB receptors: From oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell 25, 282–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubbert S.; Shannon K.; Bollag G. (2007) Hyperactive Ras in developmental disorders and cancer. Nat. Rev. Cancer 7, 295–308. [DOI] [PubMed] [Google Scholar]
- Tao Y. X. (2008) Constitutive activation of G protein-coupled receptors and diseases: Insights into mechanisms of activation and therapeutics. Pharmacol. Ther. 120, 129–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman B. J.; Rosenthal S. M.; Vargas G. A.; Fenwick R. G.; Huang E. A.; Matsuda-Abedini M.; Lustig R. H.; Mathias R. S.; Portale A. A.; Miller W. L.; Gitelman S. E. (2005) Nephrogenic syndrome of inappropriate antidiuresis. N. Engl. J. Med. 352, 1884–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bichet D. G. (2006) Nephrogenic diabetes insipidus. Advances in Chronic Kidney Disease 13, 96–104. [DOI] [PubMed] [Google Scholar]
- Calamini B.; Morimoto R. I. (2012) Protein homeostasis as a therapeutic target for diseases of protein conformation. Curr. Top. Med. Chem. 12, 2623–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles T. P.; Vendruscolo M.; Dobson C. M. (2014) The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 15, 384–396. [DOI] [PubMed] [Google Scholar]
- Valastyan J. S.; Lindquist S. (2014) Mechanisms of protein-folding diseases at a glance. Dis. Models Mech. 7, 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z.; Hussain M. D.; Yan L. J. (2014) Microglia, neuroinflammation, and β-amyloid protein in Alzheimer’s disease. Int. J. Neurosci. 124, 307–321. [DOI] [PubMed] [Google Scholar]
- Malm T. M.; Jay T. R.; Landreth G. E. (2015) The evolving biology of microglia in Alzheimer’s disease. Neurotherapeutics 12, 81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzi R. E. (2012) The genetics of Alzheimer disease. Cold Spring Harbor Perspect. Med. 2, DOI: 10.1101/cshperspect.a006296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Strooper B.; Iwatsubo T.; Wolfe M. S. (2012) Presenilins and γ-secretase: Structure, function, and role in Alzheimer disease. Cold Spring Harbor Perspect. Med. 2, a006304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F.; Gu W.; Hurles M. E.; Lupski J. R. (2009) Copy number variation in human health, disease, and evolution. Annu. Rev. Genomics Hum. Genet. 10, 451–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetten A. M.; Suter U. (2000) The peripheral myelin protein 22 and epithelial membrane protein family. Prog. Nucleic Acid Res. Mol. Biol. 64, 97–129. [DOI] [PubMed] [Google Scholar]
- Bakthisaran R.; Tangirala R.; Rao C. M. (2014) Small heat shock proteins: Role in cellular functions and pathology. Biochim. Biophys. Acta 1854, 291–319. [DOI] [PubMed] [Google Scholar]
- Macario A. J.; Conway de Macario E. (2007) Chaperonopathies and chaperonotherapy. FEBS Lett. 581, 3681–3688. [DOI] [PubMed] [Google Scholar]
- Diaz G. A. (2005) CXCR4 mutations in WHIM syndrome: A misguided immune system?. Immunol. Rev. 203, 235–243. [DOI] [PubMed] [Google Scholar]
- Guo Y.; Wei X.; Das J.; Grimson A.; Lipkin S. M.; Clark A. G.; Yu H. (2013) Dissecting disease inheritance modes in a three-dimensional protein network challenges the “guilt-by-association” principle. Am. J. Hum. Genet. 93, 78–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantschel O. (2012) Structure, regulation, signaling, and targeting of abl kinases in cancer. Genes Cancer 3, 436–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburg M. A.; Collins F. S. (2010) The path to personalized medicine. N. Engl. J. Med. 363, 301–304. [DOI] [PubMed] [Google Scholar]
- Hooker G. W.; Ormond K. E.; Sweet K.; Biesecker B. B. (2014) Teaching genomic counseling: Preparing the genetic counseling workforce for the genomic era. Journal of Genetic Counseling 23, 445–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg S.; Shen Y.; Jones S. J.; Gibson W. T. (2014) Genetic counseling in direct-to-consumer exome sequencing: A case report. Journal of Genetic Counseling 23, 742–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexov E. (2014) Advances in Human Biology: Combining Genetics and Molecular Biophysics to Pave the Way for Personalized Diagnostics and Medicine. Advances in Biology 2014, 16. [Google Scholar]
- Gong S.; Worth C. L.; Cheng T. M.; Blundell T. L. (2011) Meet me halfway: When genomics meets structural bioinformatics. Journal of Cardiovascular Translational Research 4, 281–303. [DOI] [PubMed] [Google Scholar]
- Katsonis P.; Koire A.; Wilson S. J.; Hsu T. K.; Lua R. C.; Wilkins A. D.; Lichtarge O. (2014) Single nucleotide variations: Biological impact and theoretical interpretation. Protein Sci. 23, 1650–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thusberg J.; Vihinen M. (2009) Pathogenic or not? And if so, then how? Studying the effects of missense mutations using bioinformatics methods. Hum. Mutat. 30, 703–714. [DOI] [PubMed] [Google Scholar]
- Dorfman R. (2011) Cystic Fibrosis Mutation Database (http://www.genet.sickkids.on.ca/Home.html).
- Bell S. C.; De Boeck K.; Amaral M. D. (2015) New pharmacological approaches for cystic fibrosis: Promises, progress, pitfalls. Pharmacol. Ther. 145C, 19–34. [DOI] [PubMed] [Google Scholar]
- Lukacs G. L.; Verkman A. S. (2012) CFTR: Folding, misfolding and correcting the ΔF508 conformational defect. Trends Mol. Med. 18, 81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berliner N.; Teyra J.; Colak R.; Garcia Lopez S.; Kim P. M. (2014) Combining structural modeling with ensemble machine learning to accurately predict protein fold stability and binding affinity effects upon mutation. PLoS One 9, e107353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crockett D. K.; Lyon E.; Williams M. S.; Narus S. P.; Facelli J. C.; Mitchell J. A. (2012) Utility of gene-specific algorithms for predicting pathogenicity of uncertain gene variants. Journal of the American Medical Informatics Association 19, 207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson T. A.; Doughty E.; Kann M. G. (2013) Towards precision medicine: Advances in computational approaches for the analysis of human variants. J. Mol. Biol. 425, 4047–4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potapov V.; Cohen M.; Schreiber G. (2009) Assessing computational methods for predicting protein stability upon mutation: Good on average but not in the details. Protein Eng., Des. Sel. 22, 553–560. [DOI] [PubMed] [Google Scholar]
- Thusberg J.; Olatubosun A.; Vihinen M. (2011) Performance of mutation pathogenicity prediction methods on missense variants. Hum. Mutat. 32, 358–368. [DOI] [PubMed] [Google Scholar]
- Capriotti E.; Altman R. B.; Bromberg Y. (2013) Collective judgment predicts disease-associated single nucleotide variants. BMC Genomics 14(Suppl. 3), S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.; Petukh M.; Alexov E.; Panchenko A. R. (2014) Predicting the Impact of Missense Mutations on Protein-Protein Binding Affinity. J. Chem. Theory Comput. 10, 1770–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan C. J.; Cimermancic P.; Szpiech Z. A.; Sali A.; Hernandez R. D.; Krogan N. J. (2013) High-resolution network biology: Connecting sequence with function. Nat. Rev. Genet. 14, 865–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudha G.; Nussinov R.; Srinivasan N. (2014) An overview of recent advances in structural bioinformatics of protein-protein interactions and a guide to their principles. Prog. Biophys. Mol. Biol. 116, 141–150. [DOI] [PubMed] [Google Scholar]
- Yates C. M.; Sternberg M. J. (2013) The effects of non-synonymous single nucleotide polymorphisms (nsSNPs) on protein-protein interactions. J. Mol. Biol. 425, 3949–3963. [DOI] [PubMed] [Google Scholar]
- Folkman L.; Stantic B.; Sattar A. (2014) Feature-based multiple models improve classification of mutation-induced stability changes. BMC Genomics 15(Suppl. 4), S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromiha M. M. (2007) Prediction of protein stability upon point mutations. Biochem. Soc. Trans. 35, 1569–1573. [DOI] [PubMed] [Google Scholar]
- Khan S.; Vihinen M. (2010) Performance of protein stability predictors. Hum. Mutat. 31, 675–684. [DOI] [PubMed] [Google Scholar]
- Tian J.; Wu N.; Chu X.; Fan Y. (2010) Predicting changes in protein thermostability brought about by single- or multi-site mutations. BMC Bioinf. 11, 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainreb G.; Wolf L.; Ashkenazy H.; Dehouck Y.; Ben-Tal N. (2011) Protein stability: A single recorded mutation aids in predicting the effects of other mutations in the same amino acid site. Bioinformatics 27, 3286–3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Chen B.; Tan G.; Vihinen M.; Shen B. (2013) Structure-based prediction of the effects of a missense variant on protein stability. Amino Acids 44, 847–855. [DOI] [PubMed] [Google Scholar]
- Arviv O.; Levy Y. (2012) Folding of multidomain proteins: Biophysical consequences of tethering even in apparently independent folding. Proteins 80, 2780–2798. [DOI] [PubMed] [Google Scholar]
- Batey S.; Nickson A. A.; Clarke J. (2008) Studying the folding of multidomain proteins. HFSP J. 2, 365–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara R. M.; Srinivasan N. (2011) Stability of domain structures in multi-domain proteins. Sci. Rep. 1, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z.; Bowie J. U. (2012) Shifting hydrogen bonds may produce flexible transmembrane helices. Proc. Natl. Acad. Sci. U.S.A. 109, 8121–8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H.; Bowie J. U. (2011) Dramatic destabilization of transmembrane helix interactions by features of natural membrane environments. J. Am. Chem. Soc. 133, 11389–11398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R. E.; Blois T. M.; Bowie J. U. (2013) Membrane proteins can have high kinetic stability. J. Am. Chem. Soc. 135, 15183–15190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming K. G. (2014) Energetics of membrane protein folding. Annu. Rev. Biophys. 43, 233–255. [DOI] [PubMed] [Google Scholar]
- Allali-Hassani A.; Wasney G. A.; Chau I.; Hong B. S.; Senisterra G.; Loppnau P.; Shi Z.; Moult J.; Edwards A. M.; Arrowsmith C. H.; Park H. W.; Schapira M.; Vedadi M. (2009) A survey of proteins encoded by non-synonymous single nucleotide polymorphisms reveals a significant fraction with altered stability and activity. Biochem. J. 424, 15–26. [DOI] [PubMed] [Google Scholar]
- Fossbakk A.; Kleppe R.; Knappskog P. M.; Martinez A.; Haavik J. (2014) Functional studies of tyrosine hydroxylase missense variants reveal distinct patterns of molecular defects in Dopa-responsive dystonia. Hum. Mutat. 35, 880–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.; Stiers K. M.; Kain B. N.; Beamer L. J. (2014) Compromised catalysis and potential folding defects in in vitro studies of missense mutants associated with hereditary phosphoglucomutase 1 deficiency. J. Biol. Chem. 289, 32010–32019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou B.; Kazimierczuk A.; Zhang M.; Scott C. R.; Hegde R. S.; Grabowski G. A. (2006) Analyses of variant acid β-glucosidases: Effects of Gaucher disease mutations. J. Biol. Chem. 281, 4242–4253. [DOI] [PubMed] [Google Scholar]
- North C. L.; Blacklow S. C. (2000) Evidence that familial hypercholesterolemia mutations of the LDL receptor cause limited local misfolding in an LDL-A module pair. Biochemistry 39, 13127–13135. [DOI] [PubMed] [Google Scholar]
- Opefi C. A.; South K.; Reynolds C. A.; Smith S. O.; Reeves P. J. (2013) Retinitis pigmentosa mutants provide insight into the role of the N-terminal cap in rhodopsin folding, structure, and function. J. Biol. Chem. 288, 33912–33926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pey A. L.; Desviat L. R.; Gamez A.; Ugarte M.; Perez B. (2003) Phenylketonuria: Genotype-phenotype correlations based on expression analysis of structural and functional mutations in PAH. Hum. Mutat. 21, 370–378. [DOI] [PubMed] [Google Scholar]
- Westover J. B.; Goodman S. I.; Frerman F. E. (2003) Pathogenic mutations in the carboxyl-terminal domain of glutaryl-CoA dehydrogenase: Effects on catalytic activity and the stability of the tetramer. Mol. Genet. Metab. 79, 245–256. [DOI] [PubMed] [Google Scholar]
- Daiger S. P.; Sullivan L. S.; Bowne S. J. (2013) Genes and mutations causing retinitis pigmentosa. Clin. Genet. 84, 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walerych D.; Napoli M.; Collavin L.; Del Sal G. (2012) The rebel angel: Mutant p53 as the driving oncogene in breast cancer. Carcinogenesis 33, 2007–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue W. W.; Oppermann U. (2011) High-throughput structural biology of metabolic enzymes and its impact on human diseases. J. Inherited Metab. Dis. 34, 575–581. [DOI] [PubMed] [Google Scholar]
- Khafizov K.; Madrid-Aliste C.; Almo S. C.; Fiser A. (2014) Trends in structural coverage of the protein universe and the impact of the Protein Structure Initiative. Proc. Natl. Acad. Sci. U.S.A. 111, 3733–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M.; Coggill P. C.; Eberhardt R. Y.; Mistry J.; Tate J.; Boursnell C.; Pang N.; Forslund K.; Ceric G.; Clements J.; Heger A.; Holm L.; Sonnhammer E. L.; Eddy S. R.; Bateman A.; Finn R. D. (2012) The Pfam protein families database. Nucleic Acids Res. 40, D290–D301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardinoglu A.; Gatto F.; Nielsen J. (2013) Genome-scale modeling of human metabolism: A systems biology approach. Biotechnol. J. 8, 985–996. [DOI] [PubMed] [Google Scholar]
- Garcia-Alonso L.; Jimenez-Almazan J.; Carbonell-Caballero J.; Vela-Boza A.; Santoyo-Lopez J.; Antinolo G.; Dopazo J. (2014) The role of the interactome in the maintenance of deleterious variability in human populations. Mol. Syst. Biol. 10, 752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper D. N.; Krawczak M.; Polychronakos C.; Tyler-Smith C.; Kehrer-Sawatzki H. (2013) Where genotype is not predictive of phenotype: Towards an understanding of the molecular basis of reduced penetrance in human inherited disease. Hum. Genet. 132, 1077–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do R.; Kathiresan S.; Abecasis G. R. (2012) Exome sequencing and complex disease: Practical aspects of rare variant association studies. Hum. Mol. Genet. 21, R1–R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur D. G.; Manolio T. A.; Dimmock D. P.; Rehm H. L.; Shendure J.; Abecasis G. R.; Adams D. R.; Altman R. B.; Antonarakis S. E.; Ashley E. A.; Barrett J. C.; Biesecker L. G.; Conrad D. F.; Cooper G. M.; Cox N. J.; Daly M. J.; Gerstein M. B.; Goldstein D. B.; Hirschhorn J. N.; Leal S. M.; Pennacchio L. A.; Stamatoyannopoulos J. A.; Sunyaev S. R.; Valle D.; Voight B. F.; Winckler W.; Gunter C. (2014) Guidelines for investigating causality of sequence variants in human disease. Nature 508, 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szamecz B.; Boross G.; Kalapis D.; Kovacs K.; Fekete G.; Farkas Z.; Lazar V.; Hrtyan M.; Kemmeren P.; Groot Koerkamp M. J.; Rutkai E.; Holstege F. C.; Papp B.; Pal C. (2014) The genomic landscape of compensatory evolution. PLoS Biol. 12, e1001935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freudenberg-Hua Y.; Freudenberg J.; Vacic V.; Abhyankar A.; Emde A. K.; Ben-Avraham D.; Barzilai N.; Oschwald D.; Christen E.; Koppel J.; Greenwald B.; Darnell R. B.; Germer S.; Atzmon G.; Davies P. (2014) Disease variants in genomes of 44 centenarians. Mol. Genet. Genomic Med. 2, 438–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulit S. L.; Leusink M.; Menelaou A.; de Bakker P. I. (2014) Association claims in the sequencing era. Genes 5, 196–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauk C. L.; Lucas Argueso J.; Auerbach S. S.; Awadalla P.; Davis S. R.; Demarini D. M.; Douglas G. R.; Dubrova Y. E.; Elespuru R. K.; Glover T. W.; Hales B. F.; Hurles M. E.; Klein C. B.; Lupski J. R.; Manchester D. K.; Marchetti F.; Montpetit A.; Mulvihill J. J.; Robaire B.; Robbins W. A.; Rouleau G. A.; Shaughnessy D. T.; Somers C. M.; Taylor J. G. t.; Trasler J.; Waters M. D.; Wilson T. E.; Witt K. L.; Bishop J. B. (2013) Harnessing genomics to identify environmental determinants of heritable disease. Mutat. Res. 752, 6–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y.; Lu L.; Liu M.; Li X. C.; Sun R. R.; Zheng Y.; Zhang P. Y. (2014) Impact of epigenetics in the management of cardiovascular disease: A review. Riv. Eur. Sci. Med. Farmacol. 18, 3097–3104. [PubMed] [Google Scholar]
- Kukurba K. R.; Zhang R.; Li X.; Smith K. S.; Knowles D. A.; How Tan M.; Piskol R.; Lek M.; Snyder M.; Macarthur D. G.; Li J. B.; Montgomery S. B. (2014) Allelic expression of deleterious protein-coding variants across human tissues. PLoS Genet. 10, e1004304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierman H. J.; Fortney K.; Roach J. C.; Coles N. S.; Li H.; Glusman G.; Markov G. J.; Smith J. D.; Hood L.; Coles L. S.; Kim S. K. (2014) Whole-genome sequencing of the world’s oldest people. PLoS One 9, e112430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherall D. J. (2001) Towards molecular medicine; reminiscences of the haemoglobin field, 1960–2000. Br. J. Haematol. 115, 729–738. [DOI] [PubMed] [Google Scholar]
- Dorschner M. O.; Amendola L. M.; Turner E. H.; Robertson P. D.; Shirts B. H.; Gallego C. J.; Bennett R. L.; Jones K. L.; Tokita M. J.; Bennett J. T.; Kim J. H.; Rosenthal E. A.; Kim D. S.; National Heart Lung and Blood Institute Grand Opportunity Exome Sequencing; Tabor H. K.; Bamshad M. J.; Motulsky A. G.; Scott C. R.; Pritchard C. C.; Walsh T.; Burke W.; Raskind W. H.; Byers P.; Hisama F. M.; Nickerson D. A.; Jarvik G. P. (2013) Actionable, pathogenic incidental findings in 1,000 participants’ exomes. Am. J. Hum. Genet. 93, 631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]