Abstract
Many powerful tools have been created to detect and describe the similarities between nucleic acid or protein sequences. Frequently these take the form of a sequence consensus, expressing simple most popular positional identities, positional identities with allowances for varying positions or some type of statistical description of the positional frequency characteristics of the defining sequence family. Despite the fact that some provide intuitively interpretable descriptions of the consensuses themselves, they typically do not give the viewer any information about regions of the sequence that might have inter-positional dependencies, and that therefore do not obey a strict consensus behavior. Herein, we present MAVL (Multiple Alignment Variation Linker) and StickWRLD. MAVL is our web-based application for detecting and displaying both positive and negative inter-positional correlations in nucleic acid sequences. MAVL examines all positional pairs in each of a collection of pre-aligned sequences and determines any pairs that occur with either greater or lesser frequency than a positional frequency matrix would predict. These data are then composited into a StickWRLD representation and supplied back to the user as a VRML (virtual reality modeling language) file. MAVL and StickWRLD can be accessed at http://www.microbial-pathogenesis.org/stickwrld/. A tutorial that explains MAVL features and demonstrates typical user interactions with StickWRLD graphs is available at http://www.microbial-pathogenesis.org/stickwrld/tutorial/sticktut2.html. This tutorial is quite large; please be patient while it loads.
INTRODUCTION
In examining biosequences, one of the fundamental tasks is the description and display of related patterns within a group of sequences. This task has been approached in several different manners: simply cataloging the most frequently occurring residue at each position and producing a string describing each most popular residue; cataloging the frequency of each residue at each position and producing a description that contains either the single most prevalent residue or a set describing the allowable residues at each position (1); cataloging the frequency of each residue at each position and producing a positional frequency matrix (usually transformed into a positional weight matrix or log-odds matrix) instead of a simple consensus (1); or applying more advanced statistical methods to describe the sequence family in a construct such as a hidden Markov model (HMM) (2).
Unfortunately, with popular methods, as the descriptive power of the method (in terms of its ability to capture nuances in the sequence family) increases, the interpretability of the results (in terms of their being intuitively understood by the viewer) generally decreases. Figure 1a shows a small collection of aligned sequences (the complete list of sequences used and a subsequence of an Archaeal endonuclease target site are available from http://www.microbial-pathogenesis.org/data/variable_core.txt), and Figure 1b–d illustrates a range of typical depictions of the information contained in these sequences. These representations, generated by MEME (1), include a typical consensus description (Figure 1b), the consensus with positional variability indicated (Figure 1c) and a positional frequency matrix (Figure 1d). HMMs do not lend themselves to simple visual representation.
Figure 1.
Typical representations used to describe a family of sequences. (a) Shows a collection of sequences; (b) is the consensus for this entire family (including sequences not shown here); (c) is a consensus with positional variation as calculated by MEME; (d) is a positional frequency matrix as calculated by MEME; (e) is a Sequence Logo for this family of sequences.
Some of the difficulty in visually interpreting the more sophisticated descriptions lies in the fact that they contain more information about the sequence family, and therefore require more effort on the part of the viewer. Some is also due to the fact that the more powerful descriptions are designed primarily to facilitate sequence searches, rather than to convey the pattern information to a human reader. On the other hand, representations that are specifically designed to convey the content of a positional frequency matrix, such as Sequence Logos (3), can convey the information content in the matrix much more intuitively than the arrayed values of the matrix itself. Figure 1e shows a Sequence Logo for the sequences shown in Figure 1a. This provides a considerably more intuitive presentation of the positional probabilities of the sequences, including the information content at each position. However, it conveys only the positional properties of the sequence family, and not relationships between the positions.
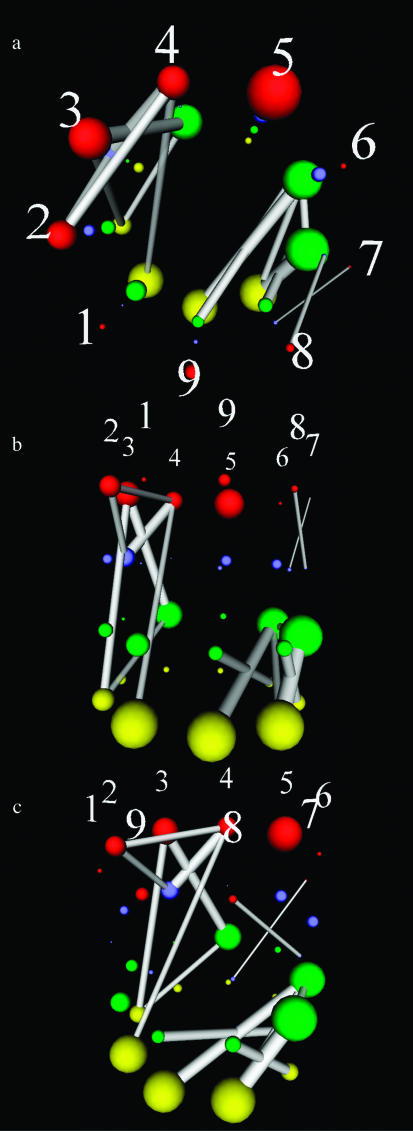
In this paper, we propose StickWRLD representations of aligned sequence patterns for nucleic acid (NA) sequences, and announce the availability of the MAVL/StickWRLD server at http://www.microbial-pathogenesis.org/stickwrld/. StickWRLD representations are three-dimensional VRML graphs (http://www.web3d.org/) that simultaneously depict the positional frequency characteristics of an NA sequence family, and any first-order interactions that appear between the identities of bases at different positions in the sequence. Figure 2 displays three views of the StickWRLD representation of the same sequence family shown in Figure 1. Positional frequencies are indicated by the size of the spheres, with red, blue, green and yellow balls corresponding to A, T, G and C nucleotides respectively. Positions whose identities show a relationship that is not explained by the consensus are connected by a line. The thickness of the line indicates the strength of the relationship, and the color indicates a positive (white) or negative (red) relationship between the identities at the positions. As can be seen, the co-occurrence of G and C at the sixth and ninth (and also seventh and eighth) positions is clearly shown, as well as other fine structure in the positional identities. In the live VRML graph the diagram can be scaled, translated and rotated.
Figure 2.
StickWRLD graph representations of the sequence family shown in Figure 1a. (a) Shows an ‘overhead’ view of the StickWRLD graph, while (b) and (c) show other orientations of the VRML graph. The overhead view is useful for detecting stem–loop type motifs, while the side views are better for examining detail in the position-to-position relationships.
StickWRLD representations are created by plotting an array of spheres that represent the scores in a positional frequency/weight matrix. The diameters of the spheres are scaled to the percentage frequency of the corresponding nucleotide at each position, and the array is wrapped around a cylinder. Visually, this is an exact analog of a positional frequency matrix, presented graphically in a form where there is an unobstructed line of sight between any two positions in the matrix. Lines are then plotted between the spheres based on the overpopulation or underpopulation of sequences sharing these nucleotides as compared with the consensus expectation. The transparency of the lines can optionally be scaled to represent the significance of the observed relationship (the probability that this relationship has occurred by chance).
Scale markers can optionally be displayed, to assist with visual orientation, and to assist in visually determining the quantitative differences in positional or link populations.
ALGORITHM
Conceptually, MAVL calculates residuals for the membership of each possible shared pair of sequence positions and identities. The line connecting these positions/identities is scaled to the magnitude of the normalized residual, and the color is set by the sign. The observed value ![]() for base N at position i, and base M at position j, used for calculating the residual is the number of sequences that share N at i and M at j. The expected value
for base N at position i, and base M at position j, used for calculating the residual is the number of sequences that share N at i and M at j. The expected value ![]() is the number of sequences that would be predicted to share these bases if the identity of the bases at each position were defined solely by the consensus. That is,
is the number of sequences that would be predicted to share these bases if the identity of the bases at each position were defined solely by the consensus. That is, ![]() is the probability of finding (N at i) multiplied by the probability of finding (M at j) multiplied by the total number of sequences TS under observation.
is the probability of finding (N at i) multiplied by the probability of finding (M at j) multiplied by the total number of sequences TS under observation.
The significance ![]() of each link is defined by the probability of finding, given the number of sequences examined, the observed number of members (or more) sharing the positions, in a population where these sequences are expected to constitute
of each link is defined by the probability of finding, given the number of sequences examined, the observed number of members (or more) sharing the positions, in a population where these sequences are expected to constitute ![]() of the complete population. If the identities of the bases at each position are linearly separable, then the number of sequences sharing specific bases at specific positions may be modeled as a binomial random variable with probability
of the complete population. If the identities of the bases at each position are linearly separable, then the number of sequences sharing specific bases at specific positions may be modeled as a binomial random variable with probability ![]() . Therefore, the significance of a link,
. Therefore, the significance of a link, ![]() , is the sum of the binomial probabilities from
, is the sum of the binomial probabilities from ![]() to TS successes in TS trials, where the probability of success is
to TS successes in TS trials, where the probability of success is ![]() per trial. For negative residuals, the calculation is identical, with the exception that the sum is for finding the observed number or fewer sequences sharing the positions.
per trial. For negative residuals, the calculation is identical, with the exception that the sum is for finding the observed number or fewer sequences sharing the positions.
Despite the fact that any particular level of overpopulation and an identical level of underpopulation in a link may have identical levels of significance, there is an asymmetry in the intuitive utility of displaying these features identically. This is due to the fact that quite small outlier populations that do exist are frequently indicative of interesting sequence features (or of mis-alignments in the input sequences), while the non-existence of other small outlier populations, statistically significant though they may be, is less intuitively interpretable. On the other hand, certain patterns of multiple links (most prevalently indicative of shifted regions in the alignment of one or a few sequences) provide useful visual clues to the user, even when each link individually falls below any arbitrary significance cutoff. To accommodate this, the system provides several ways for the user to impose his or her viewing preferences on the system. A significance level cutoff A can be applied to each link. Links with significance ![]() worse than A are not shown. The transparency of each link can be scaled to the significance calculated for it.
worse than A are not shown. The transparency of each link can be scaled to the significance calculated for it. ![]() would be a completely opaque link. A total residual, Tr, cutoff can be selected, and is applied to the normalized residuals, restricting the display to links representing residuals where
would be a completely opaque link. A total residual, Tr, cutoff can be selected, and is applied to the normalized residuals, restricting the display to links representing residuals where ![]() . A partial positive residual Pr value can be selected that additionally enables display of links where
. A partial positive residual Pr value can be selected that additionally enables display of links where ![]() . This is useful for displaying small positive residuals, where the residual is a large portion of the observed population of the link. Finally, a negative residual cutoff, Nr, can be selected, enabling the display of links where
. This is useful for displaying small positive residuals, where the residual is a large portion of the observed population of the link. Finally, a negative residual cutoff, Nr, can be selected, enabling the display of links where ![]() . The use of all three residual controls simultaneously allows the user to select between a display where positive and negative relationships are weighted and displayed equally, and one where either positive relationships are masked, allowing better visualization of the negative relationships, or negative relationships are masked, allowing better visualization of the positive ones.
. The use of all three residual controls simultaneously allows the user to select between a display where positive and negative relationships are weighted and displayed equally, and one where either positive relationships are masked, allowing better visualization of the negative relationships, or negative relationships are masked, allowing better visualization of the positive ones.
The results of the three residual cutoff selectors are Boolean ORed together. The significance level cutoff is absolute, and is ANDed with the residual cutoff results.
A recommended starting point for analyzing sequences of unknown character is to use a relatively stringent significance cutoff, with relaxed residual magnitude selector settings, to find major motifs that might be of interest, and then to relax the significance cutoff and increase the stringency of the magnitude selectors to maintain a similar level of detail in the display. This practice often allows the visual identification of many links that are consistent and collectively supportive of a structural hypothesis, even though none can pass the significance cutoff individually.
ACCESS
MAVL and StickWRLD are available through the http://www.microbial-pathogenesis.org/stickwrld/ web interface. Sequences must be pre-aligned and submitted in raw format to MAVL, with one sequence per line, spaces indicating gaps. There is no algorithmic restriction on the length of the sequences; however, practical considerations with respect to the visualization limit useful submissions to the neighborhood of 200 bp aligned length. The VRML for the StickWRLD graph is returned directly to the user's web browser, and requires that the user have one of the freely available VRML viewers installed to view it. The Cosmo (http://www.cosmosoftware.com/), Cortona (http://www.parallelgraphics.com/) and FreeWRL (http://freewrl.sourceforge.net/) viewers have been tested. Each has minor differences in how it renders and navigates VRML files, and the user is encouraged to test each to find the one that best suits his or her personal needs. Mac users should be aware that Cortona does not currently handle text properly on Mac OS X.
RESULTS AND DISCUSSION
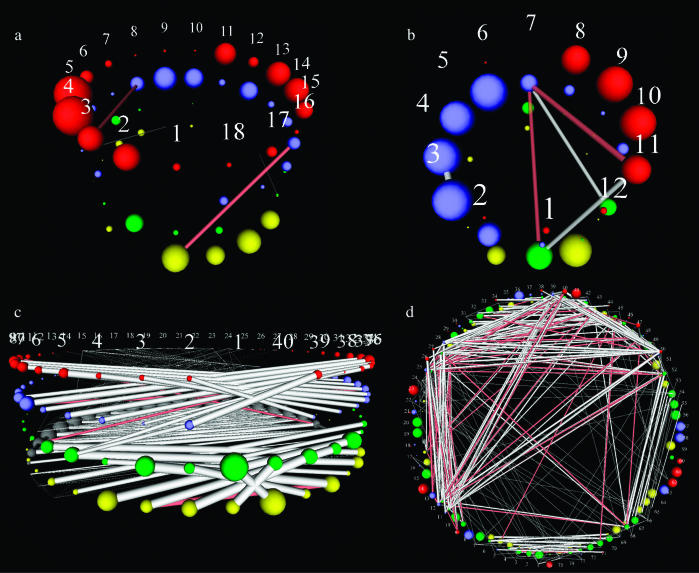
After examining a number of sequence families, it becomes clear that this representation makes certain types of sequence motif properties visually obvious and suggests interesting relationships that cannot be otherwise conveniently identified. Figure 3 shows StickWRLD representations that were calculated by MAVL for several common sequence families. Families that appear to be defined purely by positional identity frequency, such as the TATA-box promoter region of Archaeal tRNA promoters [Figure 3a, ‘clean upstreams’ from Archaeoglobus fulgidus (4) tRNA genes, as extracted by PACRAT (http://www.biosci.ohio-state.edu/~pacrat/(5)], appear as a visual analog to their positional frequency matrix, with few positions diverging from a pure consensus with sufficient frequency to show up as having any inter-positional dependency. Significantly, even sequence families such as T-tract transcriptional terminators, which frequently display poly(T) followed by poly(A) motifs, but that are not structurally palindromes, are revealed to have no inter-positional relationships (Figure 3b, PACRAT ‘clean downstreams’ from A.fulgidus tRNAs). Sequence families that have strong structure components, however, such as rho-independent terminators, show distinct cross-positional relationships, with the structural pairing requirements of the hairpin clearly shown by the positive and negative relationship links in the diagram [Figure 3c, 391 predicted rho-independent terminators with length <40 from the Escherichia coli K12 (GenBank accession no. NC_000913) data available at http://www.tigr.org/software/transterm.html]. Even complex structural inferences can be drawn from the detected relationships. Figure 3d shows a StickWRLD diagram of aligned A.fulgidus tRNA sequences (those without introns or extended variable arms), with the acceptor stem at the bottom of the image, and the sequence proceeding 5′ to 3′ in a clockwise direction around the image (the TΨC motif is the pair of blue spheres followed by a yellow sphere at the right of the image). The hairpin pairing for each of the four stems can be easily detected, as well as relationships between positions in the D arm into the variable arm/bottom of the TΨC arm, and from the D arm into the anti-codon region. Some of these relationships are explained by the complex three-dimensional structure of tRNAs, and are predicted by observed close contacts in the NMR and crystal structures [(6) and many others]. Others are not directly explicable and suggest areas for further examination.
Figure 3.
StickWRLD graph representations of several commonly studied sequence families. (a) Shows a family of TATA-box promoter regions from Archaeal (A.fulgidus) tRNAs; (b) displays a family of T-tract transcriptional terminators from A.fulgidus tRNA genes; (c) illustrates a family of subsequences extracted from predicted rho-independent terminators from E.coli. The base pairing requirements of the stem can be clearly seen; (d) represents similar-length (shorter) tRNAs from A.fulgidus.
This notion of examining over- and underpopulated identity pairs in a family of sequences is also useful for rejecting hypotheses regarding possible false palindromic sequence features. Many DNA-binding proteins, such as the cAMP receptor protein, bind as dimers (7). Because each subunit has identical specificity for its target, and the subunits have opposite orientation, the overall DNA motif can appear palindromic. In this case, however, there is no structural base pairing constraint, and variation in the identity of bases on one strand of the apparent palindrome does not influence the identity of the bases on the other strand. This lack of co-variation is easily detected in the StickWRLD graph.
CONCLUSION
StickWRLD diagrams as generated by MAVL are a powerful method for visually detecting situations where a simple sequence consensus, weight matrix or HMM description of a sequence family may be unable to adequately describe the properties of the family. Conversely, these diagrams are also a powerful tool for determining when a consensus-type description is adequate, and when the possibility that a motif has structural requirements may be rejected. In addition to its capabilities as a sequence visualization tool, we are currently working on an extension to the MAVL server to allow use of the StickWRLD graph as a search tool.
REFERENCES
- 1.Bailey T.L. and Gribskov,M. (1998) Combining evidence using p-values: application to sequence homology searches. Bioinformatics, 14, 48–54. [DOI] [PubMed] [Google Scholar]
- 2.Durbin R., Eddy,S., Krogh,A. and Mitchison,G. (1998) Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. Cambridge University Press, Cambridge. [Google Scholar]
- 3.Shaner M.C., Blair,I.M. and Schneider,T.D. (1993) Sequence Logos: A Powerful, Yet Simple, Tool. In Mudge, T.N., Milutinovic, V. and Hunter, L. (eds), Proceedings of the Twenty-Sixth Annual Hawaii International Conference on System Sciences, Vol. 1: Architecture and Biotechnology Computing. IEEE Computer Society Press, Los Alamitos, CA, pp. 813–821. [Google Scholar]
- 4.Klenk H.P., Clayton,R.A., Tomb,J., Quackenbush,J., White,O., Nelson,K.E., Ketchum,K.A., Dodson,R.J., Gwinn,M., Hickey,E.K. et al. (1997) The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature, 390, 364–370. [DOI] [PubMed] [Google Scholar]
- 5.Ray W.C. and Daniels,C.J. (2001) The PACRAT system: an extensible WWW-based system for correlated sequence retrieval, storage and analysis. Bioinformatics, 17, 100–104. [DOI] [PubMed] [Google Scholar]
- 6.Zagryadskaya E.I., Doyon,F.R. and Steinberg,S.V. (2003) Importance of the reverse Hoogsteen base pair 54–58 for tRNA function. Nucleic Acids Res., 31, 3946–3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Macfayden L.P. (2000) Regulation of competence development in Haemophilus influenzae: proposed competence regulatory elements are CRP-Binding Sites. J. Theor. Biol., 207, 349–359. [DOI] [PubMed] [Google Scholar]