Abstract
The identification of regulatory motifs is important for the study of gene expression. Here we present a suite of programs that we have developed to search for regulatory sequence motifs: (i) BioProspector, a Gibbs-sampling-based program for predicting regulatory motifs from co-regulated genes in prokaryotes or lower eukaryotes; (ii) CompareProspector, an extension to BioProspector which incorporates comparative genomics features to be used for higher eukaryotes; (iii) MDscan, a program for finding protein–DNA interaction sites from ChIP-on-chip targets. All three programs examine a group of sequences that may share common regulatory motifs and output a list of putative motifs as position-specific probability matrices, the individual sites used to construct the motifs and the location of each site on the input sequences. The web servers and executables can be accessed at http://seqmotifs.stanford.edu.
INTRODUCTION
Regulatory elements are short sequences of DNA (5–20 bp in length) that determine the timing, location and level of gene expression (1). In recent years, sequences that contain regulatory elements have become easily available due to the effort for large-scale sequencing of many genomes. Meanwhile, technologies such as microarray and ChIP-on-chip (or GSLA for Genome-Scale Location Analysis) make it feasible to identify potential targets of transcription factors. Incidentally, many computational methods, such as MEME (2), AlignACE (3) and Consensus (4), have been developed in the past decade to predict regulatory elements and regulatory motifs. Compared with experimental procedures to determine regulatory elements, computational motif-finding programs are fast and inexpensive. Their predictions provide biologists with valuable hypotheses for experimental validation.
Our group has developed a suite of specialized regulatory motif-finding programs. The first program is BioProspector, which is based on the original Gibbs Motif Sampler (5), but has several important improvements. It has been applied successfully to prokaryotes and lower eukaryotes such as Bacillus subtilis and yeast, respectively. In higher eukaryotes, upstream regions are usually longer and noisier. CompareProspector, with a comparative genomics component on top of BioProspector, takes in cross-species sequence comparison information to help guide the search in higher eukaryotes. We also developed MDscan, a program for motif finding from ChIP-on-chip targets. The algorithms for these three programs have been tested and published. We have since developed them into interactive web-based applications available as a public resource at http://seqmotifs.stanford.edu. Also on the website is comprehensive information about the program, format for input/output and stand-alone executables for local use. These programs are complementary in nature, and together they provide a useful resource for the study of gene expression regulation.
DESCRIPTION OF THE PROGRAMS
BioProspector
BioProspector (http://seqmotifs.stanford.edu or http://bioprospector.stanford.edu) is a program that uses Gibbs sampling to search a list of sequences (e.g. promoters of co-regulated genes) for potential regulatory motifs (6). Gibbs sampling first initializes the motif matrix using w mers (w being the width of the motif) randomly selected from the input. Then it samples from all w mers in the input sequences to update the motif matrix. The probability of selecting a w mer is proportional to the likelihood of generating the w mer from the current motif matrix over the likelihood of generating it from the non-motif background. The motif matrix is updated until convergence, or until a certain number of sampling iterations has been reached.
BioProspector has several significant improvements compared with the original Gibbs Motif Sampler (5). It uses a Markov model estimated from all promoter sequences in the genome to model adjacent nucleotide dependency and improve motif specificity. It also adopts two thresholds to allow each input sequence to contain zero to multiple copies of the motif. As prokaryotic motifs often occur in two blocks with a gap of variable length, BioProspector is capable of modeling motifs with two blocks or with palindromic patterns. In MDscan (7), a better motif scoring function has been identified via simulations and theoretical analyses, and adopted in the current version of BioProspector.
CompareProspector
Non-coding sequences in higher eukaryotes are typically much longer than those of prokaryotes. As a result, eukaryotic regulatory motif prediction faces the challenge of sifting for motif signals with much more noise. Based on our finding that known regulatory elements in non-coding sequences are more likely to be conserved than background non-coding sequences (8), we extended BioProspector to bias the motif search towards regions that are conserved across species. Even when only a subset of the input sequences has identifiable orthologs, CompareProspector (http://seqmotifs.stanford.edu or http://compareprospector.stanford.edu) shows improved performance over BioProspector for human/mouse or Caenorhabditis elegans/C.briggsae sequences.
To utilize cross-species sequence information, CompareProspector takes as input an array of window percentage identity values (WPIDs) for each input sequence with available ortholog. Although we calculated the WPIDs based on the global alignment of each input sequence with its ortholog generated using LAGAN (9), the WPIDs can be calculated using any alignment method the user chooses. In Gibbs sampling iterations, CompareProspector biases the motif search towards sequences conserved across species. First of all, users can specify two WPID thresholds. During initial iterations, only positions whose WPID values are above the high threshold are sampled. Subsequently, the WPID cutoff can be gradually decreased to the low conservation threshold to allow sampling of less conserved positions. In addition, the probability of selecting a w mer to update the motif matrix is weighted by sequence conservation to favor sampling of more conserved sequences. Sequences without orthologs are assigned the low conservation threshold, so as to participate in sampling only in later iterations.
MDscan
MDscan (http://seqmotifs.stanford.edu or http://mdscan.stanford.edu/) is designed for motif finding from sequences obtained from a ChIP-on-chip or GSLA experiment, a procedure now routinely used to characterize genome-wide protein–DNA interaction and transcription regulation (7). Since many of the sequences selected by ChIP-on-chip, especially highly enriched ones, often have multiple copies of the motif, MDscan first enumerates all the non-redundant w mers as seeds in the top t most enriched sequences. For each seed, MDscan constructs a candidate motif matrix using all the w mers in the top t sequences that are ‘neighbors’ of that seed. A pair of w mers is considered ‘neighbors’ if it shares at least m matched positions, where the probability for two random w mers sharing ≥m matched positions is 0.0015. All the motif matrices are evaluated using a semi-Bayesian scoring function and the best ones are saved. In the subsequent updating process, each retained motif matrix is refined by adding or removing w mers in all the sequences to increase the motif score, and the best refined motifs are reported.
We recently developed a new program, Motif Regressor (10), to better utilize mRNA expression level or ChIP-on-chip enrichment information to improve the performance of MDscan. Motif Regressor first identifies a set of non-redundant candidate motifs using MDscan, and scans the promoter region of every gene in the genome with each candidate motif to measure how well a promoter matches a motif (in terms of both the number of sites and the strength of matching). It then uses linear regression analysis to select motifs whose promoter matching scores are significantly correlated with ChIP-on-chip enrichment or downstream gene expression values. When ranking motifs by linear regression p-value, Motif Regressor automatically picks the best motif and optimal motif width.
Due to its computational intensity, Motif Regressor is not currently available as a web server. However, for interested users to explore the program locally, the program is available for download at http://www.techtransfer.harvard.edu/Software/MotifRegressor/.
INPUTS/OUTPUTS OF THE PROGRAMS
Inputs and parameters
The following inputs are required for the web servers of all three programs above.
User's email. Results of the search will be emailed to the user.
A user-defined job name, which will be sent in the result email.
A list of sequences that may share regulatory motif(s). These sequences can be obtained from microarray experiments, ChIP-on-chip experiments, manually selected sequences hypothesized to share sequence motifs or other methods.
The width(s) of the motif to search for.
Background model. Users can specify their own background model or choose from a list of pre-computed models for many genomes.
Whether the motif occurs in every sequence or just some of the sequences.
Whether to search in both strands of each sequence or just the forward strand.
Number of top motifs to report.
There are also some program-specific inputs.
To search for two-block motifs with BioProspector, users need to specify the width of the second block and the length of the gap between the two blocks.
CompareProspector requires a window percentage identity value file. If LAGAN is used to align orthologous sequences, users can input a file with all the alignments in multi-fasta format, and a window percentage identity file will be automatically generated from this file. Users also need to specify a high window percentage identity threshold cutoff and low threshold cutoff (if the user chooses to decrease the threshold over the iterations).
MDscan requires users to specify the number of top-ranked sequences (default 20) to search for seed motifs and the number of candidate motifs to retain.
Outputs
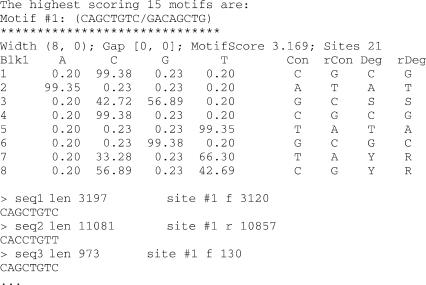
All three programs report a number of overall highest scoring motifs as position-specific probability matrices. For each motif, a list of predicted sites of the motif and their locations on the input sequence are also reported (Figure 1). For requests submitted to the servers, output will be emailed to users together with the user-specified job name, and a list of parameters used for the program.
Figure 1.
Typical output for BioProspector, CompareProspector and MDscan. Highest scoring motifs are listed. The first line for each motif lists motif width (blk 1, blk 2), gap length (min gap, max gap), motif raw score and the number of aligned sites. The motifs are shown as position-specific probability matrices. Represented as IUPAC symbols are consensus (Con)—the most abundant base, reverse complement consensus (rCon), degenerate consensus (Deg)-where all bases with >25% abundance are considered, and reverse degenerate consensus (rDeg). Also listed are sequences that have the motif, with sequence name, length of the sequence, site number, orientation and location of the site (‘f’ means the site is on the forward strand, whereas ‘r’ means the site is on the reverse strand), and the actual sequence of the site.
DISCUSSION
We present three specialized regulatory motif-finding programs and their web servers. BioProspector, a program that can search for two-block motifs with variable gap and palindromic motifs, is ideal for identifying motifs in prokaryotes and lower eukaryotes. CompareProspector is more powerful when orthologous sequences from other species are available. It gives better performance than several other motif-finding programs using datasets from both human and C.elegans (8). MDscan works best with ChIP-on-chip data, where it can take advantage of enrichment level information, or any dataset where the user can rank the sequences according to how likely motif sites are to occur.
Most of the inputs and parameters are fairly intuitive. In many motif-finding problems, the motif width is unknown. In these cases, we recommend testing different widths from 6 bp up to 17 bp. Our experiences recommend starting with motif width 8–10 bp for eukaryotes and 12–24 bp for prokaryotes. If top motifs found have highly degenerate positions (e.g. most frequent base <50% or top two frequent bases <80%) at the two ends, a shorter motif width should be used; if the consensuses of several top motifs overlap and there are conserved non-overlapping positions at either end, motif width should be increased. Recently, an algorithm named BioOptimizer has been developed that can help optimize motif width based on the conservation information (11). For CompareProspector, a high window percentage identity threshold (and a low threshold if needed) has to be specified. From our experience, 0.8 as high threshold and 0.5 as low threshold work fine for human–mouse comparisons, and 0.5 as high threshold and 0.3 as low threshold work fine for C.elegans–C.briggsae comparisons.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank Marina Sirota for her help with webpage design, Russ Altman and Stuart Kim for their insight and help during the development of CompareProspector, and Erin Conlon for her contribution in developing Motif Regressor.
REFERENCES
- 1.Pennacchio L.A. and Rubin,E.M. (2001) Genomic strategies to identify mammalian regulatory sequences. Nature Rev. Genet., 2, 100–109. [DOI] [PubMed] [Google Scholar]
- 2.Bailey T.L. and Elkan,C. (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, Vol. 2, AAA1 Press, pp. 28–36. [PubMed] [Google Scholar]
- 3.Roth F.P., Hughes,J.D., Estep,P.W. and Church,G.M. (1998) Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat. Biotechnol., 16, 939–945. [DOI] [PubMed] [Google Scholar]
- 4.Hertz G.Z., Hartzell,G.W., III and Stormo,G.D. (1990) Identification of consensus patterns in unaligned DNA sequences known to be functionally related. Comput. Appl. Biosci., 6, 81–92. [DOI] [PubMed] [Google Scholar]
- 5.Liu J.S., Neuwald,A.F. and Lawrence,C.E. (1995) Bayesian models for multiple local sequence alignment and Gibbs sampling strategies. J. Am. Stat. Assoc., 90, 1156–1170. [Google Scholar]
- 6.Liu X., Brutlag,D.L. and Liu,J.S. (2001) BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac. Symp. Biocomput., 127–138. [PubMed] [Google Scholar]
- 7.Liu X.S., Brutlag,D.L. and Liu,J.S. (2002) An algorithm for finding protein DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat. Biotechnol., 20, 835–839. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y., Liu,X.S., Wei,L., Altman,R.B. and Batzoglou,S. (2004) Eukaryotic regulatory element conservation analysis and identification using comparative genomics. Genome Res., 14, 451–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brudno M., Do,C.B., Cooper,G.M., Kim,M.F., Davydov,E., Green,E.D., Sidow,A. and Batzoglou,S. (2003) LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res., 13, 721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conlon E.M., Liu,X.S., Lieb,J.D. and Liu,J.S. (2003) Integrating regulatory motif discovery and genome-wide expression analysis. Proc. Natl Acad. Sci. USA, 100, 3339–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen S.T. and Liu,J.S. (2004) BioOptimizer: a bayesian scoring function approach to motif discovery. Bioinformatics, doi: 10.1093/bioinformatics/bth127 [DOI] [PubMed] [Google Scholar]