Abstract
One of the greatest challenges that modern molecular biology is facing is the understanding of the complex mechanisms regulating gene expression. A fundamental step in this process requires the characterization of regulatory motifs playing key roles in the regulation of gene expression at transcriptional and post-transcriptional levels. In particular, transcription is modulated by the interaction of transcription factors with their corresponding binding sites. Weeder Web is a web interface to Weeder, an algorithm for the automatic discovery of conserved motifs in a set of related regulatory DNA sequences. The motifs found are in turn likely to be instances of binding sites for some transcription factor. Other than providing access to the program, the interface has been designed so to make usage of the program itself as simple as possible, and to require very little prior knowledge about the length and the conservation of the motifs to be found. In fact, the interface automatically starts different runs of the program, each one with different parameters, and provides the user with an overall summary of the results as well as some ‘advice’ on which motifs look more interesting according to their statistical significance and some simple considerations. The web interface is available at the address www.pesolelab.it by following the ‘Tools’ link.
INTRODUCTION
Understanding the complex mechanisms governing basic biological processes requires the characterization of regulatory motifs modulating gene expression at transcriptional and post-transcriptional levels. In particular extent, chronology and cell-specificity of transcription are modulated by the interaction of transcription factors (TFs) with their corresponding binding sites (TFBSs) (1), mostly located nearby the transcription start site (TSS) of the gene (i.e. proximal promoter region) or far away (i.e. enhancers, silencers, etc.). The ever growing amount of genomic data (complemented by other sources of information such as full-length cDNA sequencing projects that permit the precise mapping of the TSS on the genome sequence) and expression data derived from microarray and other experiments open new opportunities to researchers.
The fact that transcription factor binding sites are generally short (<12–14 bp long) and degenerate oligonucleotides makes their computational discovery and large-scale annotation significantly hard—hence the need for efficient and reliable methods for detecting novel motifs significantly over-represented in the regulatory regions of sets of genes sharing common properties (e.g. similar expression profile, biological function, product cellular localization), which in turn could correspond to binding sites for common TFs regulating the genes.
We present here a web server that provides access to a previously developed enumerative pattern discovery method (2) that is able to carry out an (almost) exhaustive search of significantly conserved degenerate oligonucleotide patterns with remarkable computational efficiency.
METHODS
Nearly all the computational methods for the discovery of novel motifs in a set of sequences of co-regulated genes are based on two steps. First, one or more groups of oligonucleotides similar enough to each other (i.e. differing in some nucleotide substitutions) are detected in the sequences. Second, their presence is evaluated from a statistical point of view; that is, algorithms estimate how likely each group would be to appear in a set of sequences either picked at random from the same organism (thus very unlikely to be coregulated) or built randomly with the same nucleotide composition as the input sequences (thus very likely to present a different oligo composition). The best groups of oligos found are in turn likely to be instances of binding sites for some TF [see (3,4) for reviews on different methods applied to the problem, as well as programs and interfaces implementing them].
For the first step, two main approaches have been proposed so far: consensus-based (or pattern-driven) and profile-based (or alignment-driven) methods. In the former case, the different oligonucleotides recognized by a given TF are described by their consensus, representing, for each position, the nucleotide that appears most frequently in the binding sites. All oligos that differ from the consensus in no more than a given number of positions (usually depending on the oligo length) can in turn be supposed to be recognized by the same TF. The debate on which ‘philosophy’ is more suitable for representing and capturing TFBSs is nearly as old as the problem itself (5–7). However, comparative tests (8) have shown that the two approaches seem to be complementary, with no definite prevalence of either one. Some instances are correctly solved by representing motifs with their consensus, some others by using alignment profiles, and some by both (or neither). Thus, given a set of sequences, the best way to proceed is perhaps to try different methods based on wholly different principles, such as the one presented here and traditional alignment-driven methods such as Consensus (9), MEME (10) and the Gibbs sampler (11), and to compare the results obtained.
The key idea of consensus-based methods is to enumerate all the oligos of (or up to) a given length, in order to determine which ones appear, with possible substitutions, in a significant fraction of the input sequences, and finally to rank them according to statistical measures of significance. At first sight, this approach seems to suffer from different drawbacks. First of all, if the length of the motifs sought is m, then there are 4m candidate oligos to enumerate, with an exponential growth of the execution time according to the motif length. Then, many additional parameters are usually required by these algorithms, such as the length of the motif itself, the number of mutations allowed for its occurrences and a minimum number q of sequences the motif has to appear in (this parameter is usually called quorum). Moreover, a suitable significance measure, able to discriminate real TF binding sites from uninteresting motifs, has to be introduced, since often there are hundreds of candidate motifs satisfying the input parameters. All these factors have led to the impression that consensus-based methods are too slow (given the high number of candidates) and too difficult to use, since many different parameter combinations have to be tried.
However, as demonstrated in (2,12), the exhaustive search for motifs can be significantly accelerated if the input sequences are preprocessed and organized in a suitable indexing structure, such as a suffix tree (13), especially when their lengths fall within the usual range of TFBSs. We refer the reader to reference (2) for further details on how the exhaustive search is implemented in the Weeder algorithm. Instead, in the implementation of the interface presented here we focused on the other points just mentioned. The first task is to determine automatically which values of all the parameters needed by the algorithm are suitable for TFBSs, in order to reduce to the minimum possible the user input and any prior knowledge about the motifs to be found, as well as to make the program as simple to use as alignment-based algorithms. Then, we also propose a significance measure especially fine-tuned for TFBSs, which is used to rank the results and to find the best motifs.
User input
The Weeder Web interface (an example is shown in the online Supplementary Material) requires users to input their email address, and one or more sequences in FASTA format either by cutting and pasting the sequences or by uploading a file. Sequences can be in uppercase or lowercase letters, and can contain ambiguous IUPAC symbols. A checkbox is available in order to specify if the complementary strand of the input sequences has also to be examined by the program. Then, users must provide values of a few intuitive parameters. The first one specifies only whether the motif has to appear in all the input sequences or in some of them (analogous to the ‘zoops’ mode of alignment-based methods). The user can also select a mode in which the input is processed as a single sequence, looking for repeated oligonucleotides regardless of their distribution throughout the sequences. The second parameter needed, describing the type of analysis desired (quick, normal or thorough), simply influences the time required to obtain the results: clearly, the shorter the time, the less accurate is the analysis performed. Also, users must specify which organism their sequences come from, by selecting it from a list. This choice is fundamental for the computation of the significance of the results (see Supplementary Material), since this is based on organism-specific expected values. If the organism is not included in the list provided, users can contact the page administrators. The list of available organisms will be constantly updated and enlarged. Since the computation time in some cases might exceed an hour, the results are sent by email. The results are accessible also on a dedicated web page, whose address is communicated in the email as well. The web page will also provide a link to the original input sequences provided by the user.
In addition, if the user wants to set manually all or some of the input parameters (motif length, quorum, error and so on), an extended input form is available, which can be reached by following a link from the main page. Also, this page permits the user to submit nucleotide sequences other than upstream regulatory regions (for which the default parameter values for TFBSs might not be appropriate). At the present time, it includes human 5′ and 3′ mRNA untranslated regions. Further types of nucleotide sequences will be included in the future to broaden the applicability of the method to signals other than TFBSs.
Program runs
Once the ‘Submit’ button is clicked, if all the fields are filled in correctly the web interface automatically starts a series of runs of the Weeder algorithm, looking for motifs of lengths 6 and 8 (if launched in quick mode), or from length 6 up to 12 (in normal mode and thorough mode). The number of sequences a motif has to appear in is determined according to the user's choice (if ‘some’ is selected, the threshold is set to half of the sequences). The number of mutations allowed is one for motifs of length 6, two for length 8, three for length 10 and four for length 12. The ‘thorough’ mode performs an additional scan for motifs of length 8 with three mutations and length 10 with four mutations, and lowers the quorum choice to one-third of the input sequences. Clearly, the last parameter setting is useful when no significant motif is reported by quick and normal analyses. Experiments performed by us and by other groups [see e.g. (14,15)] have shown that these values are suitable for capturing a large class of TFBSs, even in the case of corrupted datasets including several sequences not containing instances of the motif.
The output
For each run, all the motifs satisfying the input constraints (length, error and quorum) are scored according to a statistical measure of significance (see Supplementary Material) that takes into account the number of sequences a motif appears in, how much it is conserved and the overall number of its occurrences in the input set. Then, the five most significant motifs of each run are reported to the user. However, it is sometimes difficult to judge which ones, among motifs of different lengths obtained allowing different degrees of approximation, could be more ‘interesting’ and worth further investigation, especially (as in real case studies) when the motif length is not known in advance. Clearly, if a motif reported has a score significantly higher than the others it is less likely to be the effect of random similarities. However, for this task, we have included an additional post-processing stage. The idea is best explained by an example. Suppose that the consensus for the binding sites of a given TF is A[C/G]GTAC, admitting with equal frequency either a C or a G in the second position. If all went well in the program runs, we might expect to find both ACGTAC and AGGTAC in the list of the best motifs of length 6 with one mutation. And we also expect this phenomenon to be quite frequent in real binding sites, since the different positions of the sites usually exhibit different degrees of variability, and mutations seem to appear more frequently at some positions than at others. Thus, first of all the algorithm scans each list of best motifs of length m with e mutations (the result of each single run) to see whether in each one there are motifs that differ in no more than e positions. These motifs could be two alternative consensuses for the same set of binding sites, and the higher scoring one is reported as ‘interesting’. Then, the results of different runs are also compared to each other. If a short motif is found to be a part of a longer one, then the latter might have a conserved core, another feature often encountered in real instances. Also in this case, the longer one is added to the ‘interesting’ motif list. Several experiments that we have performed on real case studies support the feasibility of these simple criteria.
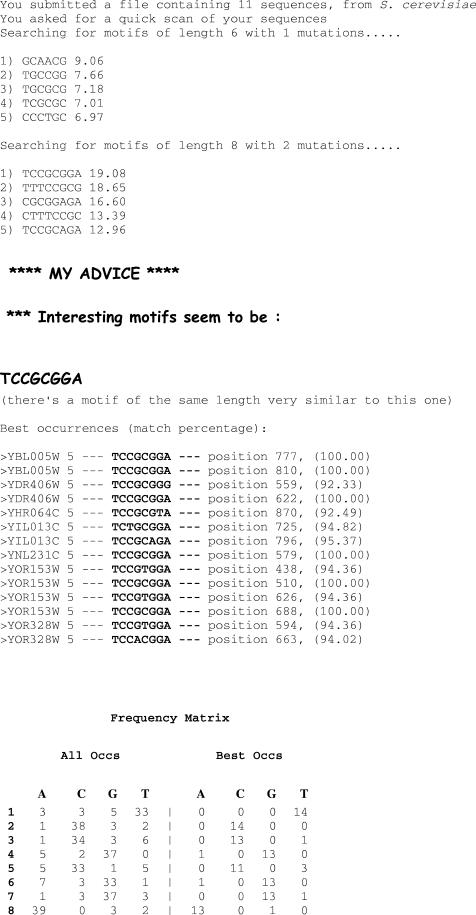
Finally, all the ‘interesting’ motifs are again listed at the bottom of the output file, under the heading ‘My Advice’. For each of these, the interface also reports a frequency matrix built by aligning all the instances of the motif found as well as a list of its best occurrences, collected from the input sequences by using the frequency matrix (see Supplementary Material). Another frequency matrix, obtained by aligning only the best occurrences of the motif, is also listed. Although motifs are discovered by allowing a predefined number of mutations e in their occurrences, the additional frequency matrix scan makes it possible to pick new instances presenting more than e mutations with respect to the motif consensus but still fitting the motif profile well, and at the same time to single out which instances are more likely to be real TFBSs. Also, even if the motif has been detected in q of the input sequences, the best instances might not be present in each of the q sequences (or else the motif might be found to appear in additional sequences). An example of the output file sent to users is shown in Figure 1. Notice that the best motif of length 6 differs significantly from the best 8mer. However, the advice of the program is to pay attention to the latter (the correct one), since there is another motif differing from it in a single position among the highest scoring motifs of length 8.
Figure 1.
An example of the output of the program. In this case, it reports the results of the runs on lengths 6 and 8 (quick mode), with the five top-scoring motifs of each run. The motif reported as ‘interesting’ corresponds to the consensus of PDR3 binding sites in yeast, as described in the SCPD database (17), obtained by analysing the same sequence set used in (14). The output lists the best occurrences of the motif in the input sequences, as well as the frequency matrices built by aligning all the instances found (left) and the best instances only (right). The match percentage value in brackets defines how well each instance fits the matrix description (see Supplementary Material).
DISCUSSION
The web interface to the Weeder program permits the analysis of a set of regulatory sequences looking for conserved motifs that in turn could represent instances of binding sites for some common TF. The interface has been designed with the 2-fold purpose of being as user-friendly as possible and of making the interpretation of the results easier. All the actual parameters that are needed by the Weeder algorithm are kept hidden and automatically set to values suitable for the discovery of TFBSs according to some intuitive indications provided by the user, who can, however, change the default values by using an extended input form. Some hints and comments on the results obtained are also output.
In the future, the interface will be constantly updated and enhanced with new features, including the graphical representation of the output and the possibility of applying the algorithm to other types of nucleotide sequences. On the algorithmic side, we plan to include the comparative analysis of regulatory sequences of orthologous genes from different species, and the possibility of detecting motifs composed of two conserved parts interrupted by a non-conserved region, as well as correlations among different motifs (i.e. combinations of two or more elements showing a conserved order, strand orientation and spacing). To this end, we have developed a suitable algorithm (16) that will be included in a further version of the server. Indeed, the search for complex regulatory modules, greatly reducing false positive rates, would make possible genome-wide promoter analyses. Finally, we are currently working on the integration of microarray expression values with sequence data, which could represent another big leap forward in promoter annotation and the discovery of novel TF binding sites. Each update will be highlighted and reported on the web page.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
REFERENCES
- 1.Levine M. and Tjian,R. (2003) Transcription regulation and animal diversity. Nature, 424, 147–151. [DOI] [PubMed] [Google Scholar]
- 2.Pavesi G., Mauri,G. and Pesole,G. (2001) An algorithm for finding signals of unknown length in DNA sequences. Bioinformatics, 17 (Suppl. 1), S207–S214. [DOI] [PubMed] [Google Scholar]
- 3.Stormo G.D. (2000) DNA binding sites: representation and discovery. Bioinformatics, 16, 16–23. [DOI] [PubMed] [Google Scholar]
- 4.Pavesi G., Mauri,G. and Pesole,G. (2001) Methods for pattern discovery in unaligned biological sequences. Brief. Bioinformatics, 2, 417–430. [Google Scholar]
- 5.Berg O.G. and von Hippel,P.H. (1988) Selection of DNA binding sites by regulatory proteins. Trends Biochem. Sci., 13, 207–211. [DOI] [PubMed] [Google Scholar]
- 6.Frech K., Quandt,K. and Werner,T. (1997) Finding protein-binding sites in DNA sequences: the next generation. Trends Biochem. Sci., 22, 103–104. [DOI] [PubMed] [Google Scholar]
- 7.Frech K., Quandt,K. and Werner,T. (1997) Software for the analysis of DNA sequence elements of transcription. Comput. Appl. Biosci., 13, 89–97. [DOI] [PubMed] [Google Scholar]
- 8.Sinha S. and Tompa,M. (2003) Performance comparison of algorithms for finding transcription factor binding sites, Third IEEE Symposium on Bioinformatics and Bioengineering. Washington DC, pp. 69–78. [Google Scholar]
- 9.Hertz G.Z. and Stormo,G.D. (1999) Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics, 15, 563–577. [DOI] [PubMed] [Google Scholar]
- 10.Bailey T.L. and Elkan,C. (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol., 2, 28–36. [PubMed] [Google Scholar]
- 11.Thompson W., Rouchka,E.C. and Lawrence,C.E. (2003) Gibbs Recursive Sampler: finding transcription factor binding sites. Nucleic Acids Res., 31, 3580–3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marsan L. and Sagot,M.F. (2000) Algorithms for extracting structured motifs using a suffix tree with an application to promoter and regulatory site consensus identification. J. Comput. Biol., 7, 345–362. [DOI] [PubMed] [Google Scholar]
- 13.Gusfield D. (1997) Algorithms on Strings, Trees, and Sequences: Computer Science and Computational Biology. Cambridge University Press, Cambridge, UK and New York. [Google Scholar]
- 14.Narasimhan C., LoCascio,P. and Uberbacher,E. (2003) Background rareness-based iterative multiple sequence alignment algorithm for regulatory element detection. Bioinformatics, 19, 1952–1963. [DOI] [PubMed] [Google Scholar]
- 15.Sinha S. and Tompa,M. (2002) Discovery of novel transcription factor binding sites by statistical overrepresentation. Nucleic Acids Res., 30, 5549–5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pavesi G., Mauri,G., Iannelli,F., Gissi,C. and Pesole,G. (2004) GeneSyn: a tool for detecting conserved gene syntenies across genomes. Bioinformatics, 10.1093/bioinformatics/bth1102 [DOI] [PubMed] [Google Scholar]
- 17.Zhu J. and Zhang,M.Q. (1999) SCPD: a promoter database of the yeast Saccharomyces cerevisiae. Bioinformatics, 15, 607–611. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.