Abstract
We describe the Conserved Domain Search service (CD-Search), a web-based tool for the detection of structural and functional domains in protein sequences. CD-Search uses BLAST® heuristics to provide a fast, interactive service, and searches a comprehensive collection of domain models. Search results are displayed as domain architecture cartoons and pairwise alignments between the query and domain-model consensus sequences. Search results may be visualized in further detail by embedding the query sequence into multiple alignment displays and by mapping onto three-dimensional molecular graphic displays of known structures within the domain family. CD-Search can be accessed at http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi.
INTRODUCTION
The accelerating growth of the amount of protein sequence data is continuing to result in an urgent need for rapid computational annotation of protein sequences. A variety of useful resources have emerged, many at the level of protein domains and/or protein families. CD-Search combines the sensitivity of carefully constructed search models based on multiple sequence alignments with the speed and significance statistics of the BLAST® software suite. The sources of CD-Search's alignment model data are imported collections, such as Pfam (1) and SMART (2), and automated alignments supplied with the Clusters of Orthologous Groups (COGs) classifications (3). Curators at the NCBI are adding in an ongoing manner models for ancient, phylogenetically widespread domain families with structure-based alignments and explicitly recorded subfamily hierarchies (4). Domain-model alignments are converted into position-specific scoring matrices (PSSMs); protein query sequences can be scanned against these PSSMs with RPS-BLAST, a variant of the Psi-BLAST algorithm (5).
We have built a web service which runs live searches against large collections of PSSM search models for protein queries supplied by users. Execution times vary with query sequence length and search database size, and typically lie in the range of a few to about twenty seconds. CD-Search uses BLAST® statistics to evaluate the significance of query–subject alignments, and returns E-values (expectation values), which can be interpreted analogously to those obtained with Psi-BLAST.
CD-Search reports graphical summaries of the search results, a tabular list of hits and individual pairwise alignments between the user query and search model consensus sequences. Consensus sequences are not used in the calculation per se. They serve as placeholders for columns in the position-specific scoring matrix, their length chosen approximately as the median length of domain sequences in the multiple alignment. The balloons showing domain footprints on the query sequence are linked to more extensive visualization. Following these links, users can embed the query sequence into individual domain alignment displays and change parameters to emphasize patterns of sequence conservation. Domain models curated at the NCBI may carry annotation of functional features, in which case specific conserved columns are highlighted. This should make it easier to understand whether key functional residues are present in a query sequence and help to rule out chance similarities in query–database hits at the borderline of significance. Many domain families are linked to three-dimensional (3D) structure. In these cases we offer alignment visualization including embedded user query sequences with NCBI's 3D structure viewer Cn3D (6).
USING CD-Search
Searches can be submitted at the following URL: http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi. The search page allows the user to set a variety of parameters.
Search database: currently we mirror some of the imported collections, as well as an all-inclusive set, the default. The default search set will be replaced with a less redundant collection in the near future.
Query sequences can be supplied as raw sequences, FASTA-formatted sequences or sequence identifiers [‘GI’ (genInfo identifier) numbers or accessions] valid in the NCBI's Entrez protein database (7).
An E-value threshold can be set to vary the search sensitivity and specificity. The default is 0.01.
A filter for low-complexity regions in the query sequence is turned on by default. This reduces the likelihood of false-positive hits.
The search heuristic can be switched from the default mode (multiple hits, one pass) to slightly more sensitive and more time-consuming options (single hit, one pass or two passes).
Output formatting options allow the user to limit the number of hits returned, reduce redundancy in the graphical results summary, modify its size and change the style of the pairwise alignments.
INTERPRETING RESULTS
Figure 1 shows the top of a CD-Search results page. The page lists the version of RPS-BLAST used in the process, repeats information about the query sequence and displays statistics on the search database used.
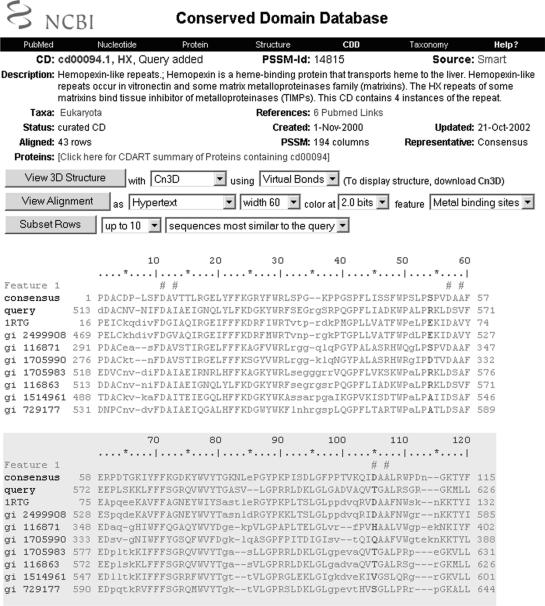
Figure 1.
Graphical summary of results and hit list. The query sequence used in this search was gi|116863 (9).
The graphical results summary comes with a text box which reports such details as accessions, short names and E-values as the mouse pointer is placed over one of the balloons representing an individual hit. The query sequence is drawn as a black bar on the top of the image, with a ruler indicating its length. Sections colored in cyan have been filtered out as low-complexity regions in the database search. RPS-BLAST, the search engine behind CD-Search, typically does not extend alignments into these regions. Alignment details are shown in individual pairwise alignment displays in the bottom part of the page, where low-complexity regions are again explicitly indicated.
The individual balloons are assigned colors according to a fixed schema. The best scoring hit is colored red; the second-best scoring hit is colored blue; for example see the online help document for details. Hits to conserved domain models that are identified as related by the CDART resource (8) are given the same color. The redundancy present in the all-inclusive search set of the CDD (Conserved Domain Database) is readily visible in this example. Balloons are drawn so that they extend from the first to the last residue of the alignment footprints on the query sequence. These alignments may contain gapped-out regions, which are visible in the pairwise alignment displays at the bottom of the page but are not indicated in the graphical summary.
Balloons may have jagged edges. A jagged edge at the N- or C-terminus of a domain footprint indicates that >20% of the domain model's extent is missing from the RPS-BLAST pairwise alignment. Pairwise alignment displays at the bottom of the page list exact percentages of the domain models that were used in the alignment. In the example shown in Figure 1, the jagged edges indicate partial hits to N- and C-terminal parts of zinc-dependent metalloprotase domains (the partial hits are caused by the insertion of additional domains).
Balloons may also have indentations, such as the best scoring hit in the example shown in Figure 1. Indentations indicate that a repeat structure has been detected algorithmically in the search model; in Figure 1, the alignment model labeled ‘HX’ spans four copies of hemopexin-repeats.
Clicking on a balloon invokes a multiple alignment view which adds the matching fragment of the query sequence, aligned according to the RPS-BLAST algorithm. An example is shown in Figure 2. The user can modify conservation thresholds used for coloring columns in the alignment to better identify conserved sites, and can select subsets of family-member sequences to be included in the display. By default, alignment rows most similar to the query sequence are chosen. In many conserved domain models curated at the NCBI, conserved functional motifs have been recorded as features of that model. A feature's address is a set of columns in the multiple alignment. Features are highlighted with hash marks printed on top of the alignment blocks. Features are recorded together with evidence, which may consist of citations or specialized 3D displays. The evidence viewer is found at the bottom of the multiple alignment display (not shown in Figure 2). These alignment display options are intended to assist the user in predicting whether a query sequence is a true and/or functional member of the domain family (and in particular to allow users to better discriminate between chance similarities and actual homology when the statistical significance places a similarity in a ‘twilight zone’).
Figure 2.
A fragment of the query sequence embedded in the domain alignment for hemopexin-like repeats. Conserved features (metal binding sites) are indicated with hash marks on top of aligned columns.
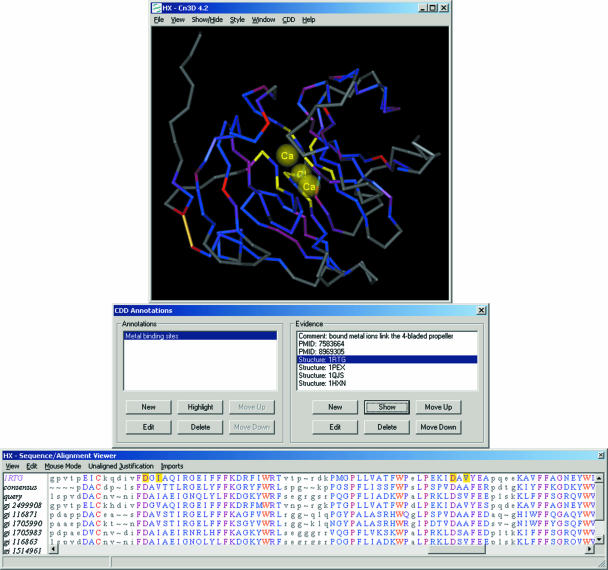
A powerful aid in studying query–subject relationships is the availability of 3D information. A large proportion of domain models can be linked to one or several 3D structures, and the NCBI's 3D structure viewer, Cn3D, can be used to interactively visualize the structure together with the multiple alignment of a domain family. Information about annotated features can be accessed from within Cn3D, and this combination provides a unique set of tools to hypothesize about the effects of sequence variation on otherwise conserved sites, such as catalytic sets of residues or binding interfaces.
An example is shown in Figure 3. The Cn3D viewer is launched by clicking the button labeled ‘View 3D Structure’ on alignment display pages (Figure 2). Cn3D needs to be installed locally as a helper application; for details follow the link labeled ‘download Cn3D’ on the alignment display pages (Figure 2). The set of sequences displayed, together with the embedded query, the consensus and a representative 3D structure, can be controlled using the options next to the ‘Subset Rows’ button. By default, Cn3D will be launched with 10 aligned rows; sequences from the alignment model are picked as the ones most similar to the query (judged by the number of identical residues, according to the aligned footprint). For domain-alignment models curated at NCBI, several 3D structures may be shown at once, with structure super-positions precalculated in the curation process. Cn3D allows the user to interactively highlight residues in either the 3D or alignment view; highlights will immediately transfer to the other view. Cn3D also allows the user to explore prerecorded annotation of conserved features, feature evidence and prerecorded links to literature.
Figure 3.
Alignment visualization in the context of 3D structure. The query sequence has been added to the domain-alignment model. In this particular view the user visualizes evidence for the metal binding site (a conserved feature of this family), which is provided by the solved 3D structure of a protein–metal complex.
ALTERNATIVE ROUTES TO CD-Search RESULTS
Conserved domain Searches have been precalculated for all but the newest proteins in the Entrez database. If a protein has been found to contain conserved domains, a link labeled ‘Domains’ is available on Entrez document summaries. Following this link, users are shown a simplified CD-Search results page which contains the graphical results summary only. From this point, the view can be expanded to give detailed search results.
By default, protein query sequences are sent to CD-Search when regular protein-BLAST searches are submitted at http://www.ncbi.nlm.nih.gov/BLAST/. While users wait for the protein-BLAST search to complete, results from the domain analysis may already be visible. BLAST's intermediate search page will show a graphical summary of the CD-Search outcome, which again can be expanded into a full view.
Users can download CD-Search databases and run RPS-BLAST locally, provided they download and install the NCBI toolkit to build local versions of the BLAST programs. Detailed instructions for downloading and configuring a set of several search databases can be found at ftp://ftp.ncbi.nih.gov/pub/mmdb/cdd/README.
FUTURE DEVELOPMENTS
Over time we plan to replace non-curated domain families with hierarchies of extensively curated models. The default display styles for CD-Search hits will become less redundant and more informative, since organizing related families in hierarchies will allow us to better understand the meaning of overlapping domain footprints. Consequently we shall be able to implement changes in display styles which may improve the resource's user-friendliness.
We will also be able to provide better support for local RPS-BLAST search database construction. Those who wish to will be able to customize specialized search sets.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the authors of the BLAST® suite of programs for developing and supporting RPS-BLAST. We thank the architects of the Pfam, SMART and COGs collections and the curators of the CDD for their invaluable contributions. We also thank the NIH Intramural Research Program for support.
REFERENCES
- 1.Bateman A., Coin,L., Durbin,R., Finn,R.D., Hollich,V., Griffiths-Jones,S., Khanna,A., Marshall,M., Moxon,S., Sonnhammer,E.L., Studholme,D.J., Yates,C. and Eddy,S.R. (2004) The Pfam protein families database. Nucleic Acids Res., 32, 138–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Letunic I., Copley,R.R., Schmidt,S., Ciccarelli,F.D., Doerks,T., Shultz,J., Ponting,C.P. and Bork,P. (2004) SMART 4.0: towards genomic data integration. Nucleic Acids Res., 32, 142–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tatusov R.L, Fedorova,N.D., Jackson,J.D., Jacobs,A.R., Kiryutin,B., Koonin,E.V, Krylov,D.M., Mazumder,R., Mekhedov,S.L., Nikolskaya,A.N., Rao,B.S., Smirnov,S., Sverdlov,A.V., Vasudevan,S., Wolf,Y.I., Yin,J.J. and Natale,D.A. (2003) The COG database: an updated version includes eukaryotes. BMC Bioinformatics, 4, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marchler-Bauer A., Anderson,J.B., DeWeese-Scott,C., Fedorova,N.D., Geer,L.Y., He,S., Hurwitz,D.I., Jackson,J.D., Jacobs,A.R., Lanczycki,C.J., Liebert,C.A., Liu,C., Madej,T., Marchler,G.H., Mazumder,R., Nikolskaya,A.N., Panchenko,A.R., Rao,B.S., Shoemaker,B.A., Simonyan,V., Song,J.S., Thiessen,P.A., Vasudevan,S., Wang,Y., Yamashita,R.A., Yin,J.J. and Bryant,S.H. (2003) CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res., 31, 383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y., Geer,L.Y., Chappey,C., Kans,J.A. and Bryant,S.H. (2000) Cn3D: sequence and structure views for Entrez. Trends Biochem. Sci., 25, 300–302. [DOI] [PubMed] [Google Scholar]
- 7.Wheeler D.L., Church,D.M., Edgar,R., Federhen,S., Helmberg,W., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Sequeira,E., Suzek,T.O., Tatusova,T.A. and Wagner,L. (2004) Database resources of the National Center for Biotechnology Information: update. Nucleic Acids Res., 32, 35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geer L.Y., Domrachev,M., Lipman,D.J. and Bryant,S.H. (2002) CDART: protein domain homology by domain architecture. Genome Res., 12, 1619–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilhelm S.M., Collier,I.E., Marmer,B.L, Eisen,A.Z., Grant,G.A. and Goldberg,G.I. (1989) SV40-transformed human lung fibroblasts secrete a 92-kDa type IV collagenase which is identical to that secreted by normal human macrophages. J. Biol. Chem., 264, 17213–17221. [PubMed] [Google Scholar]