Abstract
Objective
Histamine is an important mediator in the pathophysiology of asthma. We have previously reported that HRH1 is differentially expressed among those with asthma compared to those without asthma. Single histamine related genes have also been associated with asthma. We aimed to evaluate known single nucleotide polymorphisms (SNPs) in genes along the histamine biotransformation and response pathway and determine their association with asthma and HRH1 mRNA expression.
Methods
We enrolled children and adults (n=93) with/without asthma who met inclusion/exclusion criteria. Genotyping was performed for 9 known SNPs in the HDC, HRH1, HRH4, HNMT, and ABP1 genes. HRH1 mRNA expression was determined on RNA from buccal tissue. General linear model, Fisher's exact test, and Chi-square test were used to determine differences in allele, genotype, and haplotype frequency between subjects with and without asthma and differential HRH1 mRNA expression relative to genotype. Statistical significance was determined by p<0.05.
Results
No difference was observed in genotype/allele frequency for the 9 SNPs between subjects with and without asthma. The HNMT-1639C/ −464C/ 314C/ 3’UTRA haplotype was more frequently observed in those without asthma than those with asthma (p=0.03). We also observed genetic differences relative to race and gender. HNMT 314 genotype CT was more frequent in males with asthma compared to those without asthma (p=0.04).
Conclusions
Histamine pathway haplotype was associated with a diagnosis of asthma in our cohort but allele and genotype were not. Subgroup evaluations may also be important. Further studies are needed to determine the potential biological/clinical significance of our findings.
Introduction
Asthma is a chronic inflammatory respiratory disease characterized by episodic bronchoconstriction and airway hyper-responsiveness to a variety of stimuli including viral antigens, allergens, and environmental exposures (1). According to the National Health Interview Survey, about 25.9 million (8.5%) people in the United States had asthma in 2011 and prevalence rates are increasing (2). The underlying pathophysiology of the disease has been extensively studied but is not clearly understood. Furthermore, some potentially important pathways which may be related to disease pathogenesis, disease severity, and therapeutic treatment have not been fully explored. For example, the chemical mediator histamine (2-[4-imidazole] ethylamine) plays a key role in the inflammatory process of allergies and asthma (3-7). Histamine exposure in the airway leads to increased vascular permeability, mucus production, and contraction of airway smooth muscle cells; all of which result in airway hyper-responsiveness and obstruction. Inhalation of histamine causes direct bronchoconstriction (8). Studies have shown that histamine induces the release of epidermal growth factor receptor ligands from bronchial epithelial cells which is proposed to lead to airway remodeling (4). Furthermore, histamine causes oxidative stress in the airway through release of hydrogen peroxide by bronchial epithelial cells (9). We have observed that expression of the histamine receptor in the buccal mucosa differs between those with asthma compared to those without asthma (10).
As these studies suggest that histamine may have a significant role in the pathophysiology of asthma, targeting this amine may be important to disease treatment. HRH1 antihistamine use has been shown to reduce respiratory symptoms and need for rescue medications in children with allergic asthma (11). Antihistamines administered to atopic children and children considered high-risk for atopy may prevent the onset of asthma when compared with placebo (12, 13). However, the results of clinical trials addressing the effectiveness of antihistamines in alleviating asthma symptoms are inconclusive (14). Conflicting results from these trials may be due to variation with respect to the specific antihistamine as well as the study population under consideration (11, 14, 15). Current evidence does not support the clinical efficacy of H1-antihistamines in non-allergic asthma; however differences in histamine biotransformation and resultant exposure, along with actions at other receptors (e.g. H4 receptor), must also be considered as having a role in asthma pathogenesis. Genetic polymorphisms among genes responsible for histamine formation, degradation and response should be considered as a source of variation. The role of histamine in the pathogenesis of asthma and the basis for individual differences in response to antihistamine therapy deserves more investigation.
Histamine is synthesized by alpha-decarboxylation of L-histidine catalyzed by the enzyme L-histidine decarboxylase (HDC) (encoded by HDC on chr15q21-q2) (3, 7, 16). Histamine is metabolized for elimination from the body by two main enzymes, histamine N-methyltransferase (HNMT) and diamine oxidase (DAO) (17). HNMT (encoded by HNMT on chr2q22) is responsible for the majority of histamine degradation and is known to be expressed in the bronchial epithelium (3, 17). DAO (encoded by amiloride binding protein 1 gene ABP1, also known as amine oxidase copper containing 1 gene AOC1 on chr7q34-36) is expressed in kidney, colon, placenta, thymus and seminal vesicles (3, 18) and is mainly involved in degradation of extracellular histamine. Histamine binds to and exerts its effects on four known subclasses of histamine receptors (HRH1, HRH2, HRH3 and HRH4). HRH1 (encoded by HRH1 on chr3p25) is known to be involved in hypersensitivity reactions with activation of the receptor resulting in increased vascular permeability and smooth muscle contraction. The HRH2 receptor (encoded by HRH2 on chr5q35.2) is well-known to be associated with gastric acid secretion but activation of the receptor also results in smooth muscle relaxation in the airway and vasculature. The HRH3 receptor (HRH3 on chr20q13.33) exists on histaminergic neurons and is involved in control of neurotransmitter release. The HRH4 receptor (HRH4 on chr18q11.2) is expressed on numerous cell types and is involved in cellular chemotaxis and inflammatory mediator release (19).
Single nucleotide polymorphisms (SNPs) have been identified within the genes of the histamine pathway which may affect protein production and/or action potentially resulting in alterations in how histamine is handled in the body. Several studies have evaluated the relationship between individual genes within the histamine pathway and asthma or allergic disease in addition to various other diseases. SNPs in HDC, including HDC 92 C→T, have been studied in those with allergic rhinitis with conflicting reports as to whether genetic variants in this gene are indeed associated with allergic rhinitis (16, 20). Although, we have observed that HRH1 mRNA expression is elevated in subjects with asthma relative to those without the disease (10), studies have not found that polymorphisms in HRH1 are associated with asthma (21). Genetic variation within HRH4 has been associated with infection-induced asthma (22). The relationship of asthma and allergic diseases with SNPs in HNMT has been most widely investigated. Studies have identified associations between HNMT genetic variation and atopic dermatitis and asthma (23, 24). However, conflicting reports also exist regarding whether there is a relationship between HNMT tested variants and asthma (21, 23-28). ABP1 has been studied among those with asthma and a relationship between genetic variants within the gene and asthma has not been identified (26, 28).
These previous studies have primarily focused on few SNPs within individual genes involved with histamine biotransformation and action. This study is based on the premise that it is important to consider all aspects of histamine biotransformation and effect (i.e. production, response, and elimination) when considering the potential for excess accumulation or altered effect of histamine. However, for the majority of SNPs in genes involved with the bioavailability and response to histamine, little is known regarding the functional and clinical effects with respect to asthma. Therefore, we have included variants from both coding and non-coding regions of genes (HDC, ABP1, HNMT, HRH1, and HRH4) along the pathway according the following criteria: (1) found among African American and Caucasian individuals with allele frequency >2% (2) proposed functional significance (i.e. non-synonymous in coding regions) (3) potential association with allergic/inflammatory disease (see Table 1 in supplementary material). The primary goal of this study was to conduct a pilot investigation of the potential association of known SNPs in genes along the histamine pathway with the diagnosis of asthma in children and adults. We also aimed to explore possible associations between histamine pathway genetic polymorphisms and HRH1 mRNA expression.
Materials and methods
Study population
We utilized a convenience sample of children and adults in the study who had participated in another study to evaluate gene expression patterns among subjects with and without asthma (10). Children and adults were enrolled from Allergy, Asthma and Immunology outpatient clinics at tertiary care centers, The Children's Mercy Hospital and Truman Medical Center in Kansas City, Missouri, respectively, after written informed consent or parental permission along with child assent was obtained. The study was approved by the respective Institutional Review Boards.
The diagnosis of asthma was defined by spirometry measurement according to American Thoracic Society guidelines (29) or by clinical history in children unable to perform spirometry. Participants without asthma were also recruited from clinics at The Children's Mercy Hospital and Truman Medical Center and confirmation of non-asthma was performed clinically and reconciled with the treating physician. Participants in the non-asthma group were siblings of study participants with asthma or patients seen in the clinic with a diagnosis of allergic rhinitis and/or food allergies only. Exclusion criteria included: tobacco smokers, subjects with chronic lung disease, immunodeficiency, uncontrolled gastroesophageal reflux, pneumonia in the past 6 months, use of intranasal steroids within the past 2 weeks, use of systemic steroids within the past 6 weeks, and current immunomodulator treatment (due to concerns for alterations in gene expression in the tissue being studied).
DNA extraction and genotyping
Blood (5 ml) was collected into a glass tube containing ACD or calcium EDTA anticoagulant, mixed by repeated inversion and either stored for up to 7 days at 4°C or immediately frozen at −70°C. Genomic DNA was extracted from blood using the Illustra Blood Genomic Prep Mini Spin Kit (GE Healthcare, Piscataway, NJ). Genotyping assays were performed on genomic DNA (10-20ng) using commercially available TaqMan assays to detect SNPs of interest including HDC 92C→T (rs17740607), HRH1 -17C→T (rs901865), HRH4 413C→T (rs11665084), HNMT −1639C→T (rs6430764), HNMT −464C→T (rs2071048), HNMT 314C→T (rs11558538), HNMT 3’UTRA→T (rs1050900), ABP1 47C→T (rs10156191), and ABP1 995C→T (rs1049742) (Applied Biosystems, Foster City, CA) and KAPA Probe Fast qPCR master mix (Kapa Biosystems, Boston, MA). Genotyping was performed on an Eco™ Real-Time PCR System (Illumina, San Diego, CA) with a thermal profile of 95°C for 3 minutes followed by 40-50 cycles of 95°C for 3 seconds and 64°C for 20 seconds. Genotyping for any genomic DNA sample that revealed undetermined results was repeated with controls. Regardless of the results, at least 10% of samples selected at random were genotyped in duplicate to rule out a random error.
RNA extraction and gene expression
Buccal samples were utilized to evaluate HRH1 mRNA expression. Samples were obtained from participants by gently scraping the inside of each cheek with a sterilized, soft bristled toothbrush after rinsing the mouth. Cells were immediately immersed in Saliva Protect RNA stabilization solution (Qiagen, Valencia, CA) for 24 hours at 4°C prior to RNA extraction. RNA was extracted from 9-30 mg of buccal cells using the Illustra RNA spin Mini RNA isolation kit (GE Healthcare).
Gene expression levels for HRH1 were determined as previously described (10). Briefly, RNA (20ng) was reverse transcribed using the High Capacity RNA to cDNA kit (Applied Biosystems) and cDNA was pre-amplified using the TaqMan Pre-Amplification Kit (Applied Biosystems) according to manufacturer's protocol. Pre-amplification reactions were subjected to 14 (HRH1) or 10 cycles (GAPDH, PPIA) and diluted 1:40. Quantitative PCR was performed using PerfeCTa qPCR SuperMix (Quanta Biosciences, Gaithersburg MD) and TaqMan Gene Expression assays (Hs00911670_s1 for HRH1, 4333764F for GAPDH, and 4333763F for PPIA (Applied Biosystems)). Gene expression was normalized by the geometric mean of GAPDH and PPIA and logarithmically transformed to the base 2.
Data analysis
Concordance with Hardy-Weinberg equilibrium was confirmed for each SNP using available online software (30) and haplotypes were inferred using PHASE 15 software (31). Statistical analyses were performed using SAS 9.2 (Cary, NC). Descriptive statistics, Chi-square test, and t-test were used to compare demographics between subjects with and without asthma. Fisher's exact test and Chi-square test were used to compare the frequency distributions of genotype, allele, and haplotype in subjects with asthma to those without asthma. For genotype, we further performed dominant analysis (comparing wild type vs. non-wild type genotypes), and recessive analysis (comparing recessive genotype vs. non-recessive genotype) using the Fisher's exact test. We assessed linear effect of genotype using the Cochran-Armitage trend test. General linear model (GLM) was used to study the association of genotype with gene expression. Significance was determined by p-values <0.05. We further evaluated genotype frequency according to the subgroup stratifications of race and gender. We performed the Cochran-Mantel-Haenszel (CMH) test to compare the frequencies of genotype and allele between subjects with asthma and controls, adjusted by the confounding race effect. We also evaluated genotype frequency among those with asthma relative to asthma severity which was defined by medication step as determined by the 2007 NHLBI guidelines (1). This stratification included (i) subjects on Step 1 asthma therapy (ii) subjects on Step 2/3 asthma therapy, and (iii) subjects without asthma in the pediatric age group. Adult subjects included two stratified groups (i) subjects on Step 2/3-asthma therapy and (ii) subjects without asthma for data analysis.
Sample size calculation was performed using nQuery Advisor 7.0. This study was powered to detect a relatively large effect size in allele frequency between the two extreme groups of participants with asthma and those without the disease. For instance, a two group continuity corrected chi-square test with a 0.05 two-sided significance level will have 83% power to detect the difference between a control group proportion, 5% and the asthma group proportion, 22% (odds ratio of 5.359). The power calculation was also applicable for the comparison by gender and comparison by race (89% power to detect 5% vs. 22% proportion with two-sided significance of 0.05). The analysis was not powered for further secondary/stratified analyses.
Results
Demographics
We enrolled a total of 93 subjects in our study. Subject demographics are shown in Table 1. There were no significant differences observed between subjects with and without asthma relative to age or gender. However, 38.7% (n=36) subjects with asthma were African American compared to 12.9% (n=12) subjects without asthma, with a marginal p-value of 0.048). There were 33 subjects (22 with asthma, 11 without asthma) under 12 years and 60 subjects (40 with asthma, 20 without asthma) who were 12 years or above.
Table 1.
Demographics of study subjects and comparison of demographic characteristics among those with and without asthma
| Demographic | Total subjects | Subjects without Asthma | Subjects with Asthma |
|---|---|---|---|
| Age in years: Mean± SD | 20.5±15.3 | 20.7±15.5 | 20.3±15.3 |
| Gender % (N): | |||
| Males | 53.7 (50) | 16.1 (15) | 37.6 (35) |
| Females | 46.2 (43) | 17.2 (16) | 29.0 (27) |
| Race % (N): | |||
| *African Americans | 51.6 (48) | 12.9 (12) | 38.7 (36) |
| Caucasians | 36.6 (34) | 12.9 (12) | 23.7 (22) |
| Other | 11.8 (11) | 7.5 (7) | 4.3 (4) |
There was no significant difference in demographic characteristics except marginal difference in race between those with and without asthma (all demographic comparisons p≥ 0.05).
African Americans with asthma compared to non-asthma, p=0.048
Allele/Genotype/Haplotype results
Our study did not reveal any significant differences in allele or genotype frequency between subjects with asthma and those without asthma for any of the nine SNPs investigated (Table 2, 3). We identified 3 haplotypes for ABP1 and 9 for HNMT SNPs among our cohort. As with the individual polymorphisms, no differences in haplotype frequency relative to asthma diagnosis were initially observed for APB1 or HNMT (Table 4, 5). All results remained consistent after adjustment for race. After adjustment for race we observed one haplotype (HNMT CCCA) which was more frequent in those without asthma (24.2% vs. 32.3%, p=0.03).
Table 2.
Variant allele frequencies of studied single nucleotide polymorphisms (SNPs) among subjects with and without asthma
| SNP | Allele | Asthma subjects % (N) | Control subjects % (N) | p-value | Adjusted p-value* |
|---|---|---|---|---|---|
| HDC 92C→T rs17740607 | C | 95.9 (117) | 90.3 (56) | 0.19 | 0.1 |
| T | 4.1 (5) | 9.7 (6) | |||
| HRH1 −17C→T rs901865 | C | 80.3 (98) | 79.0 (49) | 0.84 | 0.2 |
| T | 19.7 (24) | 21.0 (13) | |||
| HRH4 413C→T rs11665084 | C | 97.6 (121) | 93.55 (58) | 0.2 | 0.2 |
| T | 2.4 (3) | 6.45 (4) | |||
| HNMT −1639C→T rs6430764 | C | 49.2 (61) | 45.2 (28) | 0.6 | 0.5 |
| T | 50.8 (63) | 54.8 (34) | |||
| HNMT −464C→T rs2071048 | C | 41.9 (52) | 41.9 (26) | 1 | 0.4 |
| T | 58.1 (72) | 58.1 (36) | |||
| HNMT 314C→T rs11558538 | C | 90.3 (112) | 93.55 (58) | 0.46 | 0.2 |
| T | 9.7 (12) | 6.45 (4) | |||
| HNMT 3’ UTR A→T rs1050900 | A | 80.65 (100) | 83.9 (52) | 0.59 | 0.4 |
| T | 19.35 (24) | 16.1 (10) | |||
| ABP1 47C→T rs10156191 | C | 58.1 (72) | 61.3 (38) | 0.67 | 1 |
| T | 41.9 (52) | 38.7 (24) | |||
| ABP1 995C→T rs1049742 | C | 91.8 (112) | 96.8 (60) | 0.34 | 0.4 |
| T | 8.2 (10) | 3.2 (2) |
The Cochran-Mantel-Haenszel (CMH) test was performed to evaluate the associations between group and allele, taking race into account.
Table 3.
Genotype frequencies of studied single nucleotide polymorphisms (SNPs) among subjects with and without asthma
| SNP | Genotype | Asthma subjects % (N) | Control subjects % (N) | p-value | Adjusted p-value* |
|---|---|---|---|---|---|
| HDC 92C→T rs17740607 | CC | 91.8 (56) | 83.9 (26) | 0.3 | 0.2 |
| CT | 8.2 (5) | 12.9 (4) | |||
| TT | 0 (0) | 3.2 (1) | |||
| HRH1 −17C→T rs901865 | CC | 65.6 (40) | 64.5 (20) | 1.0 | 0.2 |
| CT | 29.5 (18) | 29.0 (9) | |||
| TT | 4.9 (3) | 6.5 (2) | |||
| HRH4 413C→T rs11665084 | CC | 95.2 (59) | 87.1 (27) | 0.2 | 0.2 |
| CT | 4.8 (3) | 12.9 (4) | |||
| TT | 0 (0) | 0 (0) | |||
| HNMT −1639C→T rs6430764 | CC | 22.6 (14) | 22.6 (7) | 0.7 | 0.5 |
| CT | 53.2 (33) | 45.2 (14) | |||
| TT | 24.2 (15) | 32.3 (10) | |||
| HNMT −464C→T rs2071048 | CC | 14.5 (9) | 19.4 (6) | 0.7 | 0.4 |
| CT | 54.8 (34) | 45.2 (14) | |||
| TT | 30.7 (19) | 35.5 (11) | |||
| HNMT 314C→T rs11558538 | CC | 80.7 (50) | 87.1 (27) | 0.4 | 0.1 |
| CT | 19.4 (12) | 12.9 (4) | |||
| TT | 0 (0) | 0 (0) | |||
| HNMT 3’ UTR A→T rs1050900 | AA | 64.5 (40) | 71 (22) | 0.8 | 0.4 |
| AT | 32.3 (20) | 25.8 (8) | |||
| TT | 3.2 (2) | 3.2 (1) | |||
| ABP1 47C→T rs10156191 | CC | 37.1 (23) | 35.5 (11) | 0.6 | 1 |
| CT | 41.9 (26) | 51.6 (16) | |||
| TT | 21.0 (13) | 12.9 (4) | |||
| ABP1 995C→T rs1049742 | CC | 85.3 (52) | 93.6 (29) | 0.7 | 0.4 |
| CT | 13.1 (8) | 6.5 (2) | |||
| TT | 1.6 (1) | 0 (0) |
The Cochran-Mantel-Haenszel (CMH) test was performed to evaluate the associations between group and genotype, taking race into account.
Table 4.
Comparison of haplotype frequencies for ABP1 47C→T and ABP1 995C→T polymorphisms between subjects with and without asthma
| ABP1 haplotypes | All subjects |
p | Adjusted p-value* | |
|---|---|---|---|---|
| With asthma | Without asthma | |||
| CC | 58 (72) | 61.3 (38) | 0.75 | 0.97 |
| TC | 34 (42) | 35.5 (22) | 0.87 | 0.62 |
| TT | 8(10) | 3.2 (2) | 0.34 | 0.41 |
The Cochran-Mantel-Haenszel (CMH) test was performed to evaluate the associations between group and haplotype, taking race into account.
Table 5.
Comparison of haplotype frequencies for HNMT −1639C→T, −464C→T, 314C→T, and 3’UTR A→T polymorphisms between subjects with and without asthma
| HNMT haplotypes | All subjects | p | Adjusted p* | |
|---|---|---|---|---|
| With asthma | Without asthma | |||
| TTCA | 48.4 (60) | 50 (31) | 0.87 | 0.42 |
| TTCT | 0.8 (1) | 4.8 (3) | 0.11 | 0.14 |
| TTTT | 0.8 (1) | 0 (0) | 1.0 | 0.56 |
| TCCA | 0.8 (1) | 0 (0) | 1.0 | 0.46 |
| CTCA | 7.3 (9) | 1.6 (1) | 0.17 | 0.31 |
| CTCT | 0.8 (1) | 1.6 (1) | 1.0 | 0.41 |
| CCCA | 24.2 (30) | 32.3 (20) | 0.29 | 0.03 |
| CCCT | 8.1 (10) | 3.2 (2) | 0.34 | 0.38 |
| CCTT | 8.9 (11) | 6.5 (4) | 0.78 | 0.21 |
The Cochran-Mantel-Haenszel (CMH) test was performed to evaluate the associations between group and haplotype, taking race into account.
Subgroup analysis results
We also evaluated genotype and allele frequency according to subgroups of race, gender, and asthma severity.
Analysis by race
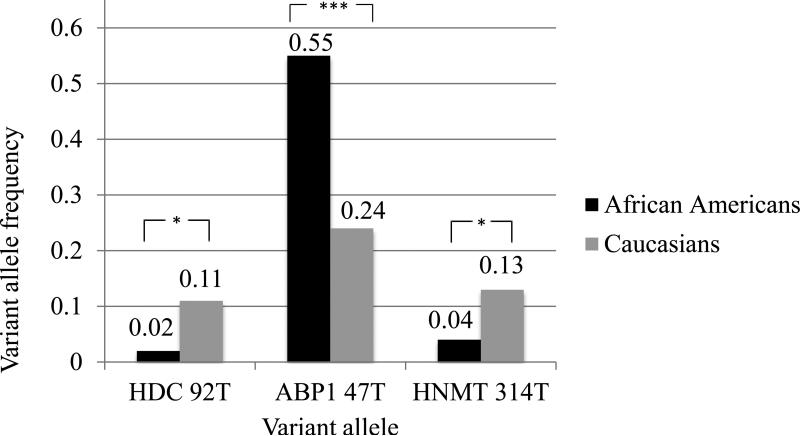
The frequency of genotypes with variant allele for ABP1 47C→T (heterozygous CT and homozygous TT) were over-represented in African American participants relative to other represented races whereby 83% (n=40) of African Americans possessed at least one T allele (CT 56% (n=27) and TT 27% (n=13)) vs. only 36% (n=12) of Caucasians possessed a T allele (CT 24% (n=8) and TT 12% (n=4)) (p<0.0001). Hence, the variant allele (T) frequency for ABP1 47C→T among African American subjects was significantly higher than among Caucasians (55% (n=53) vs. 24% (n=16), p=<0.0001) as shown in Figure 1. We also observed other differences relative to race. For HNMT 314C→T, the variant (T) allele frequency was 4% (n=4) in African Americans compared to 13% (n=9) in Caucasians (p=0.03) (Figure 1). These allele frequencies result in 8% (n=4) of African Americans with heterozygous genotype CT for HNMT 314C→T relative to 27% (n=9) of Caucasians who were heterozygous for the SNP (p=0.03). The homozygous genotype for the variant TT did not occur in our cohort. The HDC variant allele (T) was also less frequent among African Americans compared to Caucasians (2% (n=2) vs. 11% (n=7) respectively, p=0.03) (Figure 1). Race did not appear to be associated with genotype or allele frequency for the other six polymorphisms evaluated.
Figure 1.
Comparison of the variant allele frequency of single nucleotide polymorphisms ABP1 47C→T, HNMT 314C→T, and HDC 92C→T between African American and Caucasian participants
* p<0.05, *** p<0.001
Analysis by gender
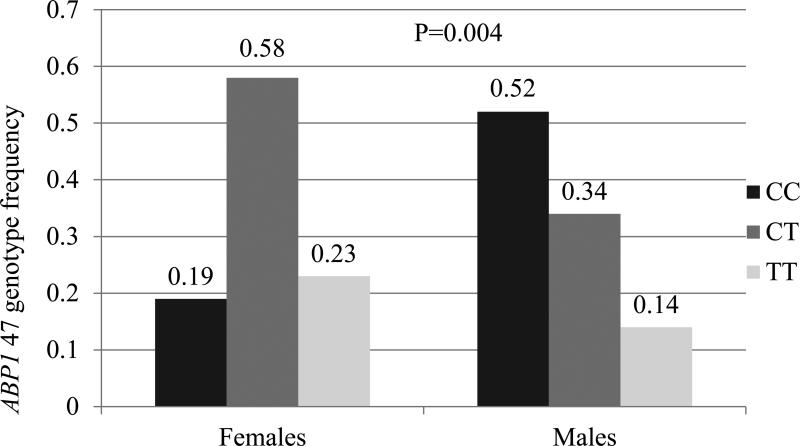
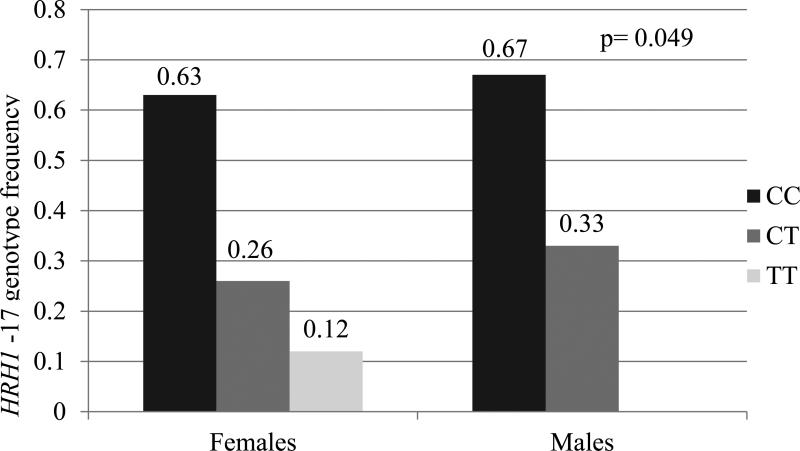
In our study population, two variants were associated with gender. Among our cohort, 23% (n=10) of females were homozygous for ABP1 47TT compared to 14% (n=7) of males who were homozygous for the variant allele (p=0.004 unadjusted; p=0.027 adjusted) (Figure 2A). For HRH1-17C→T, 11.6% (n=5) of females were homozygous for variant allele (TT) compared to no males that were homozygous for the variant (p=0.049 unadjusted; p=0.225 adjusted) (Figure 2B). There were no statistically significant differences in the allele frequency relative to gender for the other 7 SNPs evaluated. Also, the haplotype frequencies for ABP1 were similar for males and females.
Figure 2.
Comparison of genotype frequencies between males and females for HRH1 -17C→T and ABP1 47C→T
Panel A: Comparison of ABP1 47 genotype frequency between males and females; ABP1 47 TT genotype more common in females
p=0.004
Panel B: Comparison of HRH1-17 genotype frequency between males and females; HRH1-17 genotype more common in females (not present in males)
p =0.049
Analysis by asthma severity
No statistically significant difference was observed in genotype, allele, or haplotype frequency relative to asthma severity/step therapy for any of the nine SNPs. There remained no significant difference after adjustment for race.
Analysis of allele, genotype, and haplotype in subjects with and without asthma within gender and racial groups
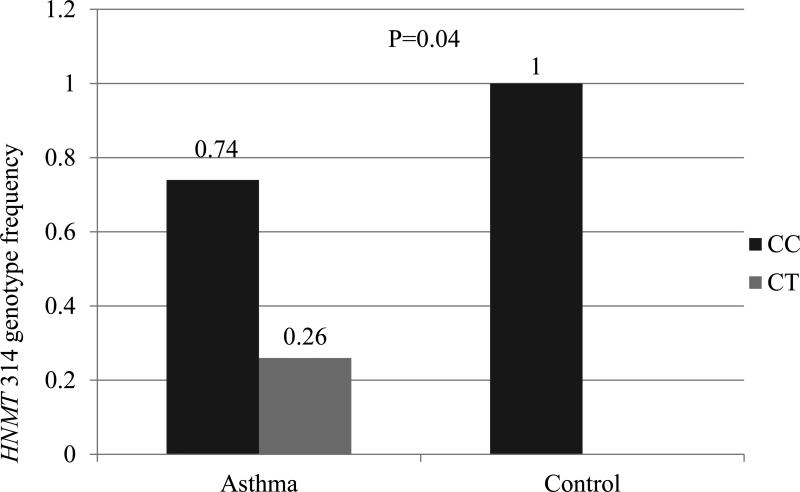
Due to our findings relative to gender and race and the recognition that disease phenotype, and thus pathophysiology, likely differs between males and females and various races, we performed further analysis to evaluate allele, genotype, and haplotype among gender and racial subgroups with and without asthma. Among males with asthma, the CT genotype for HNMT 314C→T was over-represented (26%, n=9) relative to males without asthma (0%, n=0) (p=0.04) (Figure: 3). No other SNPs showed a significant difference in allele frequency or genotype between subjects with asthma and without asthma among gender groups. Though the genotype and allele frequencies in each racial subgroup with and without asthma were similar, we found the HNMT haplotype (−1639C/ −464C/ 314C/ 3’UTRA) to be more frequent in Caucasian subjects without asthma than those with asthma (46% (n=11) vs. 18% (n=8), p=0.02).
Figure 3.
Comparison of HNMT 314C→T genotype between those with and without asthma among male participants. The CT genotype was not represented among males without asthma
p=0.04
RNA expression results
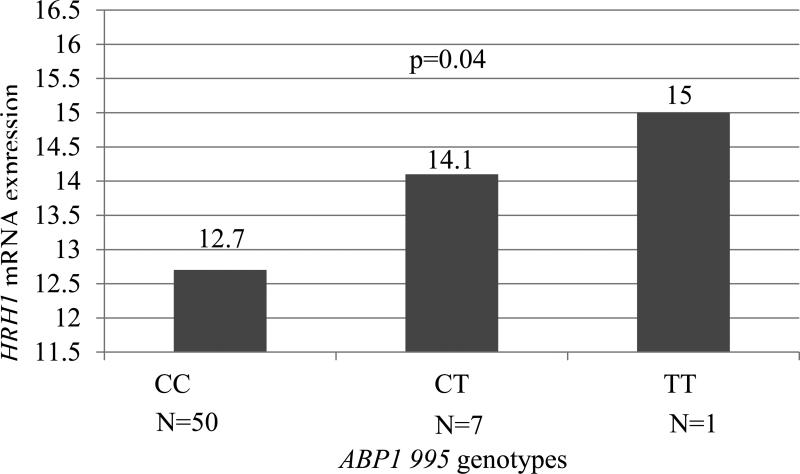
Samples previously utilized to evaluate buccal mucosal mRNA expression were also utilized in our current study to evaluate the relationship between HRH1 gene expression and genotype among participants with and without asthma (10). We did not observe a difference inHRH1 mRNA expression relative to genotype for any of the 9 SNPs evaluated including HRH1-17 SNP. We also did not observe a difference in HRH1 expression relative to gender or race. However, our previous studies have suggested that expression of HRH1 mRNA is increased in asthma subjects greater than 12 years of age relative to those without asthma (10). Therefore, further studies were performed to evaluate HRH1 expression among the older age group of participants (>12 years of age). HRH1 expression was higher in subjects who possessed the ABP1 995 variant allele T in those 12 years and older (p=0.04, Figure 4). No difference in mRNA HRH1 expression relative to genotype in this older age group was observed for the other SNPs evaluated.
Figure 4.
Comparison of Log HRH1 mRNA expression of relative to ABP1 995C→T genotypes in subjects 12 years or older. Subjects with ABP1 995 genotypes containing variant allele T (CT and TT) conferred increased HRH1 expression with an observed dose response for these genotypes.
p=0.04
Discussion
Previous studies have evaluated single nucleotide polymorphisms in genes involved with histamine metabolism and action in relation to asthma. In our study we targeted multiple polymorphisms in several individual genes along the histamine pathway in a sequential manner. Our study is the first to evaluate genetic variability involved in the production of (HDC), response to (HRH1, HRH4), and degradation of (HNMT, ABP1) histamine in relation to asthma diagnosis. Determination of genetic variation between those with asthma and non-asthma may also be important in identifying other pathways (e.g. histamine) which could be explored further among groups of patients with the disease as potential therapeutic targets and/or diagnostic predictors. As a critical first step, we felt that it was important to investigate SNPs involved with histamine between asthmatic and non-asthmatic group. Without conducting such studies one may not know whether or not some variants may be important for all patients with asthma. Although we did not observe a difference in genotype among the 9 SNPs evaluated in the 5 histamine related genes between those individuals with and without asthma, we believe our findings are important. In contrast with previous studies which have been conducted in primarily Caucasian and Asian populations, our study included a majority of African Americans, an understudied group who are disproportionately affected by asthma. We also performed a more comprehensive analysis in evaluating potential associations related to race, gender, and asthma severity. In addition, we considered haplotype structure and its potential relationship to asthma.
HNMT is the most widely studied gene among the histamine related gene group with several studies investigating the potential for association between variation within HNMT and allergic disease and asthma. HNMT 314C→T is a non-synonymous SNP leading to amino acid substitution Thr105Ile which results in altered protein structure and decreased enzyme activity (17). This SNP is among the most widely studied among the histamine pathway variants. Previous investigators have observed an association between this SNP and allergic disease and asthma; however, results are conflicting. Kennedy et al. found that the frequency of the variant T allele was higher in children with atopic dermatitis than in those without atopic dermatitis (0.12 vs. 0.06, p=0.04) (23). In Caucasians, Yan et al. observed a difference in frequency of the variant allele among those with asthma compared to a control population comprised of randomly selected Caucasian blood donors (0.08 vs. 0.14, p<0.01) (24). Szczepankiewicz et al. found that the TT genotype for HNMT 314 was more common in Caucasian children with asthma compared to those without the disease (28). But these findings were not confirmed by other studies that evaluated the same SNP in relation to asthma in German, Japanese, Indian, or Spanish-Caucasian asthma subjects (21, 25-27). Similarly, we did not identify an association between the variant and asthma among our cohort. This discrepancy may perhaps be due to the racial makeup of our study cohort which was largely African American.
Other studies that have evaluated SNPs in genes involved with histamine regulation have not observed an association with asthma. Gervasini et al. investigated HDC 92C→T (rs17740607) and did not observe a difference in variant allele (T) frequency relative to a diagnosis of allergic rhinitis with asthma vs. allergic rhinitis only in a Caucasian study population (20). Another study has evaluated a SNP (rs901865) in HRH1-17C→T in Japanese children and, similar to our findings, did not observe an association between the variant allele and asthma (21). In a study that investigated 20 variants within the HRH4 gene among Caucasian subjects, including HRH4 413C→T SNP (rs11665084), there was no relationship between genetic variation within this gene and asthma (22). Further stratification of subjects with asthma into identified asthma phenotypes (e.g. exercise-induced, allergen-induced, and infection-induced) resulted in the identification of an association between variants of HRH4 (rs527790, rs487202, rs17187619) and infection-induced asthma. In our study, we did not stratify based on asthma phenotypes other than asthma severity and did not find an association between the variant and this phenotype.
Results of our study highlight the importance of demographic characteristics, such as gender, age, and race, in the evaluation of potential genetic polymorphisms and asthma. Asthma is a heterogeneous disease in which underlying pathophysiology is known to differ relative to demographics (32). Asthma is more prevalent in boys than girls but the ratio is reversed in adults with higher prevalence in women than men, which suggests a difference in underlying pathophysiology and asthma phenotype between males and females (2). Asthma prevalence, morbidity, and mortality is higher in African Americans which is not fully accounted for by socioeconomic differences, suggesting a difference in underlying disease pathology relative to race (2). Hence our findings from gender and race analyses may be important in further understanding the differences in asthma pathophysiology between these groups (2). There was a striking difference in allele frequency for ABP1 47C→T relative to race. The majority of African Americans possessed a variant allele (T) for ABP1 47 compared to a relatively small percentage of Caucasians. Although, this variant was not more common among African Americans with asthma, our limited sample size likely affected our ability to detect such differences. Therefore, this finding warrants further investigation into the potential impact of this variant among African Americans with asthma. DAO, the enzyme which is coded for by the gene ABP1, has been shown to have differences in activity relative to genotype and gender (33, 34). To our knowledge DAO activity has not been investigated in relation to race. Our finding relative to ABP1 47C→T among African Americans may suggest decreased activity of the enzyme in this racial group which may lead to an accumulation of histamine and impact disease pathophysiology in this group. We also observed that the variant allele for HNMT 314 (T) and HDC 92 (T) are more common among Caucasians than African Americans, which is similar to what has previously been reported (35); however, according to previous literature, HNMT 314T is absent in African American populations (3, 35) while our cohort had African American subjects with T allele in low frequency.
Though we did not identify an association between HNMT variants and asthma subjects, a significantly higher number of subjects without asthma carried the HNMT haplotype -1639C/ -464C/ 314C/ 3’UTRA when compared to those with asthma. This finding suggests that the HNMT wild-type alleles may be protective in the development of asthma. The haplotype was also more common among Caucasians without asthma than those with the disease. These findings are further supported by previous studies which have shown that HNMT 314T allele was associated with asthma in Caucasian populations but also may explain the discrepancy observed between previous investigations as the relationship may in fact be related to variants that are in linkage with the HNMT 314T allele (24). Other studies have also compared HNMT haplotypes among those with and without asthma (27, 28). In a study among Indian populations, HNMT haplotypes (a total of 47 including 8 haplotypes with a frequency >5% in study population) constructed for 314C→T, Intron 5 (CA) n, 939A→G, and BV677277 (CA) n were not associated with asthma when compared to controls (27). A study among Polish Caucasian children investigated two HNMT haplotypes, -1637C→T/ -411C→T and -411C→T/ 314C→T (28). Though the −411T/ 314T haplotype was observed more frequently in asthma subjects compared to control subjects (p=0.0046), investigators were doubtful that association was due to the haplotype and more likely due to the 314C→T SNP, given the distance between the two markers and as they found that the 314C→T SNP was alone associated with a diagnosis of asthma in their cohort (28). These studies have included other loci which were not included in our study. We will need to explore other loci within the histamine pathway in future studies.
We also identified differences in genotype relative to gender. Previous studies have shown that gender may be associated with activity of DAO. Garcia-Martin et al. observed that females had higher basal levels of DAO serum activity compared to males (33). Enzyme activity has also been associated with ABP1 genotype. German adults with reduced serum DAO activity (<10 U/mL using putrescine as a substrate), are more likely to carry a variant allele for ABP1, including ABP1 47C→T (Thr16Met) and ABP1 995C→T (Ser332Phe) (34). Among subjects with reduced DAO activity, the variant allele for ABP1 47 has also been associated with “histamine intolerance”, defined as 2 or more symptoms, such as headache and flushing, when consuming histamine-rich food, alcohol, or some drugs (34). In our cohort, we found that the variant allele for ABP1 47 was over-represented in females, suggesting that they may have reduced DAO activity. However, the significance of this potentially reduced DAO function in females in our cohort requires further investigation given the opposite findings of Garcia-Martin et al (33). Chen et al. observed that HNMT activity in erythrocytes was higher in Chinese males compared to females (p<0.0001), suggesting HNMT activity may also be influenced by gender (36). Also, the average HNMT activity was significantly lower (34%) among Chinese subjects with HNMT 314CT genotype compared to those with wild type CC genotype (p<0.0001) (36). Szczepankiewicz et al. found that the HNMT 314T (Thr105Ile) allele was more common among boys with asthma. In our study we observed a possible association between the variant allele among males with asthma relative to those without asthma suggesting a potential role for the HNMT 314T allele in the pathogenesis of asthma among males via reduction in HNMT activity and histamine accumulation.
HRH1 mRNA expression
Few studies have evaluated HRH1 expression among those with asthma and/or allergic disease (37, 38). We have previously shown that HRH1 mRNA expression was significantly up-regulated in adults (p=0.04) with asthma compared to those without asthma (10). Selivanova et al. also found that HRH1 mRNA expression is significantly higher in patients with “brittle” asthma compared to those with mild to moderate asthma suggesting a possible “dose response” (39). Therefore in our study we wanted to determine if the difference in mRNA expression between those with and without asthma could be explained by genetic variation in genes that directly involve histamine. In our study population, increased HRH1 mRNA levels were associated with the variant allele for ABP1 995 but only in the older age group (>12 years of age) which parallels our previous findings relative to age. In our current study we also demonstrated a possible dose response as gene expression increased with presence of one versus two variant alleles. ABP1 995C→T, has not been shown to affect enzyme activity significantly in previous studies (18). Biological relevance of the ABP1 995C→T SNP is currently unknown. However, it is possible that ABP1 995C→T is in linkage with another polymorphism that affects the level or activity of DAO. As previously mentioned DAO enzyme activity was found to be significantly associated with five SNPs within the ABP1 gene. Four of the five SNPs, including ABP1 995C→T, were in high linkage disequilibrium (34). If one of the linked polymorphisms increases the enzyme activity/level leading to lower histamine levels, it may increase HRH1 mRNA expression to compensate for the lower histamine level by a feedback mechanism. However, the linked polymorphism may also reduce the enzyme activity/level or perhaps the ABP1 995 variant itself confers decreased histamine biotransformation leading to histamine build up and increasing expression of HRH1 by a feed-forward mechanism. These combined findings warrant further investigation to delineate the significance of ABP1 variants in relation to enzyme activity and clinical impact.
There were limitations of our study that may have affected the observed results. One significant limitation of our study is that our sample size likely prevented us from observing potential relevant associations. However, this study was conducted as a pilot study to explore potential associations for further exploration in larger and better defined cohorts. We recognize that stratification with small sample size may lead to erroneous results however a Type II error would more likely be the outcome instead of finding an association when none truly exists. Although, one of the strengths of our study is that we have included racial groups which have not been previously studied in this context, our findings may have been diluted by racial heterogeneity within our cohort. Future studies should also be conducted in larger populations of under-represented racial groups (e.g. African Americans). Disease heterogeneity may have also masked potential findings related to asthma. It is likely that genetic differences along the histamine pathway may be more directly related to specific asthma phenotypes such as an allergic vs. non-allergic phenotype. The non-asthma group in our study may have included individuals with exaggerated histamine response and/or exposure due to other underlying allergic disease states (e.g. allergic rhinitis, atopic dermatitis). As the design of this study was to identify potential genetic differences between asthma and non-asthma, we were not able to perform a comparison related to allergic desensitization. We point out that our findings relative to race and gender may instead be driven by differences in allergic sensitization among these groups. We have conducted the initial steps in investigating histamine related SNPs among those with asthma vs. those without asthma and recognize that further work is needed in well-defined asthma phenotypes (defined by allergy skin testing and clinical history). These studies are currently under way.
Conclusions
Though allele and genotype within the histamine production, response, and degradation pathway was not associated with asthma diagnosis, HNMT haplotype appeared to be protective in development of the disease. We also found an association between asthma and genetic variation within the pathway among specific subgroups of subjects based on age, gender, and race. Further studies are warranted in a larger study population with well-defined asthma phenotypes. Further studies should also focus specifically on previously under-studied populations (e.g. African Americans). Furthermore, the biologic relevance of gender-related associations needs to be further elucidated. Although enzyme activity has been associated with gender, it is not clear what the role, if any, genetics may play. Future studies investigating the functional activity of histamine receptors and enzymes are needed to clearly elucidate potential implications of gene variants in the histamine pathway on asthma and allergic disease as such findings may be important in better understanding of asthma disease pathophysiology and in guiding improved and more directed therapies for patients with asthma.
Acknowledgments
Financial Support: These studies were supported by funds from the Marion Merrell Dow Clinical Scholar Award, CMH Young Investigator Award, and partial funding from the National Heart, Lung, and Blood Institute grant #1K23HL105783
References
- 1.National Heart, Lung, and Blood Institute Expert Panel Report 3 (EPR3): Guidelines for the Diagnosis and Management of Asthma. 2007 [11/13/2013]. Available from: http://www.nhlbi.nih.gov/guidelines/asthma/asthgdln.htm.
- 2.American Lung Association Trends in Asthma Morbidity and Mortality. 2012 Sep; [12/03/2013]. Available from: http://www.lung.org/finding-cures/our-research/trend-reports/asthma-trend-report.pdf.
- 3.Garcia-Martin E, Ayuso P, Martinez C, Blanca M, Agundez JA. Histamine pharmacogenomics. Pharmacogenomics. 2009 May;10(5):867–83. doi: 10.2217/pgs.09.26. PubMed PMID: 19450133. Epub 2009/05/20. eng. [DOI] [PubMed] [Google Scholar]
- 4.Hirota N, Risse PA, Novali M, McGovern T, Al-Alwan L, McCuaig S, Proud D, et al. Histamine may induce airway remodeling through release of epidermal growth factor receptor ligands from bronchial epithelial cells. FASEB J. Apr;26(4):1704–16. doi: 10.1096/fj.11-197061. PubMed PMID: 22247333. Epub 2012/01/17. eng. [DOI] [PubMed] [Google Scholar]
- 5.Igaz P, Fitzimons CP, Szalai C, Falus A. Histamine genomics in silico: polymorphisms of the human genes involved in the synthesis, action and degradation of histamine. Am J Pharmacogenomics. 2002;2(1):67–72. doi: 10.2165/00129785-200202010-00006. PubMed PMID: 12083955. Epub 2002/06/27. eng. [DOI] [PubMed] [Google Scholar]
- 6.MacGlashan D., Jr. Histamine: A mediator of inflammation. J Allergy Clin Immunol. 2003 Oct;112(4 Suppl):S53–9. doi: 10.1016/s0091-6749(03)01877-3. PubMed PMID: 14530789. Epub 2003/10/08. eng. [DOI] [PubMed] [Google Scholar]
- 7.Moya-Garcia AA, Pino-Angeles A, Gil-Redondo R, Morreale A, Sanchez-Jimenez F. Structural features of mammalian histidine decarboxylase reveal the basis for specific inhibition. Br J Pharmacol. 2009 May;157(1):4–13. doi: 10.1111/j.1476-5381.2009.00219.x. PubMed PMID: 19413567. Pubmed Central PMCID: 2697795 Epub 2009/05/06. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Byrne PM, Inman MD. Airway hyperresponsiveness. Chest. 2003 Mar;123(3 Suppl):411S–6S. doi: 10.1378/chest.123.3_suppl.411s. PubMed PMID: 12629006. [DOI] [PubMed] [Google Scholar]
- 9.Rada B, Boudreau HE, Park JJ, Leto TL. Histamine stimulates hydrogen peroxide production by bronchial epithelial cells via histamine H1 receptor and dual oxidase. Am J Respir Cell Mol Biol. 2014 Jan;50(1):125–34. doi: 10.1165/rcmb.2013-0254OC. PubMed PMID: 23962049. Pubmed Central PMCID: 3930938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vyhlidal CA, Riffel AK, Dai H, Rosenwasser LJ, Jones BL. Detecting gene expression in buccal mucosa in subjects with asthma versus subjects without asthma. Pediatr Allergy Immunol. Mar 1; doi: 10.1111/pai.12042. 10.1111/pai.12042. PubMed PMID: 23448392. Epub 2013/03/02. Eng. [DOI] [PubMed] [Google Scholar]
- 11.Grant JA, Nicodemus CF, Findlay SR, Glovsky MM, Grossman J, Kaiser H, Meltzer EO, et al. Cetirizine in patients with seasonal rhinitis and concomitant asthma: prospective, randomized, placebo-controlled trial. J Allergy Clin Immunol. 1995 May;95(5 Pt 1):923–32. doi: 10.1016/s0091-6749(95)70090-0. PubMed PMID: 7751511. Epub 1995/05/01. eng. [DOI] [PubMed] [Google Scholar]
- 12.Bustos GJ, Bustos D, Romero O. Prevention of asthma with ketotifen in preasthmatic children: a three-year follow-up study. Clin Exp Allergy. 1995 Jun;25(6):568–73. doi: 10.1111/j.1365-2222.1995.tb01096.x. PubMed PMID: 7648464. Epub 1995/06/01. eng. [DOI] [PubMed] [Google Scholar]
- 13.Warner JO. A double-blinded, randomized, placebo-controlled trial of cetirizine in preventing the onset of asthma in children with atopic dermatitis: 18 months' treatment and 18 months' posttreatment follow-up. J Allergy Clin Immunol. 2001 Dec;108(6):929–37. doi: 10.1067/mai.2001.120015. PubMed PMID: 11742270. Epub 2001/12/14. eng. [DOI] [PubMed] [Google Scholar]
- 14.Van Ganse E, Kaufman L, Derde MP, Yernault JC, Delaunois L, Vincken W. Effects of antihistamines in adult asthma: a meta-analysis of clinical trials. Eur Respir J. 1997 Oct;10(10):2216–24. doi: 10.1183/09031936.97.10102216. PubMed PMID: 9387943. [DOI] [PubMed] [Google Scholar]
- 15.Aaronson DW. Evaluation of cetirizine in patients with allergic rhinitis and perennial asthma. Ann Allergy Asthma Immunol. 1996 May;76(5):440–6. doi: 10.1016/S1081-1206(10)63461-8. PubMed PMID: 8630718. [DOI] [PubMed] [Google Scholar]
- 16.Hirata N, Takeuchi K, Ukai K, Sakakura Y. Expression of histidine decarboxylase messenger RNA and histamine N-methyltransferase messenger RNA in nasal allergy. Clin Exp Allergy. 1999 Jan;29(1):76–83. doi: 10.1046/j.1365-2222.1999.00345.x. PubMed PMID: 10051705. Epub 1999/03/03. eng. [DOI] [PubMed] [Google Scholar]
- 17.Preuss CV, Wood TC, Szumlanski CL, Raftogianis RB, Otterness DM, Girard B, Scott MC, et al. Human histamine N-methyltransferase pharmacogenetics: common genetic polymorphisms that alter activity. Mol Pharmacol. 1998 Apr;53(4):708–17. doi: 10.1124/mol.53.4.708. PubMed PMID: 9547362 Epub 1998/05/09. eng. [DOI] [PubMed] [Google Scholar]
- 18.Ayuso P, Garcia-Martin E, Martinez C, Agundez JA. Genetic variability of human diamine oxidase: occurrence of three nonsynonymous polymorphisms and study of their effect on serum enzyme activity. Pharmacogenet Genomics. 2007 Sep;17(9):687–93. doi: 10.1097/FPC.0b013e328012b8e4. PubMed PMID: 17700358 Epub 2007/08/19. eng. [DOI] [PubMed] [Google Scholar]
- 19.Jones BL, Kearns GL. Histamine: new thoughts about a familiar mediator. Clin Pharmacol Ther. Feb;89(2):189–97. doi: 10.1038/clpt.2010.256. PubMed PMID: 21178984. Epub 2010/12/24. eng. [DOI] [PubMed] [Google Scholar]
- 20.Gervasini G, Agundez JA, Garcia-Menaya J, Martinez C, Cordobes C, Ayuso P, Cornejo JA, et al. Variability of the L-Histidine decarboxylase gene in allergic rhinitis. Allergy. Dec;65(12):1576–84. doi: 10.1111/j.1398-9995.2010.02425.x. PubMed PMID: 20608921. Epub 2010/07/09. eng. [DOI] [PubMed] [Google Scholar]
- 21.Sasaki Y, Ihara K, Ahmed S, Yamawaki K, Kusuhara K, Nakayama H, Nishima S, et al. Lack of association between atopic asthma and polymorphisms of the histamine H1 receptor, histamine H2 receptor, and histamine N-methyltransferase genes. Immunogenetics. 2000 Mar;51(3):238–40. doi: 10.1007/s002510050037. PubMed PMID: 10752634. Epub 2001/02/07. eng. [DOI] [PubMed] [Google Scholar]
- 22.Simon T, Semsei AF, Ungvari I, Hadadi E, Virag V, Nagy A, Vangor MS, et al. Asthma endophenotypes and polymorphisms in the histamine receptor HRH4 gene. Int Arch Allergy Immunol. 159(2):109–20. doi: 10.1159/000335919. PubMed PMID: 22653292. Epub 2012/06/02. eng. [DOI] [PubMed] [Google Scholar]
- 23.Kennedy MJ, Loehle JA, Griffin AR, Doll MA, Kearns GL, Sullivan JE, Hein DW. Association of the histamine N-methyltransferase C314T (Thr105Ile) polymorphism with atopic dermatitis in Caucasian children. Pharmacotherapy. 2008 Dec;28(12):1495–501. doi: 10.1592/phco.28.12.1495. PubMed PMID: 19025430 Pubmed Central PMCID: 2642612. Epub 2008/11/26. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan L, Galinsky RE, Bernstein JA, Liggett SB, Weinshilboum RM. Histamine N- methyltransferase pharmacogenetics: association of a common functional polymorphism with asthma. Pharmacogenetics. 2000 Apr;10(3):261–6. doi: 10.1097/00008571-200004000-00007. PubMed PMID: 10803682. Epub 2000/05/10. eng. [DOI] [PubMed] [Google Scholar]
- 25.Deindl P, Peri-Jerkan S, Deichmann K, Niggemann B, Lau S, Sommerfeld C, Sengler C, et al. No association of histamine- N-methyltransferase polymorphism with asthma or bronchial hyperresponsiveness in two German pediatric populations. Pediatr Allergy Immunol. 2005 Feb;16(1):40–2. doi: 10.1111/j.1399-3038.2005.00218.x. PubMed PMID: 15693910. Epub 2005/02/08. eng. [DOI] [PubMed] [Google Scholar]
- 26.Garcia-Martin E, Garcia-Menaya J, Sanchez B, Martinez C, Rosendo R, Agundez JA. Polymorphisms of histamine-metabolizing enzymes and clinical manifestations of asthma and allergic rhinitis. Clin Exp Allergy. 2007 Aug;37(8):1175–82. doi: 10.1111/j.1365-2222.2007.02769.x. PubMed PMID: 17651147. Epub 2007/07/27. eng. [DOI] [PubMed] [Google Scholar]
- 27.Sharma S, Mann D, Singh TP, Ghosh B. Lack of association of histamine-N- methyltransferase (HNMT) polymorphisms with asthma in the Indian population. J Hum Genet. 2005;50(12):611–7. doi: 10.1007/s10038-005-0302-4. PubMed PMID: 16205835. Epub 2005/10/06. eng. [DOI] [PubMed] [Google Scholar]
- 28.Szczepankiewicz A, Breborowicz A, Sobkowiak P, Popiel A. Polymorphisms of two histamine-metabolizing enzymes genes and childhood allergic asthma: a case control study. Clin Mol Allergy. 8:14. doi: 10.1186/1476-7961-8-14. PubMed PMID: 21040557. Pubmed Central PMCID: 2990726. Epub 2010/11/03. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller MR, Hankinson J, Brusasco V, Burgos F, Casaburi R, Coates A, Crapo R, et al. Standardisation of spirometry. Eur Respir J. 2005 Aug;26(2):319–38. doi: 10.1183/09031936.05.00034805. PubMed PMID: 16055882. [DOI] [PubMed] [Google Scholar]
- 30.Santiago Rodriguez TRGaINMD Hardy-Weinberg Equilibrium Testing of Biological Ascertainment for Mendelian Randomization Studies. 2008 doi: 10.1093/aje/kwn359. [09/05/2012]. Available from: http://www.oege.org/software/hwe-mr-calc.shtml. [DOI] [PMC free article] [PubMed]
- 31.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001 Apr;68(4):978–89. doi: 10.1086/319501. PubMed PMID: 11254454 Epub 2001/03/20. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rastogi D, Reddy M, Neugebauer R. Comparison of patterns of allergen sensitization among inner-city Hispanic and African American children with asthma. Ann Allergy Asthma Immunol. 2006 Nov;97(5):636–42. doi: 10.1016/S1081-1206(10)61093-9. PubMed PMID: 17165272. [DOI] [PubMed] [Google Scholar]
- 33.Garcia-Martin E, Ayuso P, Martinez C, Agundez JA. Improved analytical sensitivity reveals the occurrence of gender-related variability in diamine oxidase enzyme activity in healthy individuals. Clin Biochem. 2007 Nov;40(16-17):1339–41. doi: 10.1016/j.clinbiochem.2007.07.019. PubMed PMID: 17826755. Epub 2007/09/11. eng. [DOI] [PubMed] [Google Scholar]
- 34.Maintz L, Yu CF, Rodriguez E, Baurecht H, Bieber T, Illig T, Weidinger S, et al. Association of single nucleotide polymorphisms in the diamine oxidase gene with diamine oxidase serum activities. Allergy. Jul;66(7):893–902. doi: 10.1111/j.1398-9995.2011.02548.x. PubMed PMID: 21488903. Epub 2011/04/15. eng. [DOI] [PubMed] [Google Scholar]
- 35.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic acids research. 2001 Jan 1;29(1):308–11. doi: 10.1093/nar/29.1.308. PubMed PMID: 11125122. Pubmed Central PMCID: 29783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen GL, Wang W, Xu ZH, Zhu B, Wang LS, Zhou G, Wang D, et al. Genotype phenotype correlation for histamine N-methyltransferase in a Chinese Han population. Clin Chim Acta. 2003 Aug;334(1-2):179–83. doi: 10.1016/s0009-8981(03)00239-0. PubMed PMID: 12867290. Epub 2003/07/18. eng. [DOI] [PubMed] [Google Scholar]
- 37.Shirasaki H, Kanaizumi E, Seki N, Himi T. Localization and upregulation of the nasal histamine H1 receptor in perennial allergic rhinitis. Mediators of inflammation. 2012;2012:951316. doi: 10.1155/2012/951316. PubMed PMID: 23132961. Pubmed Central PMCID: 3486441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iriyoshi N, Takeuchi K, Yuta A, Ukai K, Sakakura Y. Increased expression of histamine H1 receptor mRNA in allergic rhinitis. Clin Exp Allergy. 1996 Apr;26(4):379–85. PubMed PMID: 8732234. [PubMed] [Google Scholar]
- 39.Selivanova PA, Kulikov ES, Kozina OV, Gereng EA, Freidin MB, Ogorodova LM. Morphological and molecular characteristics of “difficult” asthma. J Asthma. 2010 Apr;47(3):269–75. doi: 10.3109/02770900903584001. PubMed PMID: 20394512. [DOI] [PubMed] [Google Scholar]