Abstract
Short Sequence Repeat (SSR) typing of Mycobacterium avium subspecies paratuberculosis (Map) isolates is one of the most commonly used method for genotyping this pathogen. Currently used techniques have challenges in analyzing mononucleotide repeats >15 bp, which include some of the Map SSRs. Fragment analysis is a relatively simple technique, which can accurately measure the size of DNA fragments and can be used to calculate the repeat length of the target SSR loci. In the present study, fragment analysis was used to analyze 4 Map SSR loci known to provide sufficient discriminatory power to determine the relationship between Map isolates. Eighty-five Map isolates from 18 animals from the island of Newfoundland were successfully genotyped using fragment analysis. To the best of our knowledge, this is the first report on Map SSR diversity from Newfoundland dairy farms. Previously unreported Map SSR-types or combinations were also identified during the course of the described work. In addition, multiple Map SSR-types were isolated from a single animal in many cases, which is not a common finding.
Introduction
Mycobacterium avium subspecies paratuberculosis (Map) is a slow growing bacterium and is the cause of Johne’s disease, which is associated with chronic debilitating granulomatous enteritis that affects the small intestine of cattle, sheep, goats, farmed deer and {Li, 2005 #1017}other ruminants [1–6]. Johne’s disease is a major cause of concern to the dairy industry and there is also some concern regarding the association of Map with Crohn’s disease in humans [7–9]. Treatment of dairy animals infected with Map is impractical because it can be only achieved by using a combination of antibiotics, many of which are very expensive, not licensed for food animals and require long term dosing [10]. Therefore, infected animals are culled, which is also a part of the Johne’s disease control/management practice [5]. Because diagnosis is very challenging early on in disease progression, animals can still get infected by Map through exposure to other shedding asymptomatic animals and environmental contamination [11,12]. The long incubation period of Map and the non-specific clinical symptoms exhibited by infected animals makes the diagnosis, management and control of Johne’s disease difficult. To decrease the spread of Johne’s disease, surveillance programs are being established throughout the world for determining the sources of infections, the prevalence of the causative agent and the relationship(s) between Map isolates from dairy farms. Studies are also being conducted to examine the role of host genetics in determining the susceptibility of individual animals and their clinical course once infected. Such programs are vital for devising effective control strategies against this devastating disease [13].
More recently, there have been a number of reports on the molecular epidemiology of Map. In previous studies, single or combined molecular methods have been used to obtain epidemiological data regarding Map strain types [14–17]. Most of the previously used strain typing methods are expensive, time consuming, lack discriminatory capabilities and sometimes do not provide consistent results [14,15,18]. Despite these limitations, the information obtained from such studies is essential for identifying sources and transmission routes more accurately. When combined with information on host genetics, strain typing studies can also be used to determine strain pathogenicity and host resistance. Molecular techniques, especially DNA short-sequence-repeat (SSR) analysis has been shown to be a powerful tool for discriminating between Map isolates at the genetic level [5,14,15,16,18,19]. Due to differences in the numbers of nucleotide repeats associated with SSRs from different Map isolates, the relatedness and prevalence of Map strains can be monitored within/between farms and the environment [14,19]. One major problem with conventional methods for SSR analyses such as the use of Sanger sequencing, is that they are prone to artifacts and failure due to challenges associated with determining the DNA sequences of the repeats, with the most recent technology being capable of analyzing repeats up to 15 bp using a mass spectrometry based approach [18]. Therefore, there is need for developing cheap, reliable and reproducible methods for Map SSR analysis, which can accurately measure repeats over 15 bp in length. Recently, DNA fragment analysis was used for Map SSR typing [16]. PCR fragments containing the SSRs were obtained using fluorescently labeled primers and were subjected to capillary electrophoresis for determining their sizes [16]. The island of Newfoundland which situated at the eastern edge of North America has a number of dairy farms. There is significant movement of animals within the Newfoundland dairy industry, with new animals being brought onto the island for entry into the production chain. In addition, some heifers are also shipped to other Atlantic Canada provinces on the mainland for raring, before they return as adult producers, as it is economically more feasible to do so in some situations. Therefore, there is interest in analyzing the diversity Map isolates infecting animals from the island for comparison to those found elsewhere in North America. In the current study we used fragment analysis to analyze Map isolates from five Newfoundland dairy farms, the results of which are described below.
Materials and Methods
Ethics statement
The described study was carried out under a formal agreement between the Dairy Farmers of Newfoundland and Labrador (NL) and the Chief Veterinary Officer for the Province of NL (HGW). The study was approved by the Institutional Animal Care Committee (IACC, Memorial University of Newfoundland) as an “A” rated protocol because the samples used in the study were obtained from routine veterinary diagnostic submissions unrelated to this research. The report describes laboratory microbiological analysis and did not directly involve any animals.
Media, reagents and culture conditions
All reagents and media used in the study were purchased from Sigma Aldrich, Fisher Scientific or VWR International, Canada, unless otherwise mentioned. DNA oligonucleotide primers were purchased from Integrated DNA Technologies (USA). Map cultures were grown at 37°C. Fecal samples from 18 animals displaying varying clinical symptoms of Johne’s disease or suspected of being infected (S1 Table), were collected by the Animal Health Division, Department of Natural Resources, Government of NL and were sent to the Atlantic Veterinary College, University of Prince Edward Island (UPEI) for diagnosis. Trek-ESP II liquid culture using Trek ESP Para-JEM media (Thermo Scientific) was used to culture Map from bovine fecal samples as described previously [20], which were verified by acid-fast staining. To confirm the presence of Map in the cultures, chromosomal DNA was isolated using the Tetracore Map extraction system and was used as template along with the Tetracore VetAlert Johne’s Real-Time PCR kit as per the manufacturer’s instructions (Tetracore, USA). After the described analysis, the culture samples were stored as frozen glycerol stocks and were sent to the Memorial University of Newfoundland for further analysis.
The culture samples from UPEI were streaked out on to Middlebrook 7H11 agar plates supplemented with oleic acid-albumin-dextrose-catalase (OADC) and mycobactin J (2 mg/L, Allied Monitor, USA) to obtain isolated Map colonies as described previously [14]. The PANTA (polymyxin B, amphotericin B, nalidixic acid, trimethoprim and azlocillin) antibiotic mixture was also added to the medium to prevent the growth of other contaminating microorganisms [21]. The plates were incubated for 4–6 months until minute colonies were observed, which were confirmed to be Map by acid-fast staining. Three to five isolated colonies from each plate (corresponding to each animal) were then used to inoculate separate 5 mL Middlebrook 7H9 broth cultures supplemented with albumin-dextrose-catalase (ADC) and mycobactin J (2 mg/L). To avoid the clumping of cells, culture tubes contained sterile glass beads and were incubated with agitation. Growth was observed for 85 isolates (3–5 isolates sampled from each animal) after 2–3 months of incubation based on an increase in the turbidity of the cultures, which were then used to prepare glycerol stocks for storage and for chromosomal DNA isolation as described below. Acid-fast staining was performed at different stages to ensure that the cultures were axenic.
Chromosomal DNA isolation, SSR sequencing and fragment analysis
The QIAamp DNA Mini kit (Qiagen) was used for isolating chromosomal DNA from the remaining 3.5 ml 7H9 cultures from above using 0.1 mm zirconia silica beads and a SpeedMill PLUS homogenizer (Analytik Jena, Germany) according to the manufacturer’s recommendations. All PCR reactions were performed using the Phusion High-Fidelity PCR Kit along with 3% DMSO and the GC buffer (New England Biolabs, Inc. Canada). PCR products were visualized by agarose gel electrophoresis, purified using the EZ-10 Spin Column PCR Products Purification Kit (Bio Basic, Canada) and were sent for DNA sequencing or fragment analysis to the Centre for Applied Genomics (TCAG), University of Toronto, Canada. The four SSR loci (L1-L4) previously shown to provide good discriminatory power for subtyping Map isolates were chosen for analysis [5,14]. The DNA sequences of the four SSR repeats were determined for a handful of Map isolates obtained as part of a separate study as described previously [5,14]. The sequences of the 4 loci for the Map K10 strain (genome sequenced) were obtained from previous publications [14,22] and were confirmed using fragment analysis as described below. This was done to determine the exact numbers of the SSR repeats for each of the strains, which were later used as standards during fragment analysis.
For fragment analysis, four primer pairs were designed which were specific for each locus, and one primer from each pair was labeled with 6-fluorescein amidite (6-FAM) to give PCR products ranging from 127 to 255 bp. Primers that have the 6-FAM dye next to a guanine base near the 5' end can have decreased fluorescence. Therefore, either the forward or the reverse primer from each pair was labeled to avoid any complications. The DNA sequences of the primers used for obtaining PCR products for fragment analysis for each locus were as follows: L1 (F: GGTGTTCGGCAAAGTCGTT/R: TTGACGATCACCAGCCCG), L2 (F: TCGCCTCAGGCTTTACTGAT/R: CACGTAGGTCCGCTGATGA), L3 (AGGCCTTCTACGTGCACAAC/R: GAGATGTCCAGCCCTGTCTC) and L4 (F: CTCGTGGAAACCCTCGAC/R: GGTGCTGAAATCCGGTGT). Unpurified PCR products were sent to the TCAG facilities for fragment analysis using the ABI 3730XL or 3100 capillary electrophoresis instruments using the GeneScan 500 ROX Size Standard, which is capable of accurately sizing DNA fragments ranging from 35 to 500 bp (http://www.tcag.ca/facilities/geneticAnalysis.html. Accessed 2015 April 4). The Peak Scanner software v1.0 (Applied Biosystems, USA) was used to analyze the fragment profiles/peaks to determine the sizes of the DNA fragments, which were used to calculate SSR copy numbers. Comparison of the fragment sizes from Newfoundland isolates with those from the sequenced standards, which were included in every fragment analysis run, enabled the determination of the exact copy number of each repeat at the target SSR loci. Fragment analysis was repeated and the data was analyzed to obtain reproducible results as described previously [16]. SSR-types were assigned on the basis of the combinations of alleles for each locus and the information was used to build a dendrogram using the BioNumerics 7.1 program (Applied Maths, Inc., USA). The unweighted pair group method with arithmetic mean was used to create a minimum spanning tree using the same program, which portrays the level of divergence between strains utilizing pairwise genetic distances [23].
Results and Discussion
The sequencing of DNA repeats using conventional methods is often challenging and is prone to artifacts, which is further exacerbated by repeats with high GC content such as those found in Map. In addition, currently available technologies can only determine the lengths of repeats up to ~15 bp accurately and often longer repeats cannot be measured [18]. In the case of Map, it has been previously reported that the analysis of 4 SSRs (L1/L2: monucleotide, and L3/L4 trinucleotide) provides enough sequence information for strain discrimination, and that one of the mononucleotide SSRs (L1) can be >14 bp in length depending on the isolate [5,14]. In our own studies we found that the sequencing of PCR products containing SSRs ~14 bp using the Sanger method was not straight forward (results not shown). Previous reports also describe similar problems where the L1 SSR had sequencing errors during analysis, leading to the misinterpretation of repeat lengths [18]. Therefore, to overcome issues associated with the analysis of Map SSRs, we adapted fragment analysis as a method to analyze Map isolates from 5 dairy farms from Newfoundland [16]. Fecal samples were collected from 18 animals, some of which showed clinical signs of Johne’s disease, displayed an immune response against Map in milk samples collected during previous surveys or were suspected of being infected (S1 Table). The samples were processed to obtain 18 primary fecal cultures, which were then used to establish 85 axenic cultures for use in the current study.
Before using the fragment analysis based approach, a handful of Map isolates (henceforth referred to as control strains) that were obtained as part of another study were subjected to SSR sequencing using the Sanger method as described previously [5,14,15,19]. Multiple sequencing runs were carried out until we could reproducibly sequence across the SSRs. This was done to determine the exact numbers of repeats at the four SSR loci for the respective isolates for subsequent use as standards for comparisons during fragment analysis. Again, we could only obtain accurate and reliable sequences for SSRs smaller than 14 bp using Sanger method (data not shown). Next, the four loci specific sets of fluorescently labeled primers were used in separate PCR reactions along with chromosomal DNA as template from the control strains described above. Since the exact numbers of SSRs present in each PCR product were known from Sanger sequencing for the control strains, they were included as size standards in all future fragment analysis experiments. The lengths of the SSRs for the K10 strain were already known (L1:19, L2:10, L3:5 and L4:5) because its genome has been sequenced [22] and could be confirmed by using fragment analysis as described below.
Chromosomal DNA was isolated from 85 axenic Map cultures established using samples from Newfoundland, and were used as template to obtain fluorescently labeled PCR products for fragment analysis. Comparison of each SSR PCR product with the respective standards described above provided accurate data regarding the copy number of the repeats at each locus for all 85 isolates in a short period of time (S2 Table). In the current study we were also able to analyze L2, which was not possible in a previous report that also used fragment analysis [16]. Some samples were reanalyzed to rule out ambiguities. After excluding Map isolates from the same animal with identical SSR profiles, a total of 68 isolates with 40 different SSR-types (M1-M44) were identified from 18 animals from 5 different Newfoundland farms (Table 1). In addition, in many cases Map with multiple SSR-types were isolated from the same animal (S2 Table).
Table 1. Details of the 40 SSR-types that were identified using fragment analysis from Map isolated from five Newfoundland dairy farms in the current study.
| L-1 (G) a | L-2 (G) a | L-3 (GGT) a | L-4 (TGC) a | SSR-type b | No. of Animals with SSR-type c | Farm ID d |
|---|---|---|---|---|---|---|
| 12 | 10 | 5 | 5 | M1 | 6 | A(5), C(1) |
| 15 | 10 | 5 | 5 | M2 | 5 | A(4), C(1) |
| 11 | 10 | 5 | 5 | M3 | 4 | A(2), C(2) |
| 16 | 10 | 5 | 5 | M4 | 4 | A(3), C(1) |
| 13 | 11 | 5 | 5 | M5 | 3 | A(3) |
| 14 | 11 | 5 | 5 | M6 | 3 | A(2), F(1) |
| 10 | 11 | 4 | 5 | M7 | 2 | C(1), E(1) |
| 10 | 12 | 4 | 5 | M8 | 2 | C(2) |
| 10 | 10 | 5 | 5 | M9 | 2 | A(1), C(1) |
| 11 | 11 | 5 | 5 | M10 | 2 | A(1), C(1) |
| 12 | 9 | 5 | 5 | M11 | 2 | A(1), F(1) |
| 13 | 10 | 5 | 5 | M12 | 2 | A(1), E(1) |
| 14 | 10 | 5 | 5 | M13 | 2 | A(1), E(1) |
| 17 | 11 | 5 | 5 | M14 | 2 | A(1), F(1) |
| 20 | 10 | 5 | 5 | M15 | 2 | A(1), E(1) |
| 6 | 11 | 4 | 4 | M16 | 1 | C(1) |
| 6 | 14 | 4 | 4 | M17 | 1 | D(1) |
| 6 | 15 | 4 | 4 | M18 | 1 | D(1) |
| 7 | 11 | 4 | 4 | M19 | 1 | C(1) |
| 7 | 11 | 6 | 5 | M20 | 1 | E(1) |
| 7 | 10 | 5 | 5 | M21 | 1 | A(1) |
| 7 | 15 | 4 | 4 | M22 | 1 | D(1) |
| 10 | 11 | 5 | 5 | M23 | 1 | A(1) |
| 11 | 9 | 5 | 5 | M24 | 1 | A(1) |
| 11 | 12 | 4 | 5 | M25 | 1 | C(1) |
| 12 | 11 | 5 | 5 | M26 | 1 | A(1) |
| 14 | 9 | 5 | 5 | M27 | 1 | A(1) |
| 14 | 13 | 5 | 5 | M28 | 1 | C(1) |
| 15 | 11 | 5 | 5 | M29 | 1 | A(1) |
| 15 | 13 | 5 | 5 | M30 | 1 | C(1) |
| 16 | 9 | 5 | 5 | M31 | 1 | A(1) |
| 16 | 11 | 5 | 5 | M32 | 1 | A(1) |
| 16 | 12 | 5 | 5 | M33 | 1 | A(1) |
| 16 | 14 | 5 | 5 | M34 | 1 | C(1) |
| 18 | 9 | 5 | 5 | M35 | 1 | F(1) |
| 18 | 10 | 5 | 5 | M36 | 1 | A(1) |
| 18 | 11 | 5 | 5 | M37 | 1 | F(1) |
| 20 | 9 | 5 | 5 | M38 | 1 | F(1) |
| 20 | 11 | 5 | 5 | M39 | 1 | A(1) |
| 21 | 10 | 5 | 5 | M40 | 1 | A(1) |
aThe number/copies of repeats for each SSR detected in the current study are indicated.
bSSR-types were designated as M1-M40 based on the copy number of the repeats for the 4 SSR loci used in the analysis.
cThe total number of animals are indicated from which Map with the respective SSR-types (M1-M40) were isolated.
dThe assigned identity (ID) of each farm is indicated by capital letters followed by the number of animals (in parenthesis) from that farm from which Map with the specific SSR-type was isolated. For example, A(3) implies that 3 individual animals from Farm A had Map with the specific SSR-type.
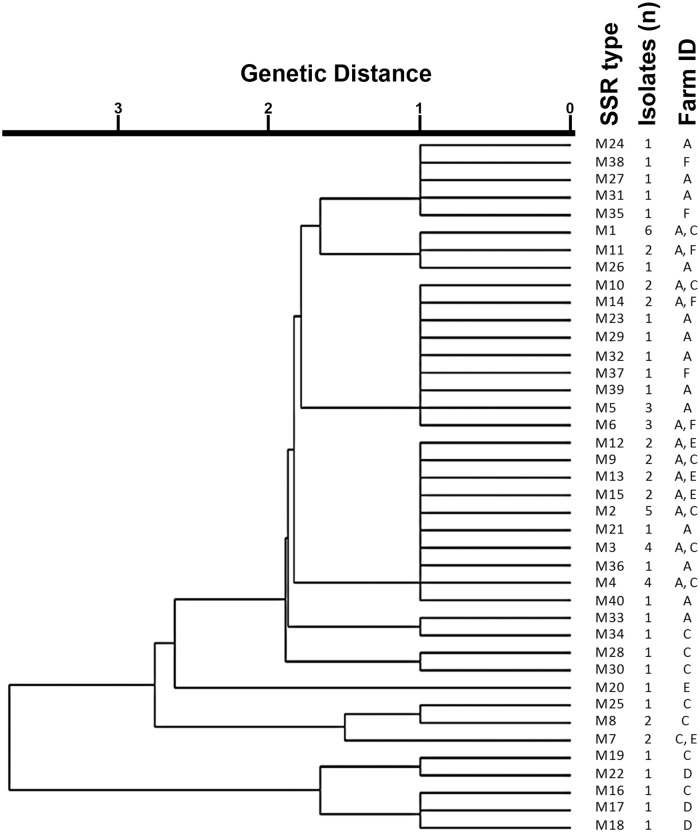
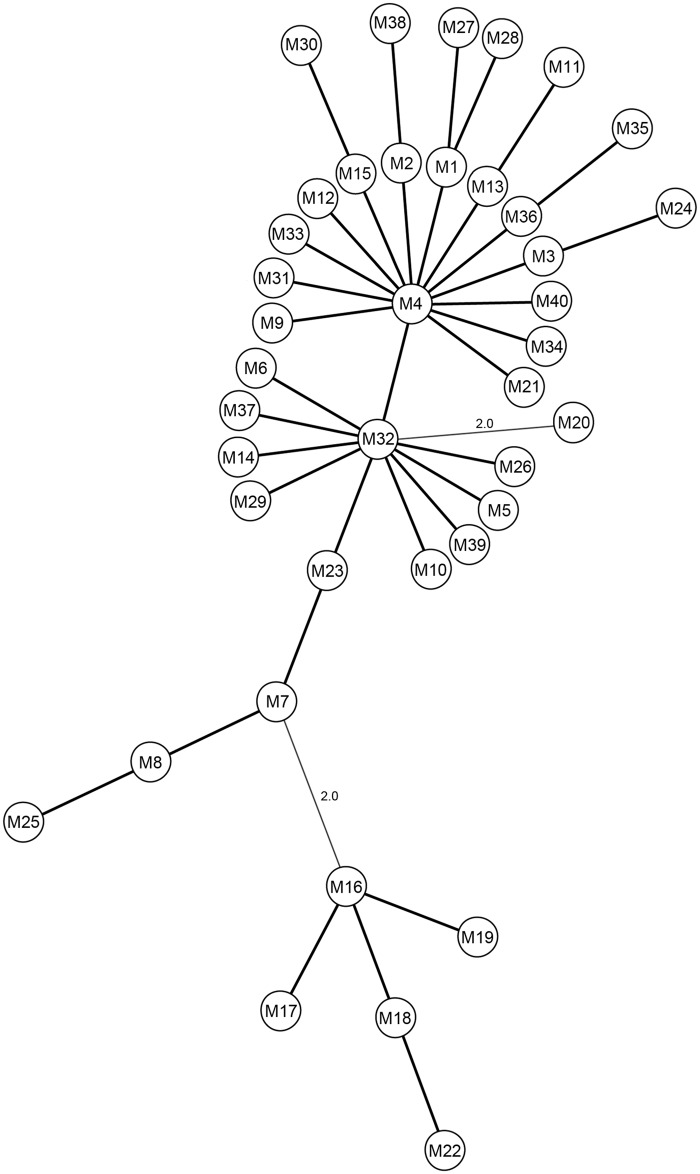
The most predominant SSR-type in the current study was M1, which was present in Map isolated from 6 separate animals from different farms (Table 1 and Fig 1). One reason for this observation could be the random distribution of Map SSR-types within farms following animal movement between farms, which is known to increase the probability of detecting similar strains on the farms involved [24]. Most SSR-types in the population were closely related to M4 and M32, differing from them in only 1–2 SSR loci (Fig 2). Overall, some farm based clustering of isolates was observed (Fig 1). A high level of diversity was seen in isolates from farm A, which alone had 25 different SSR-types out of the 40 detected. Twelve SSR-types present on farm A were also present on other farms (C, E, F) (Fig 1) suggesting probable inter herd transmission or a common source of infection. SSR-types from farm D were not detected on other farms, although they showed some level of genetic similarity with isolates from farm C (Fig 1). Animals from farm F did not exhibit any clinical signs of Johne’s disease, but tested positive for Map with unique SSR-types, in addition to SSR-types found on other farms also (Fig 1).
Fig 1. Dendrogram representing the genetic relationship between all Map isolates based on the 4 SSRs loci used in the analysis.
The dendrogram was built using the unweighted pair group method with arithmetic mean (UPGMA) using the BioNumerics 7.1 multilocus sequence typing program. Genetic distance is indicated at the top of the dendrogram. SSR-types, number of Map isolates (n) and farm ID are displayed to the right side of the dendrogram.
Fig 2. Minimum spanning tree (MST) based on the SSR profiles of the 4 loci for all 40 SSR-types identified in the current study.
The tree was generated using the BioNumerics 7.1 multilocus sequence typing program and the circles represent the M1-M40 SSR-types. Thick lines represent only one variation amidst the 4 loci, whereas thin lines represent 2 differences between the 4 loci, the latter of which is indicated.
In the current study we identified 40 distinct SSR-types, some of which have not been reported previously [5,14,18,23,26]. One reason for this finding could be the insular nature of Newfoundland, which could allow for the emergence of unique SSR-types on the island. Alternatively, animals could have acquired Map with the respective SSR-types from other herds in Atlantic Canada, since some heifers from Newfoundland are sent to New Brunswick (NB) and Nova Scotia (NS) for rearing, and are brought back when they reach adulthood. Therefore, Map with these unique SSR-types could be present in Atlantic Canada, although SSR analysis has not been reported so far for a majority of herds from NB or NS for comparison. Another explanation for the distinctive genotypes could be that all SSR-types have not been reported yet as there are only a few published Map epidemiology studies using SSR analysis [25]. In addition, mononucleotide repeats up to 21 bp could be measured using fragment analysis, but all previous studies could only resolve SSR repeats up to 15 bp [5,14,18,23,26], which could influence their results. This critical limitation of previously used technologies could underestimate Map SSR diversity despite the inclusion of a large number of isolates, as it is possible that not all loci were accurately resolved [5,19,23,25]. The only other published report which used fragment analysis for typing Map SSRs did not include the lengths of the repeats that could be analyzed [16], but it is conceivable that resolution comparable to what was observed in the current study would have been possible.
The resolution of long SSRs by fragment analysis and the recent report showing that the technique can be multiplexed for analyzing multiple SSRs [16] further demonstrates the power and versatility of this technique for typing Map isolates. Future studies using techniques with better resolution capabilities and samples from other regions of North America will help to explain if the previously unidentified Map SSRs-types reported in the current study are unique to Newfoundland or if they were not detected due to technical limitations. In addition, results from the current study also indicated Map co-infection with multiple genotypes within a single animal, which has been reported as a rare event [19]. The isolation of multiple genotypes from the same animal could also be due to evolving SSR-types due to the instability associated with long DNA repeates [27]. Based on the whole genome sequences of multiple Map isoaltes, a recent report pointed out to the limitations of certain genotyping methods for studying Map strain diversity [17]. Therefore, we are also currently in the proces sequencing the genomes of a subset of our Map isolates for more indeapth genetic analysis (data not shown). Alternately the isolation of multiple SSR-types from the same animal might represent true co- or multiple- infections, which is important in terms of source tracking and the status of the animals involved. Therefore, studies are currently underway to address the significance and implications of the described findings.
Supporting Information
(PDF)
SSR-types were designated as M1-M40 based on their unique SSR combinations.
(PDF)
Acknowledgments
We would like to thank Drs. H. Dawn Marshall and Dawn R. D. Bignell (Department of Biology, Memorial University of Newfoundland) for advice on genotyping analysis and access to specialized laboratory equipment, respectively.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The described work was funded by the Department of Natural Resources, Government of Newfoundland and Labrador (DNR-NL) and was supported by the Dairy Farmers of Newfoundland and Labrador. Equipment used in the study was purchased with funding from the Natural Sciences and Engineering Research Council of Canada (NSERC), Research & Development Corporation of Newfoundland and Labrador (RDC-NL), and the Canada Foundation for Innovation (CFI) to KT and from DNR-NL. We would also like to acknowledge NSERC and the Memorial University of Newfoundland (MUN) for additional financial assistance to MPP and SEB. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bannantine JP, Waters WR, Stabel JR, Palmer MV, Li L, Kapur V, et al. Development and use of a partial Mycobacterium avium subspecies paratuberculosis protein array. Proteomics. 2008;8: 463–474. 10.1002/pmic.200700644 [DOI] [PubMed] [Google Scholar]
- 2. Castellanos E, Aranaz A, Gould KA, Linedale R, Stevenson K, Alvarez J, et al. Discovery of stable and variable differences in the Mycobacterium avium subsp. paratuberculosis type I, II, and III genomes by pan-genome microarray analysis. Appl Environ Microbiol. 2009; 5: 676–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ferrouillet C, Wells SJ, Hartmann WL, Godden SM, Carrier J. Decrease of Johne's disease prevalence and incidence in six Minnesota, USA, dairy cattle herds on a long-term management program. Prev Vet Med. 2009;88: 128–137. 10.1016/j.prevetmed.2008.08.001 [DOI] [PubMed] [Google Scholar]
- 4. Liu X, Feng Z, Harris NB, Cirillo JD, Bercovier H, Barletta RG. Identification of a secreted superoxide dismutase in Mycobacterium avium ssp. paratuberculosis . FEMS Microbiol Lett. 2001;202: 233–238. [DOI] [PubMed] [Google Scholar]
- 5. Pradhan AK, Mitchell RM, Kramer AJ, Zurakowski MJ, Fyock TL, Whitlock RH, et al. Molecular epidemiology of Mycobacterium avium subsp. paratuberculosis in a longitudinal study of three dairy herds. J Clin Microbiol. 2011;49: 893–901. 10.1128/JCM.01107-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stabel JR. Cytokine secretion by peripheral blood mononuclear cells from cows infected with Mycobacterium paratuberculosis . Am J Vet Res. 2000;61: 754–760. [DOI] [PubMed] [Google Scholar]
- 7. Naser SA, Schwartz D, Shafran I. Isolation of Mycobacterium avium subsp paratuberculosis from breast milk of Crohn's disease patients. Am J Gastroenterol. 2000;95: 1094–1095. [DOI] [PubMed] [Google Scholar]
- 8. Prantera C, Scribano ML. Crohn's disease: the case for bacteria. Ital J Gastroenterol Hepatol. 1999;31: 244–246. [PubMed] [Google Scholar]
- 9. Ghadiali AH, Strother M, Naser SA, Manning EJ, Sreevatsan S. Mycobacterium avium subsp. paratuberculosis strains isolated from Crohn's disease patients and animal species exhibit similar polymorphic locus patterns. J Clin Microbiol. 2004;42: 5345–5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hermon-Taylor J. Treatment with drugs active against Mycobacterium avium subspecies paratuberculosis can heal Crohn's disease: more evidence for a neglected public health tragedy. Dig Liver Dis. 2002;34: 9–12. [DOI] [PubMed] [Google Scholar]
- 11. Kalis CH, Collins MT, Barkema HW, Hesselink JW. Certification of herds as free of Mycobacterium paratuberculosis infection: actual pooled faecal results versus certification model predictions. Prev Vet Med. 2004;65: 189–204. [DOI] [PubMed] [Google Scholar]
- 12. Settles M, Zanella R, McKay SD, Schnabel RD, Taylor JF, Whitlock R, et al. A whole genome association analysis identifies loci associated with Mycobacterium avium subsp. paratuberculosis infection status in US holstein cattle. Anim Genet. 2009;40: 655–662. 10.1111/j.1365-2052.2009.01896.x [DOI] [PubMed] [Google Scholar]
- 13. Manning EJ, Collins MT. Mycobacterium avium subsp. paratuberculosis: pathogen, pathogenesis and diagnosis. Rev Sci Tech. 2001;20: 133–150. [DOI] [PubMed] [Google Scholar]
- 14. Amonsin A, Li LL, Zhang Q, Bannantine JP, Motiwala AS, Sreevatsan S, et al. Multilocus short sequence repeat sequencing approach for differentiating among Mycobacterium avium subsp. paratuberculosis strains. J Clin Microbiol. 2004;42: 1694–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. El-Sayed A, Hassan AA, Natour S, Abdulmawjood A, Bulte M, Wolter W, et al. Evaluation of three molecular methods of repetitive element loci for differentiation of Mycobacterium avium subsp. paratuberculosis (MAP). J Microbiol. 2009;47: 253–259. 10.1007/s12275-008-0257-1 [DOI] [PubMed] [Google Scholar]
- 16. Oakey J, Gavey L, Singh SV, Platell J, Waltisbuhl D. Variable-number tandem repeats genotyping used to aid and inform management strategies for a bovine Johne’s disease incursion in tropical and subtropical Australia. J Vet Diagn Invest. 2014;26: 651–657. 10.1177/1040638714547257 [DOI] [PubMed] [Google Scholar]
- 17. Ahlstrom C, Barkema HW, Stevenson K, Zadoks RN, Biek R, Kao R, et al. Limitations of variable number of tandem repeat typing identified through whole genome sequencing of Mycobacterium avium subsp. paratuberculosis on a national and herd level. BMC genomics. 2015;16: 161 Available: http://www.biomedcentral.com/1471-2164/16/161. 10.1186/s12864-015-1387-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ahlstrom C, Barkema HW, De Buck J. Improved Short-Sequence-Repeat Genotyping of Mycobacterium avium subsp. paratuberculosis by Using Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry. Appl Environ Microbiol. 2014;80: 534–539. 10.1128/AEM.03212-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harris NB, Payeur JB, Kapur V, Sreevatsan S. Short-sequence-repeat analysis of Mycobacterium avium subsp. paratuberculosis and Mycobacterium avium subsp. avium isolates collected from animals throughout the United States reveals both stability of loci and extensive diversity. J Clin Microbiol. 2006;44: 2970–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tiwari A, VanLeeuwen JA, McKenna SL, Keefe GP, Barkema HW. Johne's disease in Canada Part I: clinical symptoms, pathophysiology, diagnosis, and prevalence in dairy herds. Can Vet J. 2006;47: 874–882. [PMC free article] [PubMed] [Google Scholar]
- 21. Whittington RJ, Marsh IB, Saunders V, Grant IR, Juste R, Sevilla IA, et al. Culture phenotypes of genomically and geographically diverse Mycobacterium avium subsp. paratuberculosis isolates from different hosts. J Clin Microbiol. 2011;49: 1822–1830. 10.1128/JCM.00210-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li L, Bannantine JP, Zhang Q, Amonsin A, May BJ, Alt D, et al. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis . Proc Natl Acad Sci USA. 2005;102: 12344–12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sohal JS, Arsenault J, Labrecque O, Fairbrother JH, Roy JP, Fecteau G, et al. Genetic structure of Mycobacterium avium subspecies paratuberculosis population in Quebec cattle herds revealed by using a combination of multi-locus genomic analysis. J Clin Microbiol. 2014;52: 2764–2775. 10.1128/JCM.00386-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ricchi M, Barbieri G, Taddei R, Belletti GL, Carra E, Cammi G, et al. Effectiveness of combination of Mini-and Microsatellite loci to sub-type Mycobacterium avium subsp. paratuberculosis Italian type C isolates. BMC Vet Res. 2011;7: 54 Available: http://www.biomedcentral.com/1746-6148/7/54. 10.1186/1746-6148-7-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bannantine J, Li LL, Sreevatsan S, Kapur V. How does a Mycobacterium change its spots? Applying molecular tools to track diverse strains of Mycobacterium avium subspecies paratuberculosis . Lett Appl Microbiol. 2013;57: 165–173. 10.1111/lam.12109 [DOI] [PubMed] [Google Scholar]
- 26. Forde T, Kutz S, De Buck J, Warren A, Ruckstuhl K, Pybus M, et al. Occurrence, diagnosis, and strain typing of Mycobacterium avium subspecies paratuberculosis infection in Rocky Mountain bighorn sheep (Ovis canadensis canadensis) in southwestern Alberta. J Wildl Dis. 2012;48: 1–11. [DOI] [PubMed] [Google Scholar]
- 27. Kasnitz N, Köhler H, Weigoldt M, Gerlach GF, Möbius P. Stability of genotyping target sequences of Mycobacterium avium subsp. paratuberculosis upon cultivation on different media, in vitro-and in vivo passage, and natural infection. Vet Microbiol. 2013;167: 573–583. 10.1016/j.vetmic.2013.09.008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
SSR-types were designated as M1-M40 based on their unique SSR combinations.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.