Abstract
Background
Lipoxygenases (LOXs) are important dioxygenases in cellular organisms. LOXs contribute to plant developmental processes and environmental responses. However, a systematic and comprehensive analysis has not been focused on the LOX gene family in poplar. Therefore, in the present study, we performed a comprehensive analysis of the LOX gene family in poplar.
Results
Using bioinformatics methods, we identified a total of 20 LOX genes. These LOX genes were clustered into two subfamilies. The gene structure and motif composition of each subfamily were relatively conserved. These genes are distributed unevenly across nine chromosomes. The PtLOX gene family appears to have expanded due to high tandem and low segmental duplication events. Microarray analysis showed that a number of PtLOX genes have different expression pattern across disparate tissues and under various stress treatments. Quantitative real-time PCR (qRT-PCR) analysis was further performed to confirm the responses to MeJA treatment of the 20 poplar LOX genes. The results show that the PtLOX genes are regulated by MeJA (Methyl jasmonate) treatment.
Conclusions
This study provides a systematic analysis of LOX genes in poplar. The gene family analysis reported here will be useful for conducting future functional genomics studies to uncover the roles of LOX genes in poplar growth and development.
Introduction
As an important tree species of shelterbelt and timber forest in our country, poplar trees have enormous economic and ecological benefits, as well as unique biological properties of basic scientific interest [1, 2]. However, poplar trees are affected by a variety of environmental stresses, such as the rapid spread of pests, which has led to tremendous economic loss [3, 4]. Increasing tolerance to pests and insects in order to improve both the quality and quantity of available poplar wood is one of the major aims of researchers working in this area [5].
In plants, some oxylipins [6, 7] and their derivatives, such as jasmonic acid (JA) [8], leaf aldehydes [9, 10] and divinyl ethers [11], have been suggested to be involved in plant defense against pests. Biosynthetic pathways that lead to the formation of these oxylipins in plants have been investigated. Such compounds are produced via the lipoxygenase (LOX) pathway [12].
LOX proteins (EC 1.13.11.12) are monomeric, nonheme iron that contain dioxygenases; they are widely expressed in animals, plants and fungi [13]. Polysaturated fatty acids (PUFA) that contain a cis, cis-1,4-pentadiene structural unit, such as linoleic acid (LA), α-linolenic acid and arachidonic acid, are LOX substrates. Plant LOXs are generally classified according to the specificity of their action on the substrate [14]. This substrate is oxygenated at either the C-atom 9 or C-atom 13 of the fatty acid hydrocarbon backbone. Thus, plant LOXs have been divided into two classes, 9-LOXs and 13-LOXs. According to their primary structure and overall sequence similarity, plant LOXs can be grouped into two subfamilies. Type I LOX proteins harbor no transit peptides and exhibit high sequence similarity (>75%). By contrast, type II LOX proteins carry a transit peptide sequence and show only moderate overall sequence similarity (~35%). To date, these LOX forms all belong to the subfamily of 13-LOXs [15, 16].
LOX proteins and LOX-derived products are involved in a series of biological events, such as seed germination [17], tuber development [18], sex determination [19], fruit ripening [20], and plant defense responses [21, 22]. LOXs are widely known to oxygenase polyunsaturated fatty acids (PUFAs) to produce either 13- or 9-fatty acid hydroperoxides. Hydroperoxides are then rapidly converted into diverse oxylipins [23]. In plants, oxylipins contribute to insect resistance mechanisms through two pathways. Firstly, oxylipins such as jasmonic acid (JA) that are associated with plant defenses are subsequently formed from the octadecanoid pathway via the LOX-mediated oxidation of PUFA at the 13-carbon site [8, 24]. Secondly, oxylipins such as the C6 volatile are widely known as green leaf volatiles (GLVs) because of their characteristic aroma, which is often associated with the scent of freshly-cut grass. It has been observed that different forms of GLVs play an important role in plant resistance [9, 10, 24].
Extensive research has detected the resistance of pests to LOX in a wide range of plants. In Arabidopsis thaliana, the expression of AtLOX1 is stimulated by stress-related hormones and AtLOX2 has been identified as specifically involved in jasmonate biosynthesis [25]. Previous studies have indicated that TomloxD is rapidly and transiently induced by damage in tomato [26, 27]. Antisense approaches in potato have shown that the expression of LoxH1 is detectable and involved in the generation of volatile defense systems [28]. In maize, the activity of ZmLOX3 is required for normal levels of resistance to root-knot nematodes [29]. AdLOX3, AdLOX4 and AdLOX6 are candidates for the regulation of the synthesis of kiwifruit volatile compounds [30]. VvLOXO plays a part in the response of grape berries to environmental and stress-related stimuli [31]. Recent evidence from literature reviews has shown that MdLOX5 genes might be responsible for aphid tolerance or resistance [32].
Previous work on LOX activity and the expression of two members of the LOX gene family in poplar has suggested that LOXs function in stress tolerance in trees [25]. This suggested role for LOX in poplar has prompted a more detailed analysis of the poplar LOX gene family. However, no genome-wide characterization of the LOX gene family has been performed in poplar to date.
In this study, 20 putative LOX genes were identified and characterized in the poplar genome. The responses to MeJA treatment of all of these genes were investigated using qRT-PCR analyses. Our preliminary results provide insights that will be useful for the further investigation of the roles of these candidate genes in poplar stress responses.
Materials and Methods
Ethics statement
No specific permits were required for the described field studies. No specific permissions were required for these locations and activities. The location is not privately-owned or protected in any way and the field studies did not involve endangered or protected species.
Database search and sequence retrieval
Sequences of Arabidopsis LOX proteins were obtained from the Arabidopsis Information Resource (TAIR, http://www.Arabidopsis.org/, release 10.0). Sequences of Cucumis sativus, Glycine max, Hordeum vulgare, Lens culinaris, Lycopersicon esculentum, Nicotiana attenuata, Nicotiana tabacum, Oryza sativa, Pisum sativum, Phaseolus vulgaris, Solanum tuberosum, Zea mays, Vitis vinifera, Actindia deliciosa, and Triticum aestivum were downloaded from NCBI (http://www.ncbi.nlm.nih.gov/). Sequences of Populus were downloaded from Phytozome (http://www.phytozome.net/). Local Blast searching was performed using Arabidopsis LOX proteins as queries for the identification of LOX genes from poplar. For the misannotated genes, manual reannotation was performed using the online web server Pfam (http://pfam.sanger.ac.uk/)[26]. All of the sequences were further manually analyzed to confirm the presence of LOX domain and PLAT/LH2 (polycystin-1, lipoxygenase, α-toxin domain or the lipoxygenase homology) domain using the InterProScan program (http://www.ebi.ac.uk/Tools/InterProScan/) [27].
Phylogenetic analysis
Eighty-four plant lipoxygenase amino acid sequences were analyzed, including twenty PtLOXs. Conserved protein regions were aligned using ClustalX 2.0 software [28] and adjusted manually with BioEdit [29]. NJ trees were generated using MEGA 5.0 [30], with 1000 bootstrap replicates. MrBayes software [31] was used to construct BI trees, using the WAG model of evolution, after running for 1,000,000 generations, with four Markov chains sampled every 1000 generations.
Gene structure analysis and identification of conserved motifs
The exon/intron organization of LOX genes was generated online using Gene structure display server (GSDS; http://gsds.cbi.pku.edu.cn/) by alignment of the cDNAs with their corresponding genomic DNA sequences [32]. Structural motif annotation was performed using the MEME program [33]. The maximum number of motifs was set at 20, and the optimum motif widths were set at between six and 200 residues. Structural motif annotation was performed using the Pfam program. Subcellular localization was predicted using the TargetP 1.1 (http://www.cbs.dtu.dk/services/TargetP/) server, ProtComp 9.0 (http://linux1.softberry.com/berry.phtml), Cell-PLoc (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc/) and WoLF PSORT program (http://wolfpsort.org/) [34].
Chromosomal location and gene duplication
Information about the chromosome locations of the genes was obtained from the Phytozome database. The distribution of PtLOX gene family members throughout the poplar genome was generated using MapInspect software. All these genes were compared by pairwise BLASTP (E-value<10-10) analysis [35]. Data from the recently identified duplicated blocks were obtained [36], and the segment duplication coordinates of the target genes were obtained from the Poplar JGI browser. Genes separated by five or fewer gene loci in a 60-kb region were considered to be tandem duplicates.
Calculation of Ka/Ks values
Amino acid sequences from segmentally duplicated pairs were first aligned with Clustal 2.0, and the aligned sequences were subsequently transferred into original cDNA sequences using the PAL2NAL program (http://www.bork.embl.de/pal2nal/) [37], which uses the CODEML program of PAML to estimate synonymous (Ks) and nonsynonymous (Ka) substitution rates. Divergence time (T) was calculated using a synonymous mutation rate of one substitution per synonymous site per year as T = Ks/2 λ (λ = 9.161029 for Populus) [38].
Microarray analysis
A record of gene expression in tissues was obtained from the Poplar eFP Browser (http://bar.utoronto.ca/efppop/cgi-bin/efpWeb.cgi)[39]. The genome-wide microarray data were obtained from the Gene Expression Omnibus database at the National Center for Biotechnology Information under series accession numbers GSE13990, GSE16786 and GSE23726. The data were then imported into Cluster (version3.0) to generate heatmaps. Probe sets corresponding to the putative poplar LOX genes were identified using an online Probe Match tool available at the NetAffx Analysis Center (http://www.affymetrix.com/). For genes with more than one probe sets, the median of expression values were considered. When several genes have the same probe set, then they are considered to have same level of transcript abundance [40–43].
Plant material collection
Young leaves were harvested of 1-year-old Nanlin-95 (Populus× euramericana cv. ‘Nanlin95’) plants that were grown in a greenhouse (16 h light/8 h dark, 25°C–28°C). MeJA stress treatment was conducted following a previously published method with minor modifications [25], the amount was controlled such that all leaves were densely covered with the suspension. MeJA (Sigma, 95%) was diluted 1:10 with 95% ethanol, followed by a further dilution with MilliQ water containing 0.1% Triton X-100, resulting in a final concentration of 100 μmol/L MeJA. As above, prior to each treatment, leaves were harvested for the controls. Three replicates from three independent plants were collected. Treated leaves were then harvested at 0.5, 1, 3, 6, 9, 12 and 24 h post-treatment, frozen quickly in liquid nitrogen, and stored at -70°C.
RNA extraction and qRT-PCR analysis
Total RNA from young leaves was extracted using TRIzol reagent (Shenggong, Shanghai, China) according to the manufacturer’s instructions. RNA integrity was verified by 2% agar gel electrophoresis. RNA was treated with Recombinant DNase I (RNase-free) (TaKaRa) to romove potentially contaminating genomic DNA. The first-strand cDNA was then synthesized using a PrimerScriptTM RT Reagent Kit (TaKaRa). Primers were designed using Primer5.0, and their specificity was checked using information obtained from the NCBI website (S1 Table). Subsequently, by performing analysis of melting curves and analysis of visualization of amplicon fragments, we found primers were gene-specific only when corresponding melting curves generated a single sharp peak and the primers demonstrated an electrophoresis pattern of a single amplicon with the correct predicted length. The qRT-PCR analysis was conducted using 2×SYBR Premix Ex Taq (TaKaRa). The reactions were prepared in a total volume of 25 μl containing 3 μl of template, 12.5 μl of 2×SYBR Premix, 1 μl of each specific primer (10 μM), and 7.5 μl ddH2O. The reactions were performed under the following conditions: 95°C for 30 s, 40 cycles of 5 s at 95°C, and 55°C for 34s. Every treatment included three replicates, and each reaction was run in triplicate. Relative gene expression with respect to the internal reference gene UBQ10 was determined as described previously [40–42, 44]. Standard curve analysis of all the primer pairs were performed (S1 Fig). The information of the primers and amplification efficiency of all the primer pairs were list in S1 Table. The relative mRNA level for each gene was calculated as 2-ΔΔCT values in comparison to that of unstressed leaves[45]. The relative expression levels were calculated as 2-ΔΔCT [ΔCT = CT, Target—CT, UBQ10. ΔΔCT = ΔCT, treatment- ΔCT, CK (0 h)]. The relative expression levels (2-ΔΔCT, CK (0 h)) in the untreated control plants were normalized to 1 as described previously [45–47]. If an efficiency of amplification was less than 2, the result was proofread. Statistical analyses were conducted using GraphPad Prism 5.01 software [48].
Results and Discussion
Identification of LOX gene family in poplar
A comprehensive genome-wide search of the Populus database using the AtLOX protein sequences resulted in the identification of 20 LOX genes [49]. All of the deduced proteins had features of the plant LOX gene family as described previously [15]. We excluded several genes that encoded for proteins containing only one lipoxygenase domain [50, 51] or a PLAT/LH2 domain [52, 53].
The number of LOX genes in Populus trichocarpa is approximately three-fold that of Arabidopsis [49], which is not consistent with the ratio of 1.4–1.6 putative poplar homologs per Arabidopsis gene that was predicted from comparative genomics studies. According to the present study, the number of Populus LOX genes was greater than that of Arabidopsis, which was also the case for the cucumber [54], grape [55], and apple [56] LOX gene families. The poplar LOX genes were designated PtLOXs according to nomenclature proposed in a previous study.
Detailed information about the LOX family of genes in Populus, including accession numbers and similarities to their Arabidopsis orthologs, is listed in Table 1. The length of PtLOX proteins was constant at between 796 and 927 amino acids, and the predicted open reading frames ranged from 2,391 to 2,784 bp. The calculated molecular masses of the 20 PtLOXs ranged from 90.9 to 105.6 kDa, with PtLOX14 being the largest. Sequence comparison revealed that the genes shared high levels of sequence homology at the nucleotide level (44.1% to 99.4% similartiy) within the coding region and at the amino acid level (39.9% to 99.3% similartiy) (S2 Table). Variations in the PI values of PtLOX proteins indicated that individual PtLOX proteins may require different ionic strengths, pH, and the presence of a reducing agent for their optimal performance.
Table 1. Detailed information about the LOX gene family in Populus.
| Name | Locus name | Location coordinates | Protein | Chr. | ORF length | Arabidopsis ortholog locus | ||
|---|---|---|---|---|---|---|---|---|
| (5'–3') | Length (a.a.) | Mol.Wt. (Da) | PI | (bp) | ||||
| PtLOX1 | Potri.001G015300 | 1076313–1081197 | 898 | 102243 | 7.01 | 1 | 2697 | AT3G45140 |
| PtLOX2 | Potri.001G015400 | 1090420–1098069 | 902 | 102293.9 | 6.29 | 1 | 2709 | |
| PtLOX3 | Potri.001G015500 | 1105670–1110895 | 898 | 102645.8 | 5.86 | 1 | 2697 | |
| PtLOX4 | Potri.001G015600 | 1118168–1123930 | 898 | 102654.8 | 5.71 | 1 | 2697 | |
| PtLOX5 | Potri.001G167700 | 14106872–14112847 | 923 | 104071 | 8.12 | 1 | 2772 | AT1G17420AT1G67560 |
| PtLOX6 | Potri.003G067600 | 9576888–9583048 | 925 | 104519.6 | 7.37 | 3 | 2778 | AT1G17420AT1G67561 |
| PtLOX7 | Potri.005G032400 | 2425802–2431106 | 866 | 98665.9 | 5.47 | 5 | 2601 | |
| PtLOX8 | Potri.005G032600 | 2435033–2439658 | 796 | 90918.2 | 5.48 | 5 | 2391 | |
| PtLOX9 | Potri.005G032700 | 2451619–2456194 | 866 | 98659.7 | 5.34 | 5 | 2601 | |
| PtLOX10 | Potri.005G032800 | 2462946–2469256 | 863 | 98546 | 5.51 | 5 | 2592 | |
| PtLOX11 | Potri.008G151500 | 10276751–10281394 | 880 | 100532.8 | 6.04 | 8 | 2643 | AT3G22400 |
| PtLOX12 | Potri.008G178000 | 12146645–12151320 | 927 | 105043.9 | 6.63 | 8 | 2784 | AT1G72520 |
| PtLOX13 | Potri.009G022400 | 3421114–3425183 | 901 | 102121.7 | 6.47 | 9 | 2706 | |
| PtLOX14 | Potri.010G057100 | 8651258–8655916 | 926 | 105641.2 | 6.63 | 10 | 2781 | |
| PtLOX15 | Potri.010G089500 | 11305668–11310367 | 881 | 100751 | 6.46 | 10 | 2646 | AT3G22400 |
| PtLOX16 | Potri.013G022000 | 1454479–1459136 | 871 | 99307.7 | 5.58 | 13 | 2616 | |
| PtLOX17 | Potri.013G022100 | 1461474–1466287 | 862 | 98766.3 | 6.14 | 13 | 2589 | |
| PtLOX18 | Potri.014G018200 | 1725218–1731715 | 860 | 98729.1 | 6.6 | 14 | 2583 | |
| PtLOX19 | Potri.014G177200 | 14542431–14547953 | 860 | 98485.7 | 6.34 | 14 | 2583 | |
| PtLOX20 | Potri.017G046200 | 3854572–3860007 | 898 | 102778.8 | 5.73 | 17 | 2697 | |
A previous study has reported 64 plant lipoxygenase amino acid sequences in 16 plant species [55–58]. To gain insights into the evolutionary relationship among plant LOX proteins, we examined all of the sequences of these LOX genes and all of the whole genome sequence information that was available. A complete list of these LOX genes is provided in S3 Table.
Evolutionary analysis of the Lipoxygenase genes
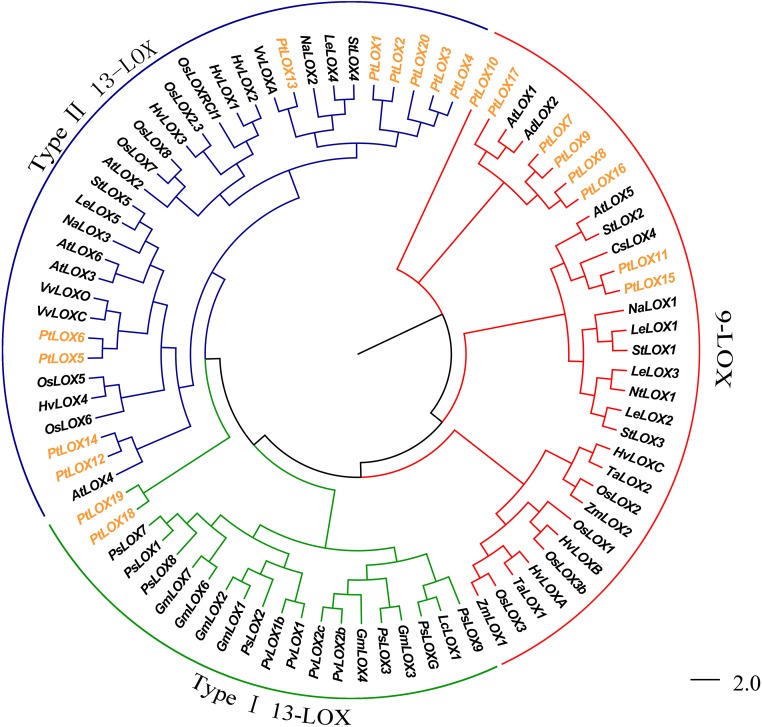
The phylogenetic analyses of LOX proteins have been reported for some plants. They could help us to uncover the evolutionary relationship of the LOX genes. Phylogenetic trees were constructed using amino acid sequence alignments of conserved regions of all predicted proteins (S4 Table) in Bayesian inference (BI) and Neighbour-Joining (NJ) methods (Fig 1 and S2 Fig). The tree topologies produced by the two methods are largely consistent. The BI tree is shown in Fig 1 and discussed below. As mentioned above, plant lipoxygenases can be divided into 9-LOX or 13-LOX subfamilies, respectively [14]. 13-LOX proteins can be further categorized into two types with respect to their protein structure. Type I LOX proteins lack plastid targeting peptides and share sequence similarities of over 75%, whereas type II LOXs possess a plastid-targeting peptide and exhibit low levels of sequence similarity.
Fig 1. An analytical view of the LOX gene family.
Bayesian estimate of phylogenetic relationships among the 84 members of LOX proteins from 17 species. Branch length is indicated by the scale bars. The tree was generated using MRBAYES program by Bayesian method and viewed in FigTree; Gene classes were indicated with different colors. Taxon labels are depicted in red for the 13-LOX clade, in blue for type II 13-LOX clade, in green for type I 13-LOX clade. Members of LOX protein from Populus were denoted in yellow.
As shown in Fig 1, PtLOX7-11 and PtLOX15-17 were grouped with the characterized 9-LOX. The remaining LOXs were placed in the type I 13-LOXs group, with the exception of PtLOX 1–4, PtLOX12-14 and PtLOX20, which belonged to the type II 13-LOX group. Upon closer inspection, we found that LOXs in 9-LOX subfamilies from monocotyledons and dicotyledons formed two exclusive clusters, respectively, which indicated that these LOX genes might have diverged after the monocotyledon/dicotyledon separation. In this subfamily, we found that PtLOX genes were closely aligned to AtLOX1, AtLOX5, StLOX2 and CsLOX4. AtLOX1 represents a regulatory point in the synthesis of jasmonic acid or other octadecanoids in response to pathogen attack [59]. AtLOX5 had been proven to be involved in lateral root development and defense responses in Arabidopsis [60]. CsLOX4 is primarily induced upon mechanical wounding and its product exhibits fungicidal activity [61]; StLOX2 was involved in herbivore-induced JA biosynthesis [62], which suggests that eight PtLOXs (PtLOX7-11, PtLOX15-17) genes may participate in the formation of JA. However, the detailed underlying mechanism of this function requires further research and confirmation.
Meanwhile, in the 13-LOX subfamily, the LOX genes exhibited an alternating distribution of monocots and eudicots, which suggested that an ancestral set of LOX genes may already have existed before the monocot-eudicot divergence. PtLOX12 and PtLOX14 were found to be closely related to AtLOX4. AtLOX4 plays an important role during seed development in Arabidopsis [63]. Genes with similar functions often are closely related and this has been confirmed in previous reports. Such a trend is also found in the LOX genes. For instances, an exclusive cluster that includes LeLOX4 [64], StLOX4 [65], NaLOX2 [66], and VvLOXA [55], all of which play important roles in protecting plants from biotic attack by producing defensive compounds. So we conjectured that PtLOX1-4, PtLOX13, and PtLOX20 could serve a similar function. Other members in this subfamily, LeLOX5 [67], StLOX5 [67], NaLOX3 [68], and VvLOXO [55] can induce plant defense mechanisms that are effective against pathogens. We can clearly see that PtLOX18 and PtLOX19 have a far-reaching genetic relationship with other members in this subfamily. It is possible that these genes have specialized roles in poplar.
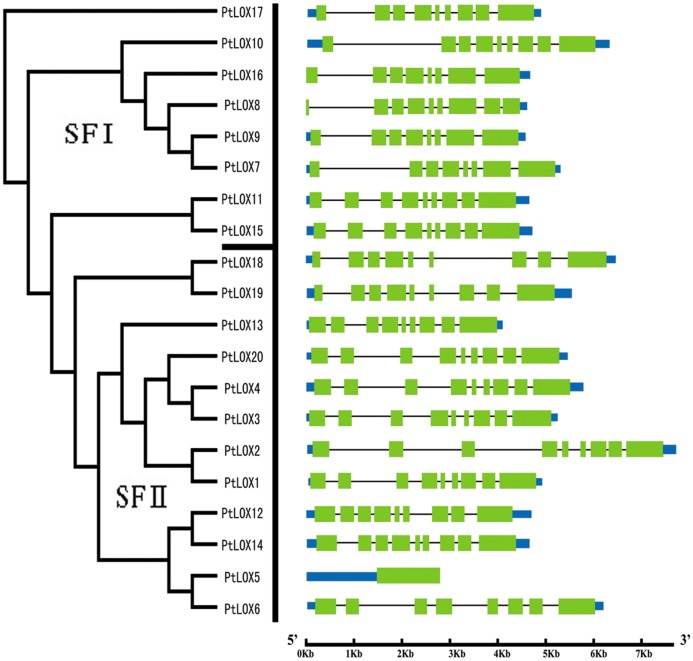
Phylogenetic and structural analysis of PtLOX genes
To gain further insight into the structural diversity of poplar LOX genes, we constructed a separate phylogenetic tree using only the poplar LOX protein sequences and compared the exon/intron organization in the coding sequence of each poplar LOX gene (Fig 2). The PtLOX genes were divided into two subfamilies; class I contains eight members, which are all 9-LOX genes, whereas class II consists of 12 members, all of which are 13-LOX genes.
Fig 2. Phylogenetic relationship and gene structure of Populus LOX genes.
The phylogenetic tree was constructed by the Neighbor-Joining (NJ) method with 1,000 bootstrap replicates. The two PtLOX phylogenetic subfamilies were designated as I–II. Exons and introns are represented by green boxes and gray lines, respectively. The sizes of exons and introns are proportional to their sequence lengths.
In our study, most members of the poplar LOX gene family were found to share similar exon/intron structures either in terms of intron numbers or exon length. As shown in Fig 2, most of the LOX genes harbor eight to nine introns in each of their open reading frames. Remarkably, only PtLOX5 lacks an intron. The emergence of a gene with no introns might result from a recent intron acquisition [69].
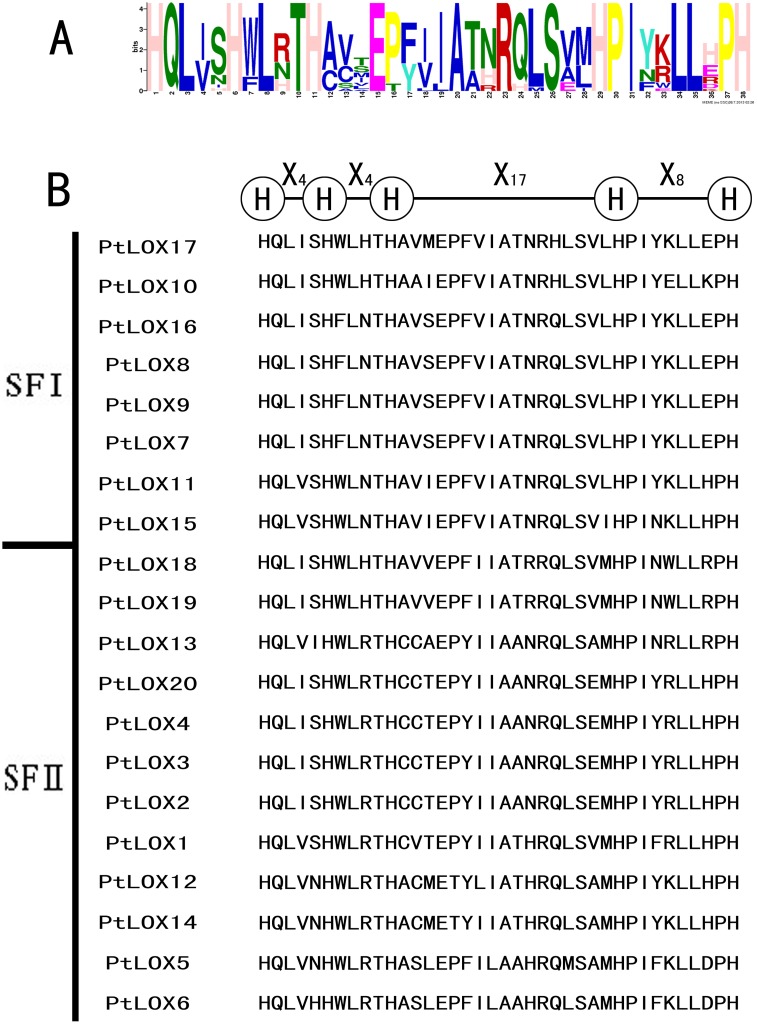
To reveal conserved motifs shared among related proteins within the family, we predicted and annotated the putative motifs. Ultimately, 20 distinct motifs were identified in the poplar LOX proteins (Fig 3 and S5 Table). As expected, most of the closely related members had common motif compositions, suggesting functional similarities among LOX proteins within the same subfamily. Based on the results, we determined that 14 of the motifs (motif 1–3, 5–6, 8, 10–11, and 13–19) are shared among all of the LOX proteins. A region rich in histamines residues was previously observed in the primary structure of soybean LOX isozymes, and further investigation has shown that His499, His504, His690, Asn694, and Ile839 are essential for the binding of iron in the C-terminal amino acid. A conservative region was also identified in 20 PtLOX members. Motif 1, which includes the representative 38 amino acid residues [24, 50, 51, 70], was in accordance with the structure His-(X)4-His-(X)4-His-(X)17-His-(X)8-His identified in a previous study; this motif is highly conserved (Fig 4). This motif plays an important role in enzyme stability and substituting any of the residues in this motif alters the activity of this enzyme.
Fig 3. Schematic representation of the conserved motifs in Populus LOX proteins elucidated using publicly available information.
Each colored box represents a motif in the protein, with the motif number indicated in the middle of the box. The length of the protein and motif can be estimated using the scale at the bottom.
Fig 4. A 38-residue motif among Populus LOX sequences.
A. The logo was created with 20 Populus LOX sequences. The overall height of each stack indicates the sequence conservation at that position and the height of each residue letter indicates the relative frequency of the corresponding amino acid residue. Below the logo is the consensus sequence. The conserved histidines (H) are marked with boldface letters. B. Sequence alignments of 38-residue motifs in Populus LOX members.
Notably, some specific motifs were present in LOXs from specific subfamilies. For instance, motif 4 was only detected in subfamily I LOX genes. Subfamily II LOXs contain the common motifs as well as other motifs (7, 9, 12, 13, and 20). Using Pfam[71], we determined that most of these motifs were LOX [50, 51]. However, whether the two novel motifs (19, 20) confer unique functional roles to LOXs remains to be investigated. In addition, we used four different kinds of software to predict the subcellular localization of these LOXs (S6 Table). Within the motif 20, proteins with extended N-termini (Fig 4) were predicted to be chloroplast-localized (S6 Table). The N-terminal regions were less conserved in both groups, harboring putative transit peptides for sub-cellular targeting.
In any case, the presence of conserved motifs in the LOX proteins from the same class provides additional support to the results of the phylogenetic analyses. On the other hand, the divergence in motif composition among different classes may indicate that the classes are functionally diversified.
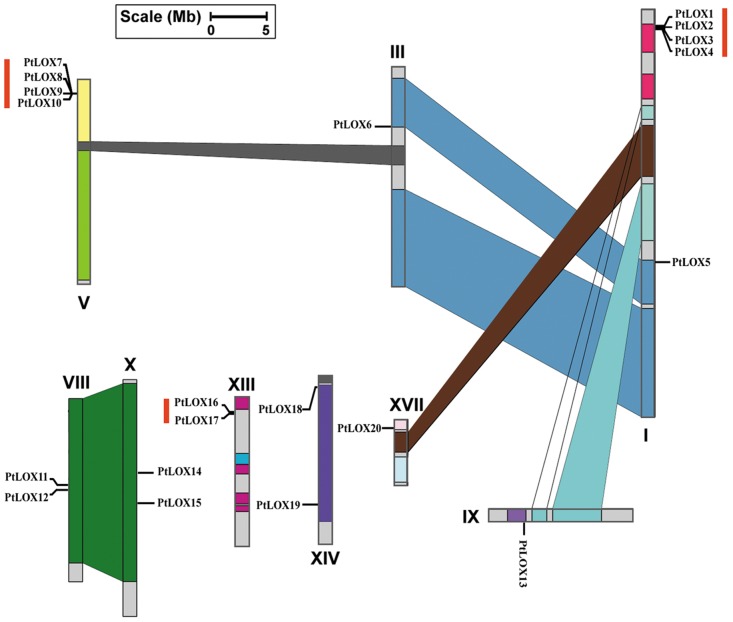
Chromosomal location and gene duplication of poplar LOX genes
Chromosomal locations were determined based on the microsatellite-based genetic map for P. trichocarpa × P. deltoides. As shown in Fig 5, PtLOX genes are present on 9 of the 19 chromosomes (Fig 5; S7 Table). The distribution of poplar LOX genes among the chromosomes appears to be uneven; chr. 3, 8, 9, 10, 13, 14, and 17 harbor one or two LOX genes, while relative high densities of LOX genes were observed in some locations on chr. 1 and chr. 5. The number of LOX genes on these chromosomes is 5 and 4, respectively.
Fig 5. Chromosomal locations of the Populus LOX genes.
Twenty genes were mapped to 9 of 19 Linkage Groups (LG). The schematic representation of genome-wide chromosome organization that arose from the whole-genome duplication event in Populus was obtained from Tuskan et al. Segmental duplicated homologous regions are shown with the same color. The duplication blocks containing LOX genes are connected with lines. Eight tandemly duplicated genes within 60 kb (shown in the red box) were organized into three clusters. Scale at the bottom represents a 5-Mb chromosomal distance.
A genome-wide duplication event occurred approximately 65 million years ago (Mya)[36]. Identification of homeologous chromosome segments resulting from whole-genome duplication events has been reported. Considering the complexity of LOX regulation, evolutionary systems biology studies are needed to explore the reasons for and implications of diversification. Based on the database from the VISTA browser, we found that three duplicate pairs (PtLOX5/6, PtLOX11/15, and PtLOX12/14) are each located in segmental duplication regions. These results suggest that the three PtLOX gene pairs (30% of the genes) are associated with the recent salicoid duplication event [36]. Additionally, comparisons of the 60-kb flanking genomic regions of the paralogous genes PtLOX1/2/3/4, PtLOX7/8/9/10 and PtLOX16/17 showed that these duplicate gene pairs were formed by a tandem duplication event. These ten PtLOX genes (50% of total) are represented in three distinct tandem duplicate gene clusters, with two clusters containing four tandem genes and one cluster containing two tandem genes. In addition, there are still four PtLOX genes that lack the corresponding duplicates, suggesting that dynamic changes may have occurred following gene duplication, which resulted in the loss of some genes.
Previous studies have shown that approximately 14,000 of the 45,000 (~32%) predicted genes are retained in duplicated pairs resulting from the salicoid duplication event [72]. In the current study, the retention rate for PtLOX family genes was found to be 30%. With such a high proportion of replication, 50% of the genes are represented in tandem clusters; this proportion is much higher than that of chromosome-level segmental duplication events. These observations suggest that the PtLOX family consists of genes originating primarily from high tandem duplication events and secondarily from low segmental duplication events. The amplification of LOX gene may due to environmental changes adaption [73].
To analyze the positive or negative selection of specific amino acid sites within the full-length sequences of the LOX proteins in the different LOX groups, we calculated the substitution rate ratios of nonsynonymous (Ka) versus synonymous (Ks) mutations [74]. The ratio of nonsynonymous versus synonymous substitutions (Ka/Ks) is an indicator of the history of selection acting on a gene or gene region. Ratios significantly <0.5 suggest purifying selection for both duplicates. A summary of Ka/Ks values for seven LOX paralogous pairs is shown in Table 2. The results suggest that all gene pairs except PtLOX18/19 have evolved mainly under the influence of purifying selection. In this study, the Ka/Ks ratio of seven putative paralogous gene pairs identified were calculated to reveal the divergence fate after duplication of poplar LOX and its closely related genes. Most Ka/Ks ratios (87.5%) of all paralogous pairs were no larger than 0.4 (Table 2). We therefore conclude that the poplar LOX gene family has undergone great purifying selection pressure and the LOX genes are slowly evolving at the protein level [75]. In addition, based on the divergence rate of 9.1×10-9 synonymous mutations per synonymous site year proposed for poplar[76], duplications of the paralogous gene pairs were estimated to occur between 0.80 and 30.87 Mya (Table 2).
Table 2. Divergence between paralogous LOX gene pairs in Populus.
| Paralogous pairs | Ka | Ks | Ka/Ks | Duplication date (MY) |
|---|---|---|---|---|
| PtLOX1–PtLOX2 | 0.1418 | 0.5618 | 0.2524 | 30.86813187 |
| PtLOX3–PtLOX4 | 0.0029 | 0.0146 | 0.2005 | 0.802197802 |
| PtLOX5–PtLOX6 | 0.0732 | 0.0288 | 0.254 | 1.582417582 |
| PtLOX7–PtLOX9 | 0.0072 | 0.0181 | 0.3982 | 0.994505495 |
| PtLOX11–PtLOX15 | 0.0425 | 0.1884 | 0.2253 | 10.35164835 |
| PtLOX12–PtLOX14 | 0.0578 | 0.2396 | 0.2412 | 13.16483516 |
| PtLOX18–PtLOX19 | 0.0224 | 0.0327 | 0.6847 | 1.796703297 |
Expression profiling of poplar LOX genes
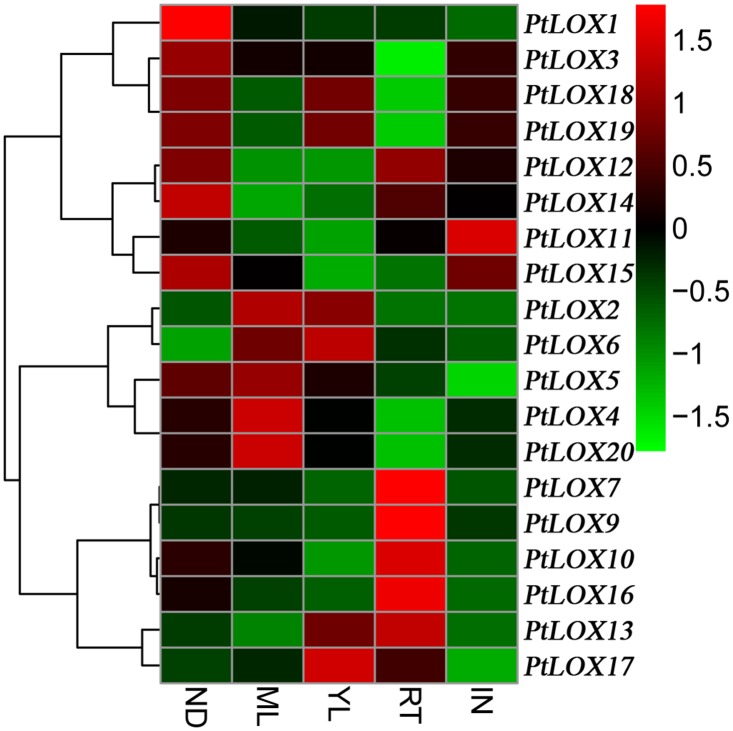
In order to understand the expression patterns of LOX genes, we data mined the publicly available microarray data. Firstly, we analyzed the expression of LOX genes in various tissues/organs. The microarray data set was obtained from PopGenIE [39]. Distinct expression profiles were identified for a total of 19 LOX genes (Fig 6 and S8 Table), whereas no corresponding probe was available for PtLOX8. Under normal growing conditions, the majority of LOX genes were expressed by only one or two tissues. There is no gene highly expressed in all tested tissues. For example, PtLOX7, -9, -10, -13, and -16 exhibited high levels of expression in roots. PtLOX2, 4–6, and -20 were present at high levels in the mature leaves, while PtLOX1 and 14–15 were mainly expressed in nodes. Thus, LOX genes showed divergent mRNA expression patterns across different tissues.
Fig 6. Expression profiles of Populus LOX genes across different tissues.
Background-corrected expression intensities were log-transformed and visualized as heatmaps (see Materials and Methods). YL, young leaves; ML, mature leaves; RT, roots; IN, internodes; ND, nodes.
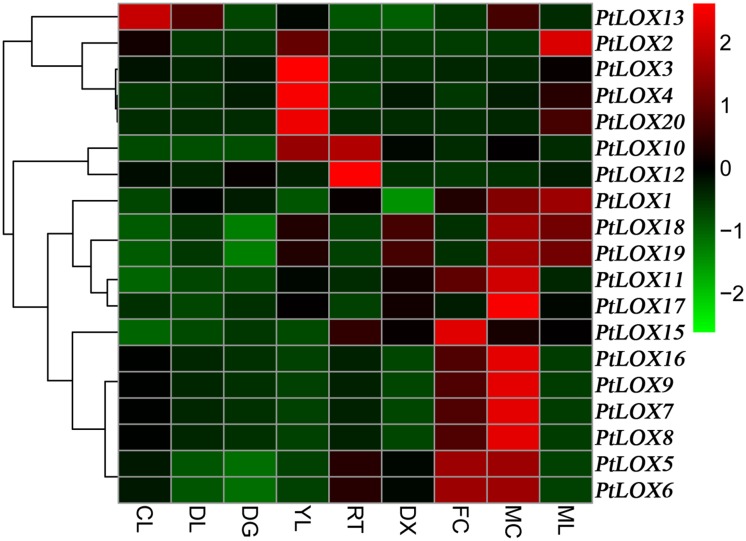
To gain further insights into the expression profiles of LOX genes, we performed a comprehensive expression analysis using some of the publicly available microarray data for Populus. Nineteen LOX genes were included on GSE13990, and only one gene (PtLOX14) was absent. Microarray data indicate that almost all LOX genes have minimal changes to each other when present in seedlings under specific conditions (CL, continuous lightgrown seedling; EL, etiolated dark-grown seedling transferred to light for 3 h; DS, dark-grown seedlings, Etiolated seedling) (Fig 7 and S9 Table). It appears that the duration of sunshine played little or no role in the distribution of LOX genes. The largest fraction of LOX genes was preferentially expressed in mature leaves (ML; 10/19), and male catkins (MC; 14/19). A relatively large fraction of LOX genes was expressed in female catkins (FC; 8/19), young leaves (YL; 10/19) and differentiating xylem (DX; 7/19). Higher expression levels of LOX genes in these parts might contribute to many developmental and biological processes. According to the expression patterns of Populus genes and the functions of their Arabidopsis orthologs, we are able to hypothesize the possible functions of these genes in Populus. For example, PtLOX15 is orthologous to AtLOX5[60], showed high expression levels in root. AtLOX5 has strong levels of biological activity and is likely to help regulate lateral root development. Therefore, PtLOX15 might have a function that is similar to AtLOX5 in the regulation of Populus root development. However, some LOX genes appear not to follow this trend. For example, PtLOX5 and PtLOX6 displayed low levels of expression in seed. However, the ortholog of PtLOX5 and PtLOX6 in Arabidopsis is AtLOX3[77], which plays an important role during seed development. This discrepancy in LOX gene regulation may suggest that LOXs adopt different physiological functions in these two plant species.
Fig 7. Hierarchical clustering of expression profiles of Populus LOX genes in different tissues.
Background-corrected expression intensities were log-transformed and visualized as heatmaps (see Materials and Methods). YL, young leaves; ML, mature leaves; RT, roots; FC, female catkins; MC, male catkins; XL, xylem. CL, continuous lightgrown seedling; DL, etiolated dark-grown seedling transferred to light for 3 h; DG, dark-grown seedlings, Etiolated seedling.
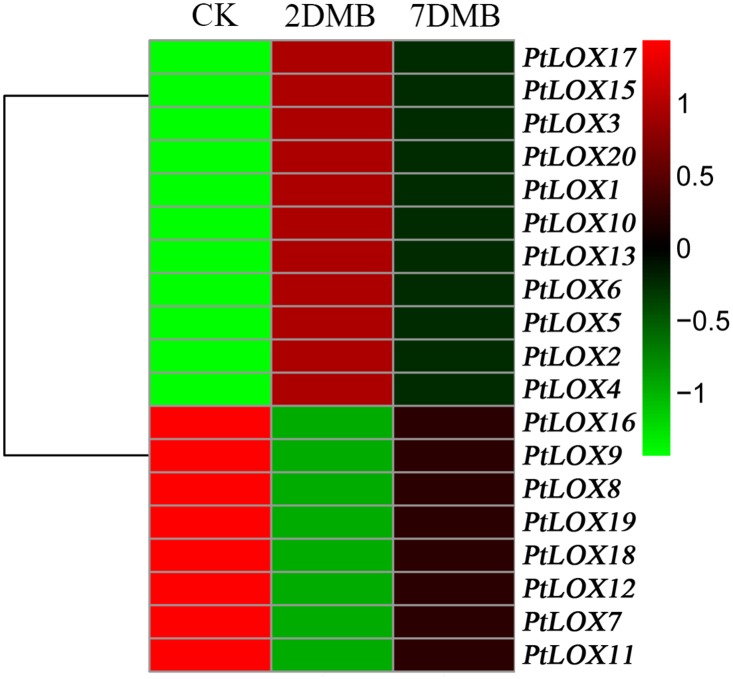
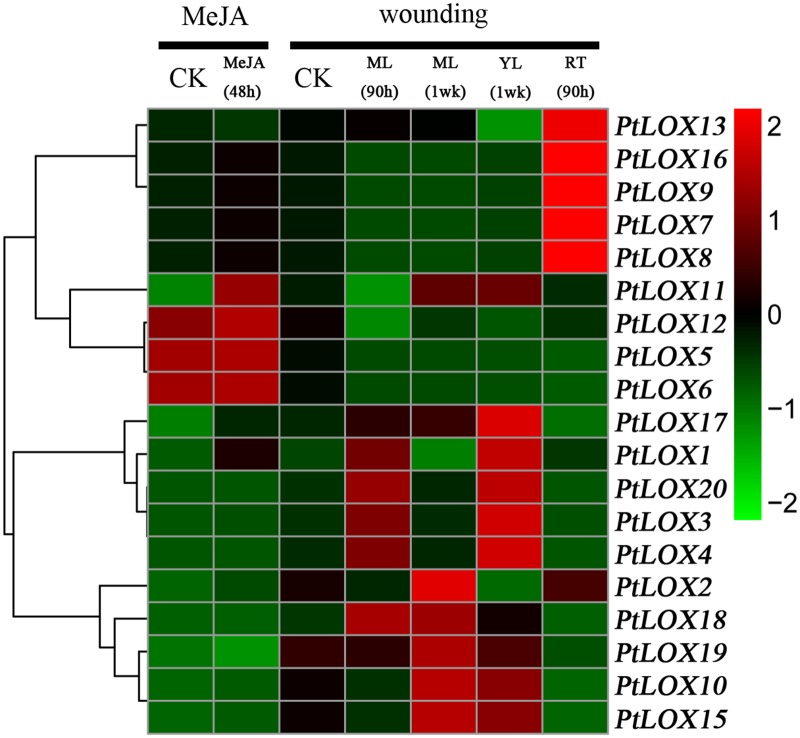
The quality of poplar is strongly affected by environmental cues during the development of the plant. Therefore, we examined the expression patterns of the Populus LOX genes under different stress conditions including infection with Marssonina brunnea (Fig 8 and S10 Table), Methyl Jasmonate (MeJA) treatment, and mechanical wounding (Fig 9 and S11 Table). The gene expression of most LOX genes is induced or suppressed under these biotic and abiotic stresses. With Marssonina brunnea infections (Fig 8), the expression levels of some genes, such as PtLOX1-6 -10, -13, -15, -17 and -20, initially increased and then decreased. Reversely, the expression levels of PtLOX7-9, 11–12, -16, -18 and -19 increased after they decreased. In response to MeJA treatments of cells in culture (Fig 9), LOX genes including PtLOX3, -4, -10, -13 and -18 were unresponsive to this stress. PtLOX15, -19 was shown to be slightly down-regulated whereas the transcripts of 12 genes (PtLOX1-2, 5–9, 11–12, 16–17, and -20) showed up-regulation. Mechanical wounding stress caused commonly down-regulation of two genes (PtLOX5 and 6) at 90 h and/or one week after wounding in young leaves, expanding leaves and root tips. In addition, a subset of genes showed up- or down-regulation only under mechanical wounding stress with root. Similarly, transcripts of a considerable proportion of genes were either enhanced or repressed following mechanical wounding in young leaves or mature leaf, respectively(Fig 9). Duplicated genes may have different evolutionary fates, which are indicated by the divergence of their expression patterns.
Fig 8. Differential expression of Populus LOX genes under different stress treatments.
Expression is indicated as fold-change of experimental treatments relative to control samples and visualized in heatmaps (see Materials and Methods). Green represents low levels and red indicates high levels of transcript abundance. Microarray data under the series accession number GSE23726 were obtained from the NCBI GEO database. CK: 0 days after inoculation with Marssonina brunnea; 2DMB: 2 days after inoculation with Marssonina brunnea; 7DMB: 7 days after inoculation with Marssonina brunnea.
Fig 9. Differential expression of Populus LOX genes under different stress treatments.
Expression is indicated as fold-change of experimental treatments relative to control samples and visualized in heatmaps (see Materials and Methods). Green represents low levels and red indicates high levels of transcript abundance. Heatmap showing hierarchical clustering of Populus LOX genes under leaf wounding and methyl jasmonate treatment. Microarray data under the series accession number GSE16786 were obtained from the NCBI GEO database. Tissues analyzed included: YL, young leaves; ML, mature leaves; RT, root tips; C, suspension cell cultures. Stress treatments included: MeJA, Methyl Jasmonate elicitation; wounding, sampled either one week or 90 hours after wounding.
Duplicated genes may have different evolutionary fates, which are indicated by the divergence of their expression patterns. Of the seven paralogous pairs of LOX genes, one gene had no corresponding database. For the remaining six paralogous pairs of LOX genes, the expression analysis of two pairs of paralogs (PtLOX5 and PtLOX6, PtLOX7 and PtLOX9) showed that they had similar expression profiles. On the contrary, one gene pair (PtLOX11 and PtLOX15) shared disparate expression patterns (Figs 8 and 9), which indicated that substantial neofunctionalization may have occurred during the evolution of one pair of genes. Beyond that, four genes (PtLOX1-4) derived from tandem duplication shared partial identical expression patterns, which suggests the functional diversification of duplicated genes [78, 79]. Such a process could increase the adaptability of duplicated genes to environmental changes, and thus improve product quality and military benefits.
Examination of LOX gene expression by qRT-PCR
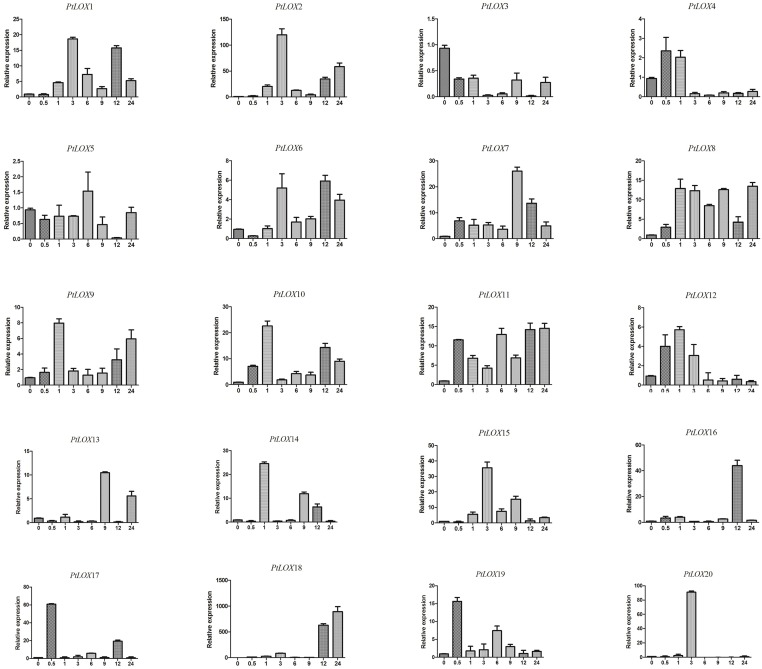
Transcript levels of the poplar LOX genes response to MeJA treatments were investigated using qRT-PCR. As shown in Fig 10A, one gene, PtLOX3, had low expression levels throughout the period, while the expression level of two other genes (PtLOX4 and -5) had no significant difference during each period. In addition to the three genes, 17 out of the 20 genes (PtLOX1-2 and -6-20) (Fig 10) were upregulated in the leaves of MeJA-treated poplar, and one gene (PtLOX18) (Fig 10) showed a significant level of upregulation after 24 h. PtLOX2 expression was upregulated within 3 h, and then its level of expression level decreased after MeJA treatment; however, the expression level of this gene showed an increasing trend after 9 h; the expression pattern of PtLOX9 (Fig 10A) and PtLOX13 (Fig 10) was similar to this. The expression of PtLOX1 and PtLOX6 (Fig 10) were also increased by MeJA treatment, reaching their maximum levels of expression after 3 h; after this time point, levels of expression was maintained above the basal levels for up to 24 h. Transcripts of PtLOX14, -16, -17, and -20 (Fig 10) were strongly induced by MeJA treatment, which peaked at some point in time, and this expression declined over time. Besides, the expression of PtLOX7, PtLOX8, PtLOX11 and PtLOX15 (Fig 10) were induced by the MeJA treatment and showed no significant changes in transcript levels during each time point after treatment. However, unlike those genes, the expression of PtLOX12 (Fig 10) reached a peak value at 3 h post-treatment. With increasing time, expression levels were lower than baseline levels at 6 h after treatment.
Fig 10. Expression analysis of PtLOX genes after MeJA induction.
Sampling occurred 0, 0.5, 1, 3, 6, 9 12 and 24 h after treatment, and the relative expression levels were analyzed. Untreated sample expression levels = 1. X-axes represent time points after MeJA treatment. Y-axes represent relative gene expression values normalized to reference gene UBQ10. Bars indicate standard deviations (SD) from three biological replicates.
By comparing the duplicated pairs of LOX genes, we observed that PtLOX11/-15 and PtLOX16/-17 (Fig 10) showed that they have similar expression profiles. This finding indicated that the responses of paralogs to stress conditions did not undergo a significant level of divergence along with the evolution of each gene after duplication. By contrast, other gene pairs PtLOX1/-2/-3/-4, PtLOX5/-6 and PtLOX12/-14 (Fig 10) shared disparate expression patterns. This is a sign of differentiation and may aid in the selection of appropriate candidate genes for further research.
Conclusions
Considerable effort has been made to characterize LOXs in many plants, including cucumber, rice, and Arabidopsis, amongst others, but such efforts have not previously been directed towards poplar. This study identified the suites of LOX genes in the poplar genome including their phylogeny, chromosomal location, gene structure, conserved motifs, and expression profiling. Finally, a total of 20 full-length LOX genes in poplar genome were identified. We searched the NCBI database for candidate poplar LOX genes and compared their protein sequences with the sequences of 64 LOX genes from 16 other plant species using phylogenetic analysis. All of the LOX genes in the draft poplar genome were verified and classified into two classes. Poplar LOX genes were clustered into two distinct subfamilies based on phylogenetic analysis. In each subfamily, the exon/intron structure and motif compositions of the LOXs were highly similar, which is indicative of their functional conservation. All PtLOX proteins contain a conventional His-(X)4-His-(X)4-His-(X)17-His-(X)8-His motif, indicating that these conserved motifs may play critical roles in family-specific functions. Chromosome distribution analysis revealed that the putative LOX genes are dispersed in 9 of 19 chromosomes. A high proportion of LOX genes are distributed preferentially at duplicated blocks, suggesting that tandem duplications contributed significantly to the expansion of this gene family. Expression analysis revealed that these PtLOX genes are expressed at higher levels in leaf tissues than in other tissues. LOXs play an important role in plant responses to multiple stresses. Furthermore, expression profiles by quantitative real-time PCR revealed that the identified PtLOXs have different patterns of expression in response to stresses. This result may reflect the involvement of these genes in specific stress-related functions.
This comprehensive analysis represents an important starting point for future efforts to elucidate the functional roles of all LOX proteins in poplar. The new information presented in this study may help in the selection of appropriate candidate genes for further functional characterization of basic aspects of the functional contexts of LOX proteins.
Supporting Information
A: UBQ10; B:PtLOX1; C:PtLOX2; D:PtLOX3; E:PtLOX4; F: PtLOX5; G:PtLOX6; H: PtLOX7; I: PtLOX 8; J: PtLOX9; K: PtLOX10; L: PtLOX11; M: PtLOX12; N: PtLOX13; O: PtLOX 14; P: PtLOX15; Q: PtLOX16; R: PtLOX17; S: PtLOX18; T: PtLOX19; U: PtLOX20.
(TIF)
Protein Neighbor-Joining tree: The unrooted tree, constructed using ClustalX 2.0, summarizes the evolutionary relationship among the 84 members of LOX proteins from 17 species. The Neighbor-Joining tree was constructed using aligned full-length amino acid sequences. The proteins are named according to their gene names (see Table 1 and S3 Table). The tree shows the two major phylogenetic classes (named 9-LOX, 13-LOX and marked with different colors) with high predictive value. The blue circle surrounds.
(TIF)
(XLS)
(XLSX)
(XLSX)
(XLSX)
Conserved motifs and the sequence logos were generated using the MEME search tool.
(XLS)
(XLSX)
Genomic sequences were obtained from Phytozome (http://www.phytozome.net/).
(XLS)
YL, young leaves; ML, mature leaves; RT, roots; IN, internodes; ND, nodes.
(XLSX)
YL, young leaves; ML, mature leaves; RT, roots; FC, female catkins; MC, male catkins; XL, xylem. CL, continuous lightgrown seedling, EL, etiolated dark-grown seedling transferred to light for 3 h, DS, dark-grown seedlings, Etiolated seedling.
(CSV)
CK: 0 days after inoculation with Marssonina brunnea; 2DMB: 2 days after inoculation with Marssonina brunnea; 7DMB: 7 days after inoculation with Marssonina brunnea.
(CSV)
CK: cell culture, control; MeJA, methyl jasmonate elicitation; wounding, sampled at 90 hours (W90h) or 1 week (W1W) after wounding.
(CSV)
Acknowledgments
We thank the members of the Laboratory of Modern Biotechnology for their assistance in this study.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by fundings from National Natural Science Foundation (31370561), Specialized Research Fund for the Doctoral Program of Higher Education (20133418110005), Anhui Provincial Natural Science Foundation (1308085MC36), Anhui Agricultural University disciplinary construction Foundation (XKTS2013001). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Dai L, Zhao F, Shao G, Zhou L, Tang L. China’s classification-based forest management: procedures, problems, and prospects. Environmental management. 2009;43(6):1162–73. 10.1007/s00267-008-9229-9 [DOI] [PubMed] [Google Scholar]
- 2. Fortier J, Truax B, Gagnon D, Lambert F. Hybrid poplar yields in Québec: implications for a sustainable forest zoning management system. The Forestry Chronicle. 2012;88(4):391–407. [Google Scholar]
- 3. Kreuzwieser J, Hauberg J, Howell KA, Carroll A, Rennenberg H, Millar AH, et al. Differential response of gray poplar leaves and roots underpins stress adaptation during hypoxia. Plant Physiology. 2009;149(1):461–73. 10.1104/pp.108.125989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Philippe RN, Bohlmann J. Poplar defense against insect herbivores This review is one of a selection of papers published in the Special Issue on Poplar Research in Canada. Botany. 2007;85(12):1111–26. [Google Scholar]
- 5. Riemenschneider DE, Stanton BJ, Vallée G, Périnet P. Poplar breeding strategies. Poplar Culture in North America. 2001;(Part A):43–76. [Google Scholar]
- 6. Prost I, Dhondt S, Rothe G, Vicente J, Rodriguez MJ, Kift N, et al. Evaluation of the antimicrobial activities of plant oxylipins supports their involvement in defense against pathogens. Plant Physiology. 2005;139(4):1902–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shah J. Plants under attack: systemic signals in defence. Current opinion in plant biology. 2009;12(4):459–64. 10.1016/j.pbi.2009.05.011 [DOI] [PubMed] [Google Scholar]
- 8. Zarate SI, Kempema LA, Walling LL. Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiology. 2007;143(2):866–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Allmann S, Baldwin IT. Insects betray themselves in nature to predators by rapid isomerization of green leaf volatiles. Science. 2010;329(5995):1075–8. 10.1126/science.1191634 [DOI] [PubMed] [Google Scholar]
- 10. Matsui K. Green leaf volatiles: hydroperoxide lyase pathway of oxylipin metabolism. Current opinion in plant biology. 2006;9(3):274–80. [DOI] [PubMed] [Google Scholar]
- 11. Itoh A, Howe GA. Molecular Cloning of a Divinyl Ether Synthase Identification as a CYP74 Cytochrome P-450. Journal of Biological Chemistry. 2001;276(5):3620–7. [DOI] [PubMed] [Google Scholar]
- 12. Howe GA, Schilmiller AL. Oxylipin metabolism in response to stress. Current opinion in plant biology. 2002;5(3):230–6. [DOI] [PubMed] [Google Scholar]
- 13. Hildebrand DF. Lipoxygenases. Physiologia Plantarum. 1989;76(2):249–53. [Google Scholar]
- 14. Brash AR. Lipoxygenases: occurrence, functions, catalysis, and acquisition of substrate. Journal of Biological Chemistry. 1999;274(34):23679–82. [DOI] [PubMed] [Google Scholar]
- 15. Porta H, Rocha-Sosa M. Plant lipoxygenases. Physiological and molecular features. Plant physiology. 2002;130(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rosahl S. Lipoxygenases in plants—their role in development and stress response. Zeitschrift fur Naturforschung C, Journal of biosciences. 1995;51(3–4):123–38. [DOI] [PubMed] [Google Scholar]
- 17. Bailly C, Bogatek-Leszczynska R, Côme D, Corbineau F. Changes in activities of antioxidant enzymes and lipoxygenase during growth of sunflower seedlings from seeds of different vigour. Seed Science Research. 2002;12(1):47–56. [Google Scholar]
- 18. Kolomiets MV, Hannapel DJ, Chen H, Tymeson M, Gladon RJ. Lipoxygenase is involved in the control of potato tuber development. The Plant Cell Online. 2001;13(3):613–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chuck G. Molecular mechanisms of sex determination in monoecious and dioecious plants. Advances in botanical research. 2010;54:53–83. [Google Scholar]
- 20. Barry CS, Giovannoni JJ. Ethylene and fruit ripening. Journal of Plant Growth Regulation. 2007;26(2):143–59. [Google Scholar]
- 21. Blée E. Impact of phyto-oxylipins in plant defense. Trends in plant science. 2002;7(7):315–22. [DOI] [PubMed] [Google Scholar]
- 22. Moran PJ, Thompson GA. Molecular responses to aphid feeding in Arabidopsis in relation to plant defense pathways. Plant Physiology. 2001;125(2):1074–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee D-S, Nioche P, Hamberg M, Raman C. Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes. Nature. 2008;455(7211):363–8. 10.1038/nature07307 [DOI] [PubMed] [Google Scholar]
- 24. Feussner I, Wasternack C. The lipoxygenase pathway. Annual review of plant biology. 2002;53(1):275–97. [DOI] [PubMed] [Google Scholar]
- 25. Cheng Q, Zhang B, Zhuge Q, Zeng Y, Wang M, Huang M. Expression profiles of two novel lipoxygenase genes in< i> Populus deltoides. Plant science. 2006;170(6):1027–35. [Google Scholar]
- 26. Finn RD, Mistry J, Schuster-Böckler B, Griffiths-Jones S, Hollich V, Lassmann T, et al. Pfam: clans, web tools and services. Nucleic acids research. 2006;34(suppl 1):D247–D51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, et al. InterProScan: protein domains identifier. Nucleic acids research. 2005;33(suppl 2):W116–W20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Larkin M, Blackshields G, Brown N, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–8. [DOI] [PubMed] [Google Scholar]
- 29. Hall T. BioEdit: An important software for molecular biology. GERF Bull Biosci. 2011;2(1):60–1. [Google Scholar]
- 30. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution. 2011;28(10):2731–9. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic biology. 2012;61(3):539–42. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guo A-Y, Zhu Q-H, Chen X, Luo J-C. GSDS: a gene structure display server]. Yi chuan = Hereditas/Zhongguo yi chuan xue hui bian ji. 2007;29(8):1023 [PubMed] [Google Scholar]
- 33. Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic acids research. 2006;34(suppl 2):W369–W73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Horton P, Park K-J, Obayashi T, Fujita N, Harada H, Adams-Collier C, et al. WoLF PSORT: protein localization predictor. Nucleic acids research. 2007;35(suppl 2):W585–W7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yu H, Luscombe NM, Lu HX, Zhu X, Xia Y, Han J-DJ, et al. Annotation transfer between genomes: protein—protein interologs and protein—DNA regulogs. Genome research. 2004;14(6):1107–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science. 2006;313(5793):1596–604. [DOI] [PubMed] [Google Scholar]
- 37. Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic acids research. 2006;34(suppl 2):W609–W12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290(5494):1151–5. [DOI] [PubMed] [Google Scholar]
- 39. Sjödin A, Street NR, Sandberg G, Gustafsson P, Jansson S. The Populus Genome Integrative Explorer (PopGenIE): a new resource for exploring the Populus genome. New phytologist. 2009;182(4):1013–25. 10.1111/j.1469-8137.2009.02807.x [DOI] [PubMed] [Google Scholar]
- 40. Chai G, Hu R, Zhang D, Qi G, Zuo R, Cao Y, et al. Comprehensive analysis of CCCH zinc finger family in poplar (Populus trichocarpa). BMC genomics. 2012;13:253 Epub 2012/06/20. 10.1186/1471-2164-13-253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hu R, Qi G, Kong Y, Kong D, Gao Q, Zhou G. Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC plant biology. 2010;10:145 Epub 2010/07/16. 10.1186/1471-2229-10-145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hu R, Chi X, Chai G, Kong Y, He G, Wang X, et al. Genome-wide identification, evolutionary expansion, and expression profile of homeodomain-leucine zipper gene family in poplar (Populus trichocarpa). PloS one. 2012;7(2):e31149 Epub 2012/02/24. 10.1371/journal.pone.0031149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cao J, Huang J, Yang Y, Hu X. Analyses of the oligopeptide transporter gene family in poplar and grape. BMC genomics. 2011;12:465 Epub 2011/09/29. 10.1186/1471-2164-12-465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zuo R, Hu R, Chai G, Xu M, Qi G, Kong Y, et al. Genome-wide identification, classification, and expression analysis of CDPK and its closely related gene families in poplar (Populus trichocarpa). Molecular biology reports. 2013;40(3):2645–62. Epub 2012/12/18. 10.1007/s11033-012-2351-z . [DOI] [PubMed] [Google Scholar]
- 45. Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2< sup>− ΔΔCT Method. methods. 2001;25(4):402–8. [DOI] [PubMed] [Google Scholar]
- 46. Chen X, Chen Z, Zhao H, Zhao Y, Cheng B, Xiang Y. Genome-Wide Analysis of Soybean HD-Zip Gene Family and Expression Profiling under Salinity and Drought Treatments. PloS one. 2014;9(2):e87156 10.1371/journal.pone.0087156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Feng L, Chen Z, Ma H, Chen X, Li Y, Wang Y, et al. The IQD Gene Family in Soybean: Structure, Phylogeny, Evolution and Expression. PloS one. 2014;9(10):e110896 10.1371/journal.pone.0110896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bryfczynski SP, Pargas RP, editors. GraphPad: a graph creation tool for CS2/CS7 ACM SIGCSE Bulletin; 2009: ACM. [Google Scholar]
- 49. Umate P. Genome-wide analysis of lipoxygenase gene family in Arabidopsis and rice. Plant signaling & behavior. 2011;6(3):335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Boyington JC, Gaffney BJ, Amzel LM. The three-dimensional structure of an arachidonic acid 15-lipoxygenase. Science. 1993;260(5113):1482–6. [DOI] [PubMed] [Google Scholar]
- 51. Steczko J, Donoho GP, Clemens JC, Dixon JE, Axelrod B. Conserved histidine residues in soybean lipoxygenase: functional consequences of their replacement. Biochemistry. 1992;31(16):4053–7. [DOI] [PubMed] [Google Scholar]
- 52. Bateman A, Sandford R. The PLAT domain: a new piece in the PKD1 puzzle. Current biology. 1999;9(16). [DOI] [PubMed] [Google Scholar]
- 53. Ponting C, Hofmann K, Bork P. A latrophilin/CL-1-like GPS domain in polycystin-1. Current biology. 1999;9(16):R585–R8. [DOI] [PubMed] [Google Scholar]
- 54. Liu S, Liu X, Jiang L. Genome-wide identification, phylogeny and expression analysis of the lipoxygenase gene family in cucumber. Genetics and molecular research: GMR. 2010;10(4):2613–36. [DOI] [PubMed] [Google Scholar]
- 55. Podolyan A. A study of the green leaf volatile biochemical pathway as a source of important flavour and aroma precursors in Sauvignon blanc grape berries: Lincoln University; 2010. [Google Scholar]
- 56. Vogt J, Schiller D, Ulrich D, Schwab W, Dunemann F. Identification of lipoxygenase (LOX) genes putatively involved in fruit flavour formation in apple (Malus× domestica). Tree Genetics & Genomes. 2013;9(6):1493–511. [Google Scholar]
- 57. Acosta IF, Laparra H, Romero SP, Schmelz E, Hamberg M, Mottinger JP, et al. tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science. 2009;323(5911):262–5. 10.1126/science.1164645 [DOI] [PubMed] [Google Scholar]
- 58. Zhang B, Chen K, Bowen J, Allan A, Espley R, Karunairetnam S, et al. Differential expression within the LOX gene family in ripening kiwifruit. Journal of experimental botany. 2006;57(14):3825–36. [DOI] [PubMed] [Google Scholar]
- 59. Zheng S-J, van Dijk JP, Bruinsma M, Dicke M. Sensitivity and speed of induced defense of cabbage (Brassica oleracea L.): dynamics of BoLOX expression patterns during insect and pathogen attack. Molecular Plant-Microbe Interactions. 2007;20(11):1332–45. [DOI] [PubMed] [Google Scholar]
- 60. Vellosillo T, Martinez M, Lopez MA, Vicente J, Cascon T, Dolan L, et al. Oxylipins produced by the 9-lipoxygenase pathway in Arabidopsis regulate lateral root development and defense responses through a specific signaling cascade. The Plant cell. 2007;19(3):831–46. Epub 2007/03/21. 10.1105/tpc.106.046052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yang X-Y, Jiang W-J, Yu H-J. The Expression Profiling of the Lipoxygenase (LOX) Family Genes During Fruit Development, Abiotic Stress and Hormonal Treatments in Cucumber (Cucumis sativus L.). International journal of molecular sciences. 2012;13(2):2481–500. 10.3390/ijms13022481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Basso B, Giribaldi MG, Mizzi L, Righi M. Cloning and sequencing of the genomic locus for a novel potato (Solanum tuberosum) lipoxygenase. Plant Biosystem. 1995;129(4):935–6. [Google Scholar]
- 63. Caldelari D, Wang G, Farmer EE, Dong X. Arabidopsis lox3 lox4 double mutants are male sterile and defective in global proliferative arrest. Plant molecular biology. 2011;75(1–2):25–33. 10.1007/s11103-011-9744-6 [DOI] [PubMed] [Google Scholar]
- 64. Gasen JB, Morecroft JF. Hemispheric Lateralization and Programming Ability. Journal of Educational Computing Research. 1990;6(1):17–27. [Google Scholar]
- 65. Göbel C. Untersuchungen zur Funktion von Oxylipinen bei der Pathogenantwort in Solanum tuberosum L: Universitäts-und Landesbibliothek; 2001. [Google Scholar]
- 66. VanDoorn A, Baldwin IT, Bonaventure G. Lipoxygenase-mediated modification of insect elicitors. Plant signaling & behavior. 2010;5(12):1674–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Acosta IF, Dellaporta SL. Methods for Controlling Flower Development in Plants. Google Patents; 2010. [Google Scholar]
- 68. Heidel AJ, Barazani O, Baldwin IT. Interaction between herbivore defense and microbial signaling: bacterial quorum-sensing compounds weaken JA-mediated herbivore resistance in Nicotiana attenuata. Chemoecology. 2010;20(2):149–54. [Google Scholar]
- 69. Takashima Y, Ohtsuka T, González A, Miyachi H, Kageyama R. Intronic delay is essential for oscillatory expression in the segmentation clock. Proceedings of the National Academy of Sciences. 2011;108(8):3300–5. 10.1073/pnas.1014418108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Shibata D, Steczko J, Dixon J, Hermodson M, Yazdanparast R, Axelrod B. Primary structure of soybean lipoxygenase-1. Journal of Biological Chemistry. 1987;262(21):10080–5. [PubMed] [Google Scholar]
- 71. Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, et al. The Pfam protein families database. Nucleic acids research. 2004;32(suppl 1):D138–D41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science. 2006;313(5793):1596–604. Epub 2006/09/16. 10.1126/science.1128691 . [DOI] [PubMed] [Google Scholar]
- 73. Kondrashov FA. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proceedings Biological sciences / The Royal Society. 2012;279(1749):5048–57. Epub 2012/09/15. 10.1098/rspb.2012.1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Nekrutenko A, Makova KD, Li W-H. The KA/KS ratio test for assessing the protein-coding potential of genomic regions: an empirical and simulation study. Genome research. 2002;12(1):198–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351(6328):652–4. [DOI] [PubMed] [Google Scholar]
- 76. Hurst LD. The Ka/ Ks ratio: diagnosing the form of sequence evolution. TRENDS in Genetics. 2002;18(9):486–7. [DOI] [PubMed] [Google Scholar]
- 77. Bannenberg G, Martínez M, Hamberg M, Castresana C. Diversity of the enzymatic activity in the lipoxygenase gene family of Arabidopsis thaliana. Lipids. 2009;44(2):85–95. 10.1007/s11745-008-3245-7 [DOI] [PubMed] [Google Scholar]
- 78. Blanc G, Wolfe KH. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. The Plant Cell Online. 2004;16(7):1667–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tattersall EA, Grimplet J, DeLuc L, Wheatley MD, Vincent D, Osborne C, et al. Transcript abundance profiles reveal larger and more complex responses of grapevine to chilling compared to osmotic and salinity stress. Functional & integrative genomics. 2007;7(4):317–33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A: UBQ10; B:PtLOX1; C:PtLOX2; D:PtLOX3; E:PtLOX4; F: PtLOX5; G:PtLOX6; H: PtLOX7; I: PtLOX 8; J: PtLOX9; K: PtLOX10; L: PtLOX11; M: PtLOX12; N: PtLOX13; O: PtLOX 14; P: PtLOX15; Q: PtLOX16; R: PtLOX17; S: PtLOX18; T: PtLOX19; U: PtLOX20.
(TIF)
Protein Neighbor-Joining tree: The unrooted tree, constructed using ClustalX 2.0, summarizes the evolutionary relationship among the 84 members of LOX proteins from 17 species. The Neighbor-Joining tree was constructed using aligned full-length amino acid sequences. The proteins are named according to their gene names (see Table 1 and S3 Table). The tree shows the two major phylogenetic classes (named 9-LOX, 13-LOX and marked with different colors) with high predictive value. The blue circle surrounds.
(TIF)
(XLS)
(XLSX)
(XLSX)
(XLSX)
Conserved motifs and the sequence logos were generated using the MEME search tool.
(XLS)
(XLSX)
Genomic sequences were obtained from Phytozome (http://www.phytozome.net/).
(XLS)
YL, young leaves; ML, mature leaves; RT, roots; IN, internodes; ND, nodes.
(XLSX)
YL, young leaves; ML, mature leaves; RT, roots; FC, female catkins; MC, male catkins; XL, xylem. CL, continuous lightgrown seedling, EL, etiolated dark-grown seedling transferred to light for 3 h, DS, dark-grown seedlings, Etiolated seedling.
(CSV)
CK: 0 days after inoculation with Marssonina brunnea; 2DMB: 2 days after inoculation with Marssonina brunnea; 7DMB: 7 days after inoculation with Marssonina brunnea.
(CSV)
CK: cell culture, control; MeJA, methyl jasmonate elicitation; wounding, sampled at 90 hours (W90h) or 1 week (W1W) after wounding.
(CSV)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.