Abstract
Molecular interaction networks, such as those involving protein–protein and protein–DNA interactions, often consist of thousands of nodes or even more, which severely limits the usefulness of many graph drawing tools because they become too slow for interactive analysis of the networks and because they produce cluttered drawings with many edge crossings. We present a new, fast-layout algorithm and its implementation called WebInterViewer for visualizing large-scale molecular interaction networks. WebInterViewer (i) finds a layout of the connected components of an entire network, (ii) finds a global layout of nodes with respect to pivot nodes within the connected components and (iii) refines the local layout of each connected component by first relocating midnodes with respect to their cutvertices and the direct neighbors of the cutvertices, and then relocating all nodes with respect to their neighbors within distance 2. The advantages of WebInterViewer over classical graph drawing methods include the facts that (i) it is an order of magnitude faster, (ii) it can visualize data directly from protein interaction databases and (iii) it provides several abstraction and comparison operations for analyzing large-scale biological networks effectively. WebInterViewer is accessible at http://interviewer.inha.ac.kr/.
INTRODUCTION
Information about molecular interactions such as those involved in protein–protein and protein–DNA interactions has greatly increased with the recent advances in high-throughput technologies, and software tools have been developed to visualize and analyze such large-scale data. These are usually visualized as a network, with molecules as nodes and molecular interactions as edges. Force-directed layout algorithms are often used to visualize molecular interaction networks, but a naive implementation of a layout algorithm encounters real difficulties when drawing large-scale graphs such as protein interaction networks. A common problem with many force-directed layout algorithms is that they are very slow when dealing with large graphs because layout adjustment at each step typically involves computation of force between every pair of nodes.
We previously developed a force-directed layout program called InterViewer (1–3) that represents protein–protein interactions as a three-dimensional network. InterViewer is much faster than Pajek (4), Tulip (5) and Cytoscape (6), all of which are recent implementations of force-directed layout algorithms. In addition to its speed, InterViewer is one of the most advanced and comprehensive visualization tools for studying protein interactions in that it can visualize data directly from protein interaction databases and provides several abstraction and comparison operations for effective analysis of large-scale protein interaction networks.
In this paper, we present a web-based application program called WebInterViewer that produces a molecular interaction network of good quality without computing force between every pair of nodes. WebInterViewer improves on InterViewer in many ways.
Whereas InterViewer produces a drawing by computing force between every pair of nodes in each iteration of the optimization process, WebInterViewer produces a more pleasing drawing without the need for such computation.
It provides several abstraction operations to reduce complex networks to simpler ones.
It can search multiple molecular interaction networks for shared molecules and for interactions shared by all or some of the networks.
It is more convenient to use since it is a web-based application program. For example, if a molecule is named by its GI number (GenInfo identifier), WebInterViewer automatically links the molecule to its entry in the NCBI database.
The rest of this paper presents the new features of WebInterViewer.
LAYOUT OF MOLECULAR INTERACTION NETWORKS
Since a molecular interaction network tends to be a disconnected graph with several connected components, we first compute a layout of connected components and then compute a layout of nodes within the connected components. Our experience is that this approach produces much better drawings in a shorter time than computing a layout of all nodes from the outset.
WebInterViewer uses a multilevel technique to draw graphs. This is composed of two steps at the top level: grouping and layout. In the grouping step, the algorithm first groups nodes of a disconnected graph into connected components, and then finds midnodes and pivot nodes in each connected component. In the layout step, the coarsest graph is an initial layout of connected components based only on their pivot nodes. The layout of each connected component is then refined locally within each connected component on the basis of its midnodes and the neighbors of each node. The details of the layout algorithm appear in (3).
If information on the functions of molecules, and on superfamilies, is available in the database, WebInterViewer can visualize molecular interaction networks as layered graphs consisting of three layers: domain, function and molecule layers (Figures 1 and 2). We describe next how WebInterViewer assigns the nodes to layers.
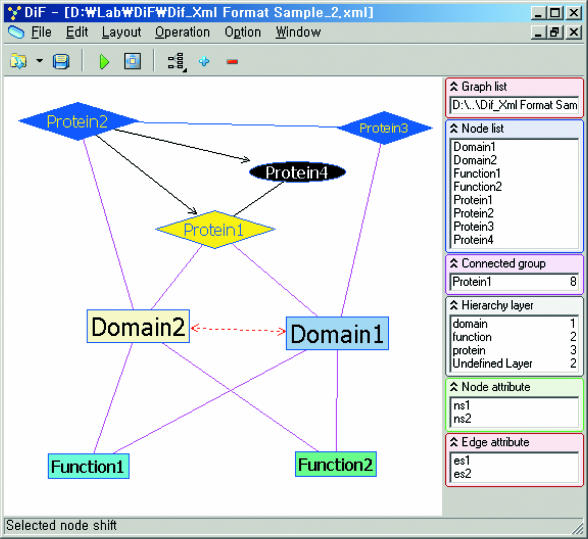
Figure 1.
Protein interaction network visualized as a layered graph with 3 layers: domain layer, function layer and protein layer.
Figure 2.
Examples of layered networks. (A) A network with three layers: domain layer (top), protein–protein interaction layer (middle) and function layer (bottom). The protein–protein interaction layer contains ENSP00000000442 and its interacting partners within distance 2. Relationships between layers are shown with cyan lines, and relationships within a layer are shown with red lines. (B) A network with two layers: function layer (top) and protein layer (bottom). The protein layer contains proteins involved in cell cycle regulation (7) and their interacting partners within distance 1. Relations between layers are shown with cyan lines, and relations within layers are shown with red lines. protein–protein interactions and functional catalogs were obtained from MIPS. Ellipses, rounded rectangles and rectangles represent functions, proteins common to genetic and physical interactions and rest proteins, respectively.
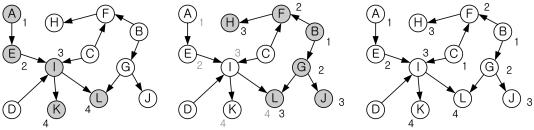
It first places all the nodes with no parent in layer L1, and then each remaining node n in layer Lp+1, where Lp is the layer of n's parent node. When the layer of a node has already been assigned, the larger layer value is assigned to that node. Node L in the middle graph of Figure 3, for example, is assigned to layer 4 from the path (A, E, I, L), but to layer 3 from the path (B, G, L). The larger value, 4, becomes the layer number of node L. The number of edges whose span > 1 should be minimized because they cause the subsequent steps of the algorithm to take longer. We place the source node of an edge whose span > 1 into a higher layer so that the span of the edge becomes 1 (the span of an edge is the difference between the layers of the end points of the edge).
Figure 3.
Example of assignment of nodes to layers. The layer numbers in gray indicate the previously assigned numbers.
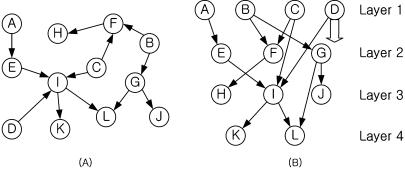
Figure 3 provides an example of initial layer assignment for the protein interaction data below. The initial layer assignment is adjusted to minimize edge spans, as shown in Figure 4.
Figure 4.
(A) Initial digraph. (B) Layered digraph. Node D is moved to layer 2 since the span of the edge (D, I) is 2.
![]()
ANALYSIS OF COMPLEX INTERACTION NETWORKS
A large number of edges and nodes in a complex molecular interaction network often reduces the readability of the network due to cluttered edges and nodes. In general, there are two ways to analyze such a complex network. One is to extract smaller subnetworks from the total network and analyze each subnetwork individually. For example, one can extract a subnetwork of molecules within a specified interaction distance from one or more target molecules, or a subnetwork of molecules shared by several interaction networks (Figure 5).
Figure 5.
Example of the extraction of proteins (shown in green) that interact with all the selected proteins (shown in yellow) within distance 3.
The other way is to abstract the entire network into a simpler form instead of focusing on subnetworks. WebInterViewer provides the following operations to abstract a network (2). The abstract network can then be expanded into a detailed network on demand.
Collapse a clique into a star-shaped subgraph. A clique in an undirected graph G = (V,E) is a subset of V, each pair of which is connected by an edge in E. Each clique is replaced by a star-shaped subgraph centered on a dummy node that is shown as a circle in the abstract graph.
Collapse a group of nodes with the same interactions into a composite node. A group of nodes with identical interacting partners is collapsed into a single composite node, shown as a diamond in the abstract graph. Whereas collapsing a clique into a star-shaped graph only reduces the number of edges, collapsing nodes with the same interactions into a composite node reduces the number of nodes as well as the number of edges (Figure 6).
Figure 6.
(A) A molecular interaction network with 307 nodes and 1063 edges. (B) Abstract network with 311 nodes and 700 edges constructed by replacing the cliques in (A) with star-shaped subgraphs centered at dummy nodes, shown as red circles. (C) Abstract network with 74 nodes and 583 edges made by collapsing a group of nodes with the same interacting partners in (A) into composite nodes, shown as diamonds.
RESULTS
There are three implementations of WebInterViewer: pure OCX version, OCX version, and Java version. The OCX versions can be executed within a web browser on any PC with Windows 2000/XP/Me/98/NT 4.0 as its operating system. The pure OCX version consists of OCX codes only. It does not require an additional file but only provides the basic functionality of InterViewer. On the other hand, the OCX version supports various features of InterViewer but requires an additional file. Both versions are updated automatically, so that a user always runs the latest version.
The Java version (called InterViewer_Java) works with virtually all browsers, but is slower than the OCX versions. It runs on any system that supports the Java 2 Platform, Standard Edition (J2SE), but does not work with J2SE 1.5 Beta. Thus, it runs on Windows 95/98/NT/2000, Solaris (SPARC and Intel editions), Linux/i486 and Macintosh OS X systems with J2SE installed. InterViewer_Java is executed by Java Web Start, which, depending on the user's system, ensures that the latest version of WebInterViewer, as well as the correct version of the Java Runtime Environment (JRE), is deployed.
The WebInterViewer takes data in several formats as input interaction. Data files in pid_label or pid_pos format are automatically read if they already exist when the user opens a file in pid format. It takes
data from a Microsoft Access database,
data on molecules, domains and functions in XML format,
GML: Graph Modelling Language format (http://www.uni-passau.de/Graphlet/GML),
pnm: names of a pair of interacting molecules, separated by a space or tab, in each line,
pid: a pair of interacting molecule indices, separated by a tab, in each line,
pid_label: the index of a pair of molecules and its name, separated by a tab, in each line. Each molecule index has a corresponding index in the pid file,
pid_pos: an ordered list of molecule indices and their positions (x, y and z coordinates), separated by a tab, in each line. Each molecule index has a corresponding index in the pid file,
data in XIN format from DIP.
As output WebInterViewer produces two kinds of drawing (bitmap and GML file). The molecular interaction data can also be saved in an ASCII file in any of the input formats described above. The program allows users to explore three-dimensional drawings by rotation or by zooming in and out.
WebInterViewer provides two ways of comparing multiple molecular interaction networks. One is to find a subnetwork shared by all networks being compared, the other to find a subnetwork shared by some of the networks, by coloring the networks. If a molecule is named by its GI number, WebInterViewer automatically connects to NCBI and displays the entry for the molecule. Users can also retrieve the information on molecular interactions from the WebInterViewer server via ftp and analyze the interactions from any system.
Acknowledgments
ACKNOWLEDGEMENT
This work was supported by the Ministry of Information and Communication of Korea under grant number IMT2000-C3-4.
REFERENCES
- 1.Ju B.-H., Park,B., Park,J. and Han,K. (2003) Visualization and analysis of protein interactions. Bioinformatics, 19, 317–318. [DOI] [PubMed] [Google Scholar]
- 2.Ju B.-H. and Han,K. (2003) Complexity management in visualizing protein interaction networks. Bioinformatics, 19, i177–i179. [DOI] [PubMed] [Google Scholar]
- 3.Han K. and Ju,B.-H. (2003) A fast layout algorithm for protein interaction networks. Bioinformatics, 19, 1882–1887. [DOI] [PubMed] [Google Scholar]
- 4.Batagelj V. and Mrvar,A. (2001) Pajek—analysis and visualization of large networks. Lecture Notes Comput. Sci., 2265, 477–478. [Google Scholar]
- 5.David A. (2001) Tulip. Lecture Notes Comput. Sci., 2265, 435–437. [Google Scholar]
- 6.Shannon P., Markiel,A., Ozier,O., Baliga,N.S., Wang,J.T., Ramage,D., Amin,N., Schwikowski,B. and Ideker,T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res., 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spirin V. and Mirny,L.A. (2003) Protein complexes and functional modules in molecular networks. Proc. Natl Acad. Sci. USA, 100, 12123–12128. [DOI] [PMC free article] [PubMed] [Google Scholar]