Abstract
Background
Where a neuron is positioned in the brain during development determines neuronal circuitry and information processing needed for normal brain function. When aberrations in this process occur, cognitive disorders may result. Patients diagnosed with schizophrenia have been reported to show altered neuronal connectivity and heterotopias. To elucidate pathways by which this process occurs and become aberrant, we have chosen to study the long isoform of nitric oxide synthase 1 adaptor protein (NOS1AP), a protein encoded by a susceptibility gene for schizophrenia.
Methods
To determine whether NOS1AP plays a role in cortical patterning, we knocked down or co-overexpressed NOS1AP and a GFP or TagRFP reporter in neuronal progenitor cells of the embryonic rat neocortex using in utero electroporation. We analyzed sections of cortex (ventricular zone VZ, intermediate zone IZ, and cortical plate CP) containing GFP or TagRFP positive cells and counted the percentage of positive cells that migrated to each region from at least three rats for each condition.
Results
NOS1AP overexpression disrupts neuronal migration, resulting in increased cells in IZ and less cells in CP, and decreases dendritogenesis. Knock down results in increased migration, with more cells reaching the CP. The phosphotyrosine binding region, but not the PDZ-binding motif, is necessary for NOS1AP function. Amino acids 181–307, which are sufficient for NOS1AP-mediated decreases in dendrite number, have no effect on migration.
Conclusions
Our studies show for the first time a critical role for the schizophrenia-associated gene NOS1AP in cortical patterning, which may contribute to underlying pathophysiology seen in schizophrenia.
Keywords: Schizophrenia, Cortical development, in utero electroporation, NOS1AP, Cortical neuron migration, Rodent model
Introduction
Proper brain development and function requires the correct migration and placement of neurons. Abnormal neuronal migration may cause abnormal cortical function. There is increasing evidence that altered neuronal connectivity seen in schizophrenia may be due to factors during neurodevelopment in utero (reviewed in (1–2)). Here we study the role of NOS1AP, a protein encoded by a schizophrenia susceptibility gene (3–7), during cortical neuron migration.
NOS1AP was first identified in the rat as a binding partner of neuronal nitric oxide synthase ((nNOS; 8). It competes with PSD-95 for nNOS binding and presumably reduces NMDA receptor signaling via PSD-95 and nNOS. There are at least three isoforms of NOS1AP (long (L), short (S), and short’ (S’)) that have been reported to have altered protein expression in the cortex of individuals diagnosed with schizophrenia (9). Specifically, NOS1AP-L protein expression, normalized to the housekeeping protein GAPDH, is elevated approximately 10-fold whereas the other two isoforms are elevated approximately 100-fold (9). In addition, NOS1AP-L mRNA expression is significantly decreased by 40% in postmortem brain tissue from patients treated with antipsychotics when compared to untreated patients with the same psychiatric diagnosis (4). All three NOS1AP isoforms contain a C-terminal PDZ-binding domain, which is responsible for the interaction of NOS1AP with nNOS. NOS1AP-L contains a phosphotyrosine binding (PTB) domain (8), which binds to DexRas1, synapsin, and Scribble (10–12). In addition, we recently showed that NOS1AP-L interacts with carboxypeptidase E by a domain contained in its middle region (amino acids 181–307) and that NOS1AP regulates dendrite morphology through this interaction (13).
While overexpression of NOS1AP-L, the isoform affected by administration of antipsychotic drugs, has been reported by our group (13) to alter dendrite number and by another group (12) to alter spine development, an understanding of how NOS1AP, specifically NOS1AP-L, regulates synaptic connectivity in vivo has not been fully elucidated. Here we show that NOS1AP-L negatively controls radial migration during cortical development. In addition, we identify the PTB domain as the main region of the protein involved in this role. These results suggest that upregulation of NOS1AP protein due to schizophrenia-associated alleles may promote altered connectivity seen in individuals with schizophrenia.
Materials and Methods
Antibodies
Rabbit polyclonal anti-NOS1AP antibodies (sc-9138) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse monoclonal anti-GAPDH antibody was from Millipore (Billerica, MA), rabbit polyclonal anti-hrGFP antibody was from Agilent Technologies (Santa Clara, CA), and chicken polyclonal anti-EGFP antibody was from AVES (Tigard, OR). Antichicken secondary antibody conjugated to Alexa-FluorR 488 was from Jackson ImmunoResearch (West Grove, PA).
DNA constructs
pCAG-GFP plasmid was obtained by subcloning the sequence of EGFP from pEGFP-C1 plasmid from Clontech (Mountain View, CA) into a vector containing the CMV–actin–β-globin (CAG) promoter (gift from Dennis O’Leary, Salk Institute). cDNA encoding human NOS1AP-L (referred to as NOS1AP or NP for this manuscript), NOS1AP-213-end (NPΔPTB), and NOS1AP-181–307 (NP181–307) were subcloned into pCAG-GFP plasmid to obtain according N-terminally labeled GFP fusion constructs. cDNA encoding NOS1AP and NOS1AP-1–487 (NPΔPDZ) were subcloned in pCAG-IRES-EGFP (pCIG, gift from Gabriella D’Arcangelo, Rutgers University) and pCAG-IRES-TagRFP plasmid (pCIR, gift from Marie-Catherine Tiveron, Institut de Biologie du Développement de Marseille).
For shRNA constructs, oligonucleotides were ligated into the pGE2hrGFPII vector (Agilent Technologies). The sequence of the RNAi target for NOS1AP was 5’-GGGTGACAGTTTGGATGAT-3’ (shNP (13)). As negative control, we used a sequence that did not align with any mammalian gene: 5’-GAGCATTTGTATGAGCGCG-3’ (shControl, against GST; gift from Estela Jacinto, Rutgers Medical School). We have previously reported the sequences of NOS1AP shRNA, human NOS1AP-L (hNP-L), and mouse/rat NOS1AP-L in Figure 2C of (13).
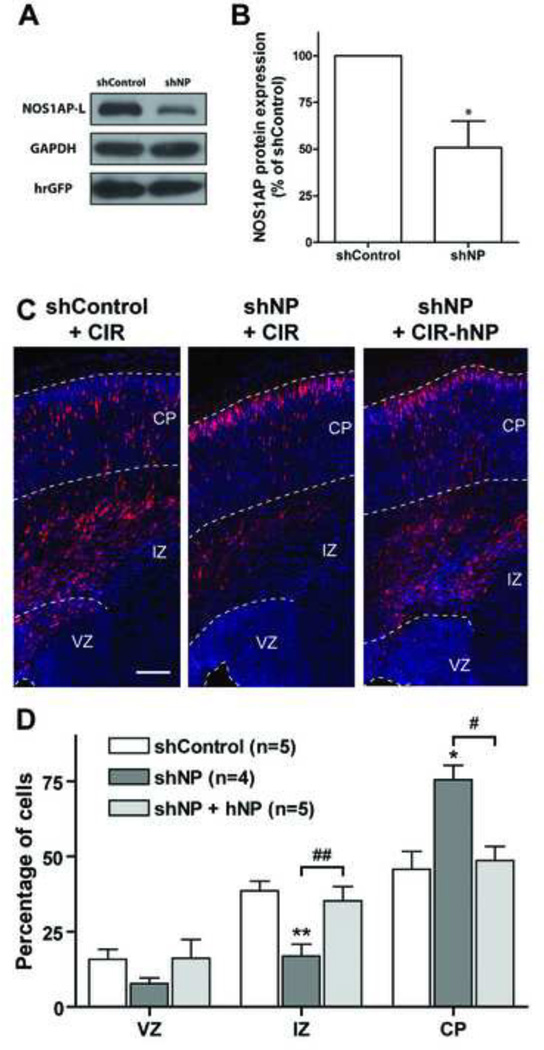
Figure 2. Knockdown of NOS1AP increases cortical neuron migration.
A, COS-7 cells were cotransfected with pEGFP-NOS1AP (mouse) and either pGE-control shRNA (shControl, negative control) or pGE-NOS1AP shRNA (shNP). After 48 h, NOS1AP, GAPDH, and hrGFP levels were detected by Western blot analysis. B, Quantification of NOS1AP levels (normalized to GAPDH expression). shNP results in a 51% decrease in NOS1AP expression (n = 3 independent experiments; p<0.05 by Student’s t-test). C, Indicated constructs were electroporated into the lateral ventricular wall at E16 and analyzed at E20. Representative images of coronal sections of rat somatosensory cortex are shown for each condition. Sections were counterstained with Hoechst (blue). D, Quantification of the percentage of transfected cells in each cortical area. Results are means ± SEM. *p<0.05 and **p<0.01 versus control; #p<0.05 and ##p<0.01 versus shNP. Scale bars: 150 µm (C).
Transfection of cultured cells
COS-7 cells were cultured in 6 well plates and transfected at 30–50% confluency with a 3:1 ratio of shNP:NOS1AP to test for shRNA efficiency for NOS1AP knockdown using LIPOFECTAMINE-2000 following the manufacturer’s protocol. (A 1:1 ratio also resulted in significant NOS1AP knockdown.)
Western Blotting
COS-7 cells were cultured in 60 mm dishes and transfected using Lipofectamine 2000 (Life Technologies). Cells were collected 2 days after transfection and lysed, and expression of NOS1AP was detected by immunoblotting after resolving proteins using SDS-PAGE gels. After electrophoresis, proteins were transferred to PVDF membranes (Immobilon-P; Millipore). After blocking with 2% bovine serum albumin (BSA) in TBST, membranes were incubated with primary antibodies overnight at 4°C: 1:250 for anti-NOS1AP (Santa Cruz), 1:1000 for anti-GAPDH (Millipore), or 1:1000 for anti hrGFP (Agilent Technologies). After washing, horseradish peroxidase linked secondary antibody was applied at 1:5.000 for one hour at RT. Immunoreactive bands were visualized using HyGlo quick spray (Denville) and quantified using Image Pro software (Media Cybernetics).
In utero electroporation
Cells were transfected in vivo by in utero electroporation. Pregnant Sprague-Dawley rats at gestation day 16 (E16) were anesthetized with Ketamine/Xylazine (75/10 mixture). The abdominal cavity was opened to expose the uterine horns. 1–3 µl of plasmids (1.5–2 µg/µl) with 1 mg/mL Fast Green (Sigma) were microinjected through the uterus into the lateral ventricles of embryos by pulled glass capillaries (Drummond Scientific, Broomall, PA). Electroporation was performed by placing heads of the embryos between tweezer-type electrodes. Square electric pulses (70 V, 50 ms) were passed five times at 1-s intervals using a CUY21 EDIT electroporator (Nepagene, Bulldog Bio, Inc., Portsmouth, NH). Embryos were allowed to develop in utero for 4 days after electroporation (until E20).
All animals used in this study were handled in accordance with a protocol approved by the Institutional Animal Care and Use Committee at Rutgers, the State University of New Jersey, and in compliance with national and international laws and policies (Council directives no. 87–848, 19 October 1987, Ministère de l'Agriculture et de la Forêt, Service Vétérinaire de la Santé et de la Protection Animale).
Histological procedures and microscopy
Embryonic rat brains (E20) were dissected and fixed for 48 h in 4% paraformaldehyde (PFA) in PBS at 4°C. Postnatal rat brains (P14) were fixed by transcardial perfusion of 4% PFA in PBS and postfixed for 3 hours in 4% PFA in PBS. Brains were then cryoprotected in 30% sucrose in PBS, frozen in OCT (Tissue-Tek) and sectioned coronally at 16 µm (E20 brains) and 30 µm and 80 µm (P14 brains) using a cryostat.
For immunofluorescence staining, sections were incubated for 1 h in antibody buffer (5% normal goat serum, 0.1% Triton X-100 in PBS) before overnight incubation with anti-EGFP antibody (1:1,000). Following washes, sections were incubated with A488 anti-chicken antibody (1:1,000) for 2 hr at room temperature. Sections were then washed, incubated for 10 min with Hoechst 33342 (Thermo Scientific, Rockford, IL) for nuclear staining and mounted using Fluoromount G (Southern Biotechnology, Birmingham, AL).
For analysis of migration at E20 and P14 and cell morphology at E20, images of immunofluorescent rat brain sections were taken on a Zeiss AxioImager M1 microscope using a 20× numerical aperture (NA) 0.75 objective (for E20 16 µm sections) or a 10× NA 0.3 objective (for P14 30 µm sections).
For analysis of neuronal morphology at P14, 40–50 µm-thick z-stacks were acquired on a Zeiss LSM 510 confocal laser-scanning microscope using a 20× NA 0.5 objective, and z-series were projected to two-dimensional representations.
In situ hybridization
For in situ hybridization (kit from Roche Applied Science), 40 µm cryostat sections from E16 and E20 rats were incubated twice in DEPC-treated PBS and twice in PBS containing 100mM glycine for 5 min at room temperature. They were then treated for 15min with PBS containing 0.3% Triton X-100 followed by two PBS washes. Sections were then permeabilized for 30min at 37°C with 2ug/ml Proteinase K, post-fixed for 5min at 4°C with PBS containing 4% paraformaldehyde, and washed twice with PBS. Sections were then incubated twice for 5 min with 0.1M triethanolamine (TEA) buffer, pH8.0, containing 0.25%(v/v) acetic anhydride, incubated in prehyridization buffer (2XSSC, 1X Denhardt’s solution, 10% dextran sulfate, 50mM phosphate buffer pH7.0, 50mM DTT, 250 µg t-RNA, 500ug/ml denatured and sheared salmon sperm DNA, 50% deionized formamide) under coverslips in a humidified chamber at 37°C for 2 hours, and incubated with hybridization buffer (2X SSC, 1X Denhardt’s solution, 10% dextran sulfate, 50 mM Phosphate buffer (pH 7.0), 50mM DTT,, 250ug t-RNA, 500ug/ml denatured and sheared salmon sperm DNA, 50% deionized formamide, 100nM digoxigenin-labeled oligonucleotide probes) in a humidified chamber at 37°C overnight. Sections were then washed in decreasing dilutions of SSC, followed by a wash in 100mM Tris-HCl (pH 7.5), 150mM NaCl (buffer 1), and then a 30 minute incubation with buffer 1 containing 0.1% Triton X-100 and 2% normal sheep serum. Slides were then incubated with buffer 1 containing 0.1% Triton X-100, 1% normal sheep serum, and 1:500 dilution of anti-DIG-alkaline phosphatase [Fab fragments] for 4 hours in a humidified chamber, washed with buffer 1, and incubated with 100mM Tris-HCl (pH 9.5), 100mM NaCl, 50mM MgCl2 and nitroblue tetrazolium (NBT)/5-bromo-4-chloro-3-indolyl-phospate (BCIP) solution. When color development was optimal, the color reaction was terminated by incubating the slides in 10mM Tris-HCl (pH 8.1), 1mM EDTA. NOS1AP-L antisense oligonucelotide sequence was 5’-AGATAGGGTCCTGCATCACCAGCATCTTGCTCTCATCCCACGTCC[DIG]-3, and sense oligonucleotide was 5’-GGACGTGGGATGAGAGCAAGATGCTGGTGATGCAGGACCCTATCT[DIG]-3’ as a negative control.
Quantification and statistical analyses
For migration analysis, GFP or TagRFP positive cells were counted using the image analysis software ImageJ 1.47e (http://rsb.info.nih.gov/ij/). The experimenter was blinded to the experimental condition. For each section analyzed, cortical regions of interest containing positive cells were manually selected using Hoechst staining of the nuclei. Then, for each region, we used a combination of ImageJ built-in Minimum and Unsharp Mask filters to enhance the signal due to fluorescent cell bodies while lowering the signal due to fluorescent processes. Cells were automatically counted as local maxima, while keeping the same level of noise tolerance for a given set of experiment (and after validation of this level by manual counting of 3–4 sections).
To normalize the analysis, we used median sections in a series of sections containing transfected cells (2 sections per brain at E20, separated by at least 48 µm) and in which the corpus callosum was present. Therefore, brains that did not meet these two criteria were discarded.
It is important to note that all experiments included control groups, and samples within each experiment were compared to the control from the same experiment. Furthermore, we attempted to minimize batch-to-batch variation by performing in utero electroporation with all constructs for a particular experiment at the same time. Since migration of sets of cells occurs at distinct times in development, any variation in timing of in utero electroporation (i.e. E16 vs E16.25 or E16.5) will show differences in control migration. This is common to in utero electroporation experiments as can be seen in the percentage of cells reaching the cortical plate in different figures after 4 days of expression of proteins/shRNA.
For analysis of neuronal morphology at P14, neurites were traced and quantified with NeuronJ software (http://rsb.info.nih.gov/ij/; (14)). Total neurite length and number of branches for each individual neuron were measured.
n values are reported as the number of brains analyzed, unless specified in the legend. Offspring were from 2–4 mothers.
Statistical analyses were performed using GraphPad Prism software (GraphPad Software, Inc., San Diego, CA). Statistical significance was calculated using Student’s t-test or with one-way ANOVA followed by Newman–Keuls post-tests for p-value adjustment for groups of more than two conditions.
Results
NOS1AP inhibits radial migration of cortical neuron
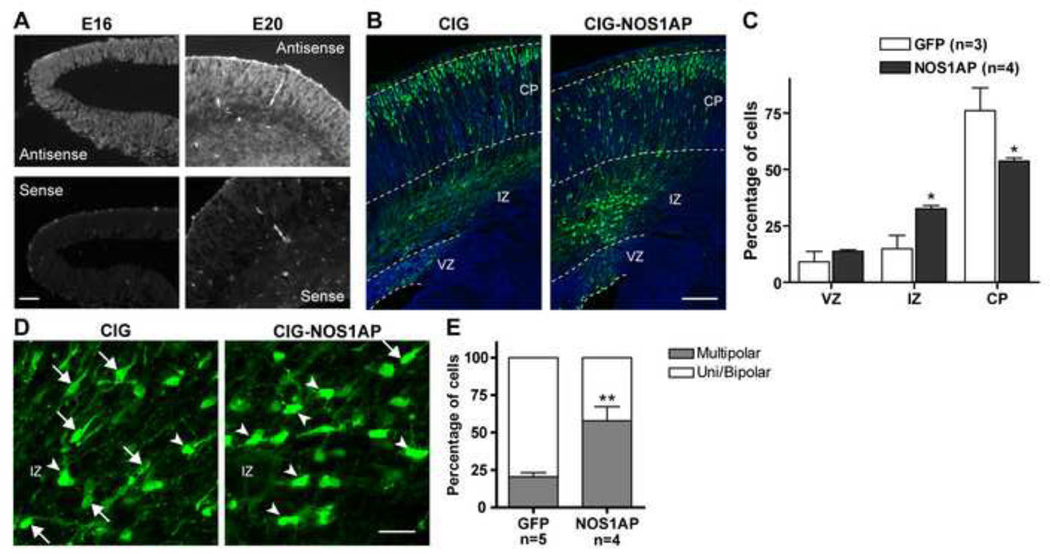
We previously showed by Western blot analysis that both NOS1AP-L and -S are expressed in the developing rat forebrain as early as embryonic day 15 (E15) (13). We were unable to perform immunohistochemistry since no antibody was available for immunostaining. (We tried both commercially available and homemade antibodies, data not shown.) Using in situ hybridization, we show that NOS1AP-L (referred to as NOS1AP) is expressed in the developing cortex (E16 and E20, Fig. 1A).
Figure 1. Effect of overexpression of NOS1AP on cortical neuron migration.
A, In situ hybridization data showing that NOS1AP is expressed in the cortex during development (representative images shown at E16 and E20). B–E, CIG or CIG-NOS1AP plasmid was electroporated into the lateral ventricular wall at E16 and analyzed at E20. B, Representative images of coronal sections of rat somatosensory cortex are shown for each condition. Sections were immunostained for GFP (green) and counterstained with Hoechst (blue). C, Quantification of the percentage of transfected cells in each cortical area. D, Representative images of transfected cells in the IZ for each condition (arrows show examples of mono/bipolar cells and arrowheads examples of multipolar cells). E, Quantification of the percentage of transfected cells within the IZ with mono/bipolar or multipolar morphology. VZ = ventricular zone, IZ = intermediate zone, CP = cortical plate. Results are means ± SEM. *p<0.05 and **p<0.01 versus control. Scale bars: 50 µm (A), 150 µm (B), 25 µm (D).
NOS1AP expression is significantly increased in the dorsolateral region of the prefrontal cortex in patients with schizophrenia (9). To investigate the role of NOS1AP overexpression during early cortical development, we performed in utero electroporation (IUE) experiments to introduce a construct expressing NOS1AP into neural progenitor cells of the embryonic rat neocortex. The construct or a control plasmid was electroporated at E16, and neuronal migration was analyzed four days later. At this stage, most cells in control condition have reached the cortical plate (CP). However, a significant amount of cells overexpressing NOS1AP is found in the intermediate zone (IZ), showing a deficit in radial migration (Fig. 1B, C). In the IZ, newborn neurons convert from multipolar to monopolar/bipolar morphology before undergoing radial glial cell-guided migration towards the CP. To investigate the role of NOS1AP overexpression on this transition, we analyzed the morphology of transfected neurons located in the IZ. As shown in Figure 1D and E, we observed that a large majority of control transfected cells found in the IZ display a uni/bipolar shape; however, more than half of cells overexpressing NOS1AP in the IZ are multipolar, suggesting that NOS1AP inhibits the transition from multipolar to bipolar morphology, necessary for efficient migration of cortical neurons towards the cortical layers.
To investigate the physiological role of NOS1AP during cortical neuron migration, we used specific shRNAs to knock down NOS1AP protein expression. Co-expression of an shRNA directed to the coding sequence of NOS1AP with mouse NOS1AP in COS-7 cells for 48 h results in a 51% (shNP) decrease in NOS1AP protein levels when compared to cells expressing control shRNA (Fig. 2A, B). We then co-electroporated the plasmids expressing the shRNA together with a construct expressing TagRFP (CIR) in cortical neuron precursors to facilitate the detection of transfected cells. Interestingly, we observed that knockdown of NOS1AP between E16 and E20 results in a significant increase in the percentage of transfected neurons reaching the cortical plate (Fig. 2 C, D).
To demonstrate the specificity of shNP, we co-expressed the rat/mouse-specific shNP with human NOS1AP in neurons. Human NOS1AP contains four differences in the sequence corresponding to the shNP construct for mouse/rat NOS1AP; therefore, it is shNP-resistant (13) and should rescue NOS1AP expression and function. As expected, neuronal repartition between ventricular zone (VZ), IZ, and CP returned to control levels in this rescue experiment (Fig. 2C, D). These data, combined with our overexpression data, strongly suggest that NOS1AP plays a role in regulating radial migration of cortical neuron during development and that aberrant expression of NOS1AP results in altered neuronal connectivity.
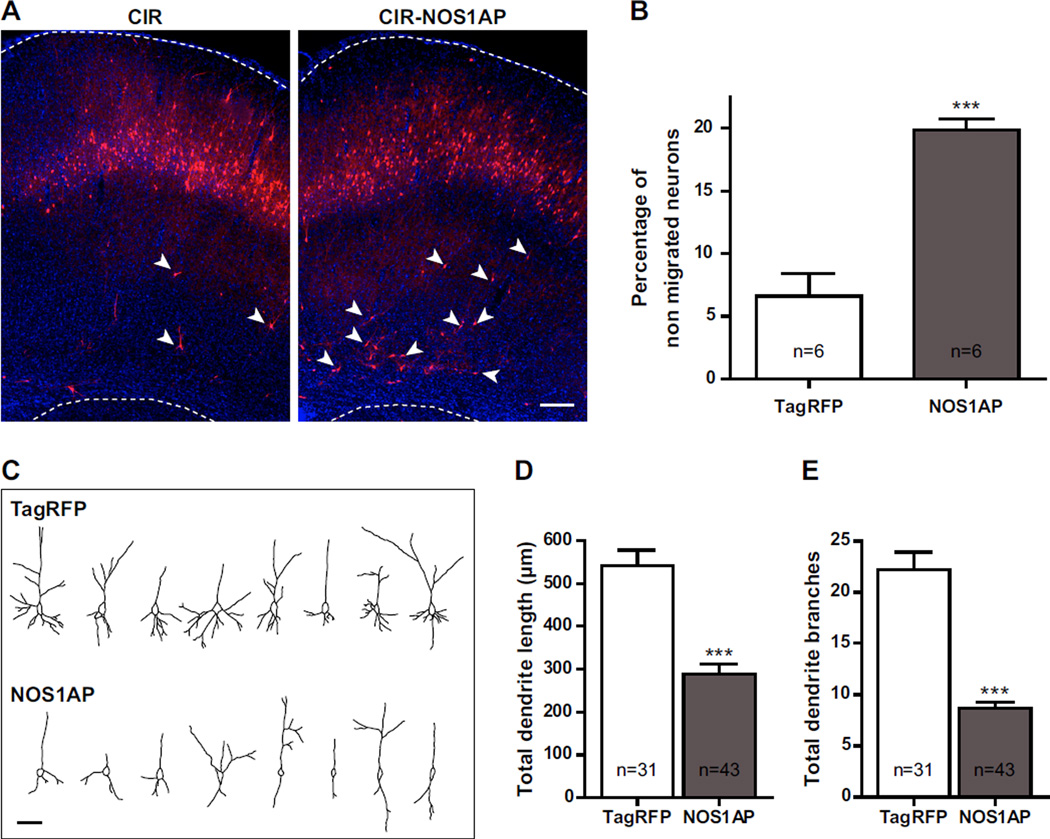
To test whether this role of NOS1AP is transitory during development or long-lasting, we electroporated neuronal precursors with a NOS1AP or a control construct at E16 and analyzed neuronal lamination three weeks later at postnatal day 14 (P14). By that time, almost all neurons in control conditions reached layers II/III of the neocortex whereas a significant percentage of neurons overexpressing NOS1AP remained in deeper layers (Figure 3A, B).
Figure 3. Overexpression of NOS1AP affects cortical neuron migration and morphology.
CIR or CIR-NOS1AP plasmid was electroporated into the lateral ventricular wall at E16 and brains were analyzed at P14. A, Representative images of coronal sections of rat somatosensory cortex are shown for each condition. Sections were counterstained with Hoechst dye (blue). B, Quantitation of the percentage of transfected cells that did not complete their migration at P14 (arrowheads in A show examples of non-migrated cells in each condition). C, Representative NeuronJ tracing of transfected cells in layers II/III for each condition. D–E, Quantification of the total dendritic length (D) and of the total number of dendrite branches (E) of transfected cells in layers II/III. n values in D and E refer to the number of analyzed cells, from 5 different brains per condition. Results are means ± SEM. ***p<0.001 versus control. Scale bars: 250 µm (A), 50 µm (C).
We also observed that NOS1AP overexpression has a marked effect on the morphology of neurons that correctly reach layers II/III. Indeed, neurons overexpressing NOS1AP exhibit less dendrites and an overall shorter dendritic tree than neurons transfected with a control construct (Figure 3C, D). These results are in agreement with our previous in vitro data showing that NOS1AP inhibits dendrite branching in hippocampal neurons (13).
Role of NOS1AP domains in cortical neuron migration
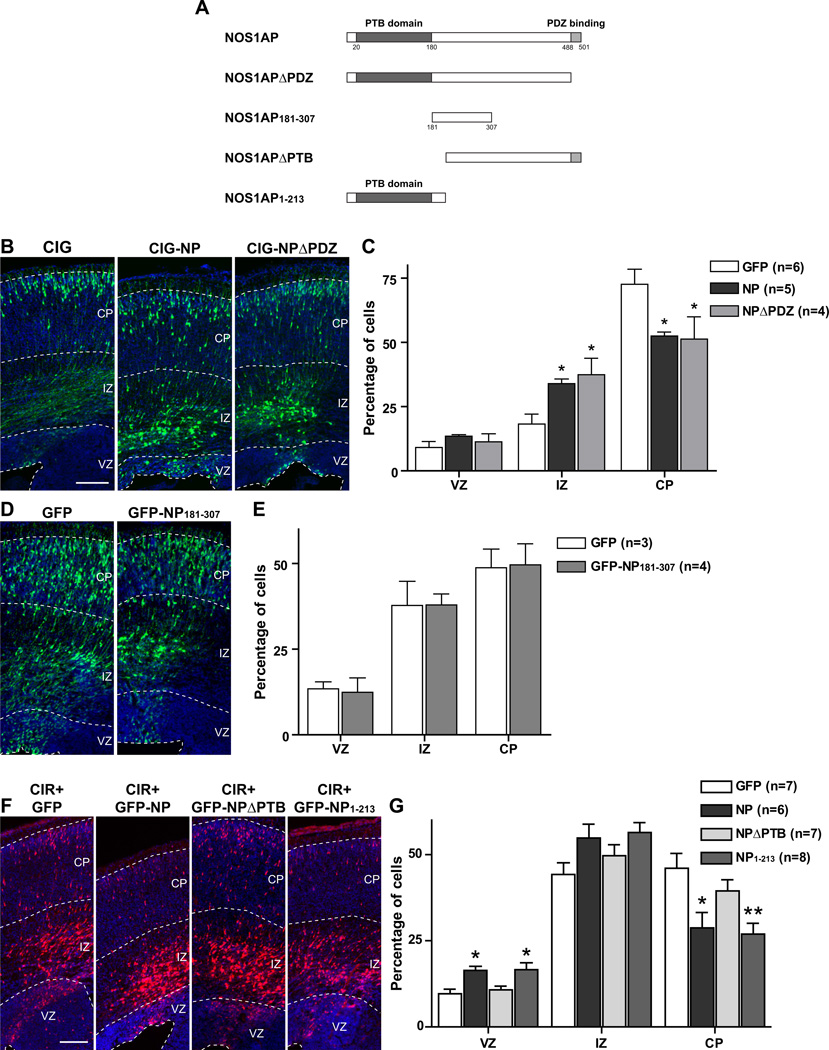
To investigate which region of NOS1AP is involved in the regulation of cortical neuron migration, we constructed several mutants of the protein (Fig. 4A) and analyzed the effects of in utero expression of these constructs in newborn cortical neurons between E16 and E20.
Figure 4. Effect of overexpression of mutant forms of NOS1AP on cortical neuron migration.
A, Four constructs encoding mutant forms of NOS1AP are illustrated. B, D, F, Indicated constructs were electroporated into the lateral ventricular wall at E16 and brains were analyzed at E20. Representative images of coronal sections of rat somatosensory cortex are shown in each condition. Sections in B and D were immunostained for GFP (green) and counterstained with Hoechst (blue). C, E, G, Quantification of the percentage of transfected cells in each cortical area. Results are means ± SEM. *p<0.05 and **p<0.01 versus control. Scale bars: 150 µm.
Deletion of the PDZ-binding domain, necessary for interaction of NOS1AP with nNOS (8), does not prevent the inhibitory effect of NOS1AP on radial migration of cortical neurons (Fig. 4B, C). We also studied the effects of a mutant containing only the central region of NOS1AP (amino acids 181–307). We previously demonstrated that this region is sufficient to mediate the effects of NOS1AP on dendrite branching through an interaction with carboxypeptidase E (13). Interestingly, overexpression of this region does not cause any migration defect of cortical neurons (Fig. 4D, E), suggesting that NOS1AP-mediated regulation of dendritogenesis occurs through a mechanism that differs from that which regulates cortical neuron migration. In contrast, deletion of the N-terminal PTB domain of NOS1AP, which binds synapsin, DexRas1, and Scribble, abolishes the inhibitory effect of NOS1AP on neuronal migration while overexpression of the PTB domain alone is sufficient to reproduce this effect (Fig. 4F, G). Taken together, these data indicate that the role of NOS1AP on radial migration does not involve its interactions with nNOS or with CPE but that it does require its N-terminal PTB domain. Furthermore, the data suggest that the effects of NOS1AP on migration and dendritic patterning do not involve the same mechanisms.
Discussion
One of the prevailing theories is that schizophrenia is a neurodevelopmental disorder (15–18). One patient with schizophrenia was reported to have ectopic gray matter (19) and additional patients ectopic neuron placement (20–22); however, whether heterotopic neurons are indicative of schizophrenia has been debated (23). Furthermore, aberrant circuitry has been thought to be to be the mechanism by which these abnormalities produce behavioral symptoms. As such, we have chosen to study NOS1AP, a protein that we have shown to be overexpressed in the postmortem dorsolateral prefrontal cortex from subjects with schizophrenia, and in this study we ask how this aberrant expression of NOS1AP affects neurodevelopment. Our results suggest that NOS1AP levels regulate cortical neuron migration, which results in aberrant neuronal connectivity, which may contribute to schizophrenia. Alterations in migration are long-lasting. We also found that the phosphotyrosine binding domain of NOS1AP is responsible for incorrect cortical neuron placement, and this domain binds other proteins thought to be involved in schizophrenia, to be detailed further in the discussion below. Our new results outlined here support the idea that the protein product encoded by a schizophrenia susceptibility gene may shape the developing brain in such a way that it is influenced by other genes or environment for the manifestation of schizophrenia.
NOS1AP as a susceptibility gene for schizophrenia
NOS1AP, encoding a regulator of the NMDA receptor pathway and nNOS activity(8), has been identified as a schizophrenia susceptibility gene (4). Several independent studies reported linkage of schizophrenia to chromosome 1q21–22, containing NOS1AP (3, 24–26). Six SNPs within NOS1AP were identified in a group of 24 medium-sized Canadian families to have significant linkage disequilibrium to schizophrenia (3). In addition, an association study found one SNP within NOS1AP and haplotypes constructed from three SNPs within NOS1AP to be associated with schizophrenia in the Chinese Han population (5). More recently, eight SNPs within NOS1AP were found to be associated with schizophrenia in a South American population isolate, two of which had been previously identified, further strengthening a link between NOS1AP and schizophrenia (6). None of the SNPs that associate with schizophrenia alter the amino acid sequence, suggesting that they may play a role in altering gene expression levels,, a hypothesis that is supported by in vitro assessment of the effects of associated SNPs on gene expression (7). While some studies failed to detect the association of NOS1AP with schizophrenia (27–28), this could be due to differences in methodology, genetic differences across populations, and the etiologic heterogeneity of schizophrenia (29).
NOS1AP was identified as a neuronal nitric oxide synthase (NOS1) binding partner (8). It competes with PSD-95 for NOS1 binding and reduces NMDA receptor signaling (8) (Fig. 5). It is also hypothesized that NMDA receptor signaling is decreased in patients with schizophrenia, known as the NMDA receptor hypofunction hypothesis (recently reviewed in (30)). Overexpression of NOS1AP, as is seen in brain tissue from patients with schizophrenia (4, 9), is consistent with this theory. Interestingly, the NMDA receptor has been implicated in the regulation of cortical (31–32) and hippocampal(33) neuron migration via a pathway that includes Disrupted-in-Schizophrenia1 (DISC1)(33) and Reelin(34). The Cajal-Retzius cells produce glutamate during the final week of gestation in the rodent (35). The glutamate released from these cells (36) then acts as a modulatory signal for other migrating neurons, stimulating NMDA (37–41) and AMPA receptors (32, 39). Later in development, the NMDA receptor and its signaling pathways regulate dendrite morphology and synaptogenesis, as is seen in both in vivo and in vitro systems of cortex and hippocampus (42–49). The fact that overexpression of NOS1AP results in a phenotype similar to that of inhibition of the NMDA receptor during development supports a potential role for NOS1AP in the NMDA receptor hypofunction hypothesis in schizophrenia.
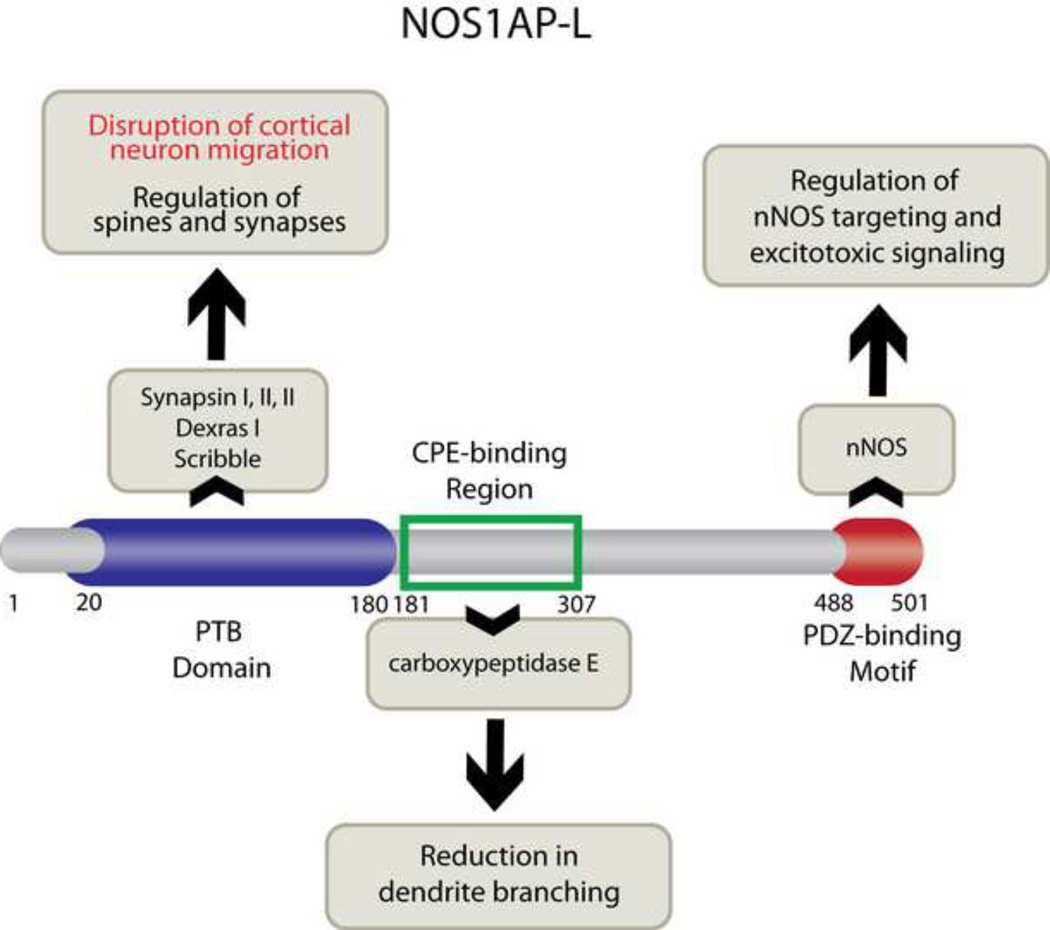
Figure 5. Model of NOS1AP function in neurons.
The long isoform of NOS1AP, NOS1AP-L, contains three domains: the N-terminal phosphotyrosine binding domain (PTB; amino acids 1–180), the carboxypeptidase E (CPE)-binding region (amino acids 181–307), and the C-terminal PDZ-binding motif (amino acids 488–501). The PTB domain is responsible for regulating cortical neuron migration (data presented in this manuscript) and binds to synapsins I, II, III (11), Dexras, (10) and Scribble (12). The CPE-binding region is responsible for NOS1AP effects on dendrite branching (13). The PDZ-binding motif binds to nNOS (also known as NOS1), competes with PSD-95 for nNOS binding, and reduces NMDA receptor-mediated neurotoxicity (8, 75).
Alterations in neuronal connectivity are seen in individuals with schizophrenia
How do our data fit in with what is known about individuals with schizophrenia? Disruptions in normal establishment and maintenance of dendrite morphology, including dendrite outgrowth, branching, and spine formation, result in functional deficits. Not surprisingly, patients with schizophrenia exhibit dendritic abnormalities in the prefrontal cortex (PFC), the area responsible for higher cortical functions. Patients with schizophrenia show a 40% decrease in basilar dendritic field size and complexity of PFC layer V pyramidal neurons (50), shorter basilar dendritic length (51–52). Individuals with schizophrenia also show altered distribution of nNOS-expressing interneurons in deeper cortical layers and underlying white matter in prefrontal and temporal cortices (53), and others show misplaced and clustered neurons in the entorhinal cortex (54). Furthermore, interstitial white matter neurons are increased in the DLPFC of patients with schizophrenia, supporting the idea that deficient migration of neurons may have occurred in these patients (55). Thus, combined reductions in dendrite length, field size, complexity, and spine number and maturity mediated by aberrant NOS1AP expression may contribute to reduced cognitive function seen in those with schizophrenia.
Importance of PTB domain in NOS1AP-mediated effects on cortical neuron migration
NOS1AP-L includes an amino-terminal PTB and a carboxy-terminal NOS1 PDZ-binding motif while NOS1AP-S contains only the carboxy-terminal NOS1 PDZ-binding motif (8) (Fig. 5). Recently, our group also identified an internal carboxypeptidase E binding region, which is only found in NOS1AP-L (13) (Fig. 5). Here, we found that the PTB domain of NOS1AP-L mediates its effects on cortical neuron migration. What is the importance of this domain, especially in regards to schizophrenia? The PTB domain of NOS1AP-L binds to the small GTPase DexRas1 and synapsins (10–11). Interestingly, the absence of synapsin isoforms in rodents has been implicated in behavior associated with schizophrenia (56–58). Furthermore, antipsychotic drugs upregulate synapsin expression (59–61), synapsin expression is decreased in postmortem brain tissue from subjects with schizophrenia(62–63), and alleles in synapsin genes have been associated with schizophrenia (64–70).
The PTB domain of NOS1AP binds to the tumor suppressor Scribble. This interaction promotes the presence of NOS1AP in a beta-Pix [beta-p21-activated kinase (PAK)-interacting exchange factor]/Git1 (G-protein-coupled receptor kinase-interacting protein)/PAK complex, thereby in turn, influencing Rac activity (12). Rac is thought to be involved in axonal guidance directed by a protein encoded by the schizophrenia susceptibility gene DISC1(71). Furthermore, mice lacking the SRGAP3 gene, in which mutations are associated with intellectual aberration and encoding a protein that regulates Rac1, show schizophrenia-related behavior (72). Furthermore, both synapsins (reviewed in (73)) and Rac1 (74) have been shown to regulate asymmetric cell division from neuronal precursors and neuronal migration. Thus, our new data elucidate an additional signaling molecule, NOS1AP, that may act in concert with synapsin and Rac1 to regulate cortical circuitry in the brain. In specific, this pathway, which includes NOS1AP and associated proteins, may contribute to the etiology of schizophrenia when aberrantly expressed. Taken together with previously published data by us (13) and others (12), NOS1AP regulates multiple aspects of neuronal development – migration, dendritogenesis, spinogenesis - all of which have been reported to be compromised in patients with schizophrenia. A model of these multiple functions for NOS1AP is shown in Figure 5.
ACKNOWLDEGEMENTS
We thank Drs. Gabriella D’Arcangelo and Joseph LoTurco for training us in in utero electroporation techniques. This work was supported by NIMH grant R01 MH062440 (to L.M.B.), a NARSAD 2012 Marion G. Nicholson Distinguished Investigator Award (to B.L.F.), and in part, by NSF grant IBN- 0919747 (to B.L.F). K.H. was supported in part by NIH IMSD Grant 2R25 GM55145, NIH Biotechnology Training Grant T32 GM008339-20, and NSF DGE 0801620. We thank Clément Léna and Rémi Proville (Institut de Biologie de l’Ecole Normale Supérieure, Paris) for insightful discussions and for their technical support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST STATEMENT: Dr. Brzustowicz serves as a consultant for the Janssen Pharmaceutical Companies of Johnson & Johnson. Drs. Firestein and Brzustowicz reported patent US 12/263,939 titled “Methods and compositions for the diagnosis and treatment of schizophrenia.” All other authors reported no biomedical financial interests or potential conflicts of interest.
References
- 1.Brown AS, Susser ES. Prenatal Nutritional Deficiency and Risk of Adult Schizophrenia. Schizophr Bull. 2008;34:1054–1063. doi: 10.1093/schbul/sbn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rehn AE, Rees SM. Investigating the neurodevelopmental hypothesis of schizophrenia. Clin Exp Pharmacol Physiol. 2005;32:687–696. doi: 10.1111/j.1440-1681.2005.04257.x. [DOI] [PubMed] [Google Scholar]
- 3.Brzustowicz LM, Simone J, Mohseni P, Hayter JE, Hodgkinson KA, Chow EW, et al. Linkage disequilibrium mapping of schizophrenia susceptibility to the CAPON region of chromosome 1q22. Am J Hum Genet. 2004;74:1057–1063. doi: 10.1086/420774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu B, Wratten N, Charych EI, Buyske S, Firestein BL, Brzustowicz LM. Increased expression in dorsolateral prefrontal cortex of CAPON in schizophrenia and bipolar disorder. PLoS Med. 2005;2:e263. doi: 10.1371/journal.pmed.0020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng Y, Li H, Qin W, Chen W, Duan Y, Xiao Y, et al. Association of the carboxyl-terminal PDZ ligand of neuronal nitric oxide synthase gene with schizophrenia in the Chinese Han population. Biochem Biophys Res Commun. 2005;328:809–815. doi: 10.1016/j.bbrc.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 6.Kremeyer B, García J, Kymäläinen H, Wratten N, Restrepo G, Palacio C, et al. Evidence for a Role of the NOS1AP (CAPON) Gene in Schizophrenia and its Clinical Dimensions: an Association Study in a South American Population Isolate. Human Heredity. 2008;67:163–173. doi: 10.1159/000181154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wratten PD, Naomi, Memoli BS, Holly, Huang MS, Yungui, Dulencin BA, Anna, Matteson PD, Paul, Cornacchia M, et al. Identification of a Schizophrenia-Associated Functional Noncoding Variant in NOS1AP. American Journal of Psychiatry. 2009;166:434–441. doi: 10.1176/appi.ajp.2008.08081266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaffrey SR, Snowman AM, Eliasson MJ, Cohen NA, Snyder SH. CAPON: a protein associated with neuronal nitric oxide synthase that regulates its interactions with PSD95. Neuron. 1998;20:115–124. doi: 10.1016/s0896-6273(00)80439-0. [DOI] [PubMed] [Google Scholar]
- 9.Hadzimichalis NM, Previtera ML, Moreau MP, Li B, Lee GH, Dulencin AM, et al. NOS1AP protein levels are altered in BA46 and cerebellum of patients with schizophrenia. Schizophrenia Research. 2010;124:248–250. doi: 10.1016/j.schres.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang M, Jaffrey SR, Sawa A, Ye K, Luo X, Snyder SH. Dexras1: a G protein specifically coupled to neuronal nitric oxide synthase via CAPON. Neuron. 2000;28:183–193. doi: 10.1016/s0896-6273(00)00095-7. [DOI] [PubMed] [Google Scholar]
- 11.Jaffrey SR, Benfenati F, Snowman AM, Czernik AJ, Snyder SH. Neuronal nitricoxide synthase localization mediated by a ternary complex with synapsin and CAPON. Proc Natl Acad Sci U S A. 2002;99:3199–3204. doi: 10.1073/pnas.261705799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richier L, Williton K, Clattenburg L, Colwill K, O'Brien M, Tsang C, et al. NOS1AP Associates with Scribble and Regulates Dendritic Spine Development. Journal of Neuroscience. 2010;30:4796–4805. doi: 10.1523/JNEUROSCI.3726-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carrel D, Du Y, Komlos D, Hadzimichalis NM, Kwon M, Wang B, et al. NOS1AP regulates dendrite patterning of hippocampal neurons through a carboxypeptidase E-mediated pathway. Journal of Neuroscience. 2009;29:8248–8258. doi: 10.1523/JNEUROSCI.5287-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- 15.Murray RM, Jones P, O'Callaghan E. Fetal brain development and later schizophrenia. Ciba Found Symp. 1991;156:155–163. doi: 10.1002/9780470514047.ch10. discussion 163–170. [DOI] [PubMed] [Google Scholar]
- 16.Benes FM. Evidence for neurodevelopment disturbances in anterior cingulate cortex of post-mortem schizophrenic brain. Schizophr Res. 1991;5:187–188. doi: 10.1016/0920-9964(91)90063-w. [DOI] [PubMed] [Google Scholar]
- 17.Bunney BG, Potkin SG, Bunney WE., Jr New morphological and neuropathological findings in schizophrenia: a neurodevelopmental perspective. Clin Neurosci. 1995;3:81–88. [PubMed] [Google Scholar]
- 18.Brent BK, Seidman LJ, Thermenos HW, Holt DJ, Keshavan MS. Self-disturbances as a possible premorbid indicator of schizophrenia risk: a neurodevelopmental perspective. Schizophr Res. 2014;152:73–80. doi: 10.1016/j.schres.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nopoulos P, Swayze V, Flaum M, Andreasen NC. Incidence of ectopic gray matter in patients with schizophrenia and healthy control subjects studied with MRI. J Neuropsychiatry Clin Neurosci. 1998;10:351–353. doi: 10.1176/jnp.10.3.351. [DOI] [PubMed] [Google Scholar]
- 20.Kiehl TR, Chow EW, Mikulis DJ, George SR, Bassett AS. Neuropathologic features in adults with 22q11.2 deletion syndrome. Cereb Cortex. 2009;19:153–164. doi: 10.1093/cercor/bhn066. [DOI] [PubMed] [Google Scholar]
- 21.Akanuma N, Saitoh O, Yoshikawa T, Matsuda H, Ishikura N, Kato M, et al. Interictal schizophrenia-like psychosis in a patient with double cortex syndrome. J Neuropsychiatry Clin Neurosci. 2002;14:210–213. doi: 10.1176/jnp.14.2.210. [DOI] [PubMed] [Google Scholar]
- 22.Pilowsky LS, Kerwin RW, Murray RM. Schizophrenia: a neurodevelopmental perspective. Neuropsychopharmacology. 1993;9:83–91. doi: 10.1038/npp.1993.46. [DOI] [PubMed] [Google Scholar]
- 23.Bernstein HG, Krell D, Baumann B, Danos P, Falkai P, Diekmann S, et al. Morphometric studies of the entorhinal cortex in neuropsychiatric patients and controls: clusters of heterotopically displaced lamina II neurons are not indicative of schizophrenia. Schizophr Res. 1998;33:125–132. doi: 10.1016/s0920-9964(98)00071-1. [DOI] [PubMed] [Google Scholar]
- 24.Hwu HG, Liu CM, Fann CS, Ou-Yang WC, Lee SF. Linkage of schizophrenia with chromosome 1q loci in Taiwanese families. Mol Psychiatry. 2003;8:445–452. doi: 10.1038/sj.mp.4001235. [DOI] [PubMed] [Google Scholar]
- 25.Brzustowicz LM, Hayter JE, Hodgkinson KA, Chow EW, Bassett AS. Fine mapping of the schizophrenia susceptibility locus on chromosome 1q22. Hum Hered. 2002;54:199–209. doi: 10.1159/000070665. [DOI] [PubMed] [Google Scholar]
- 26.Rosa A, Fananas L, Cuesta MJ, Peralta V, Sham P. 1q21-q22 locus is associated with susceptibility to the reality-distortion syndrome of schizophrenia spectrum disorders. Am J Med Genet. 2002;114:516–518. doi: 10.1002/ajmg.10526. [DOI] [PubMed] [Google Scholar]
- 27.Puri V, McQuillin A, Thirumalai S, Lawrence J, Krasucki R, Choudhury K, et al. Failure to confirm allelic association between markers at the CAPON gene locus and schizophrenia in a British sample. Biol Psychiatry. 2006;59:195–197. doi: 10.1016/j.biopsych.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 28.Fang C, Tang W, Tang RQ, Wang L, Zhou GQ, Huang K, et al. Family-based association studies of CAPON and schizophrenia in the Chinese Han population. Prog Neuropsychopharmacol Biol Psychiatry. 2008;32:1210–1213. doi: 10.1016/j.pnpbp.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 29.Brzustowicz LM. NOS1AP in schizophrenia. Curr Psychiatry Rep. 2008;10:158–163. doi: 10.1007/s11920-008-0027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laruelle M. Schizophrenia: from dopaminergic to glutamatergic interventions. Curr Opin Pharmacol. 2014;14C:97–102. doi: 10.1016/j.coph.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 31.Uchino S, Hirasawa T, Tabata H, Gonda Y, Waga C, Ondo Y, et al. Inhibition of N-methyl-D-aspartate receptor activity resulted in aberrant neuronal migration caused by delayed morphological development in the mouse neocortex. Neuroscience. 2010;169:609–618. doi: 10.1016/j.neuroscience.2010.05.024. [DOI] [PubMed] [Google Scholar]
- 32.Komuro H, Rakic P. Modulation of neuronal migration by NMDA receptors. Science. 1993;260:95–97. doi: 10.1126/science.8096653. [DOI] [PubMed] [Google Scholar]
- 33.Namba T, Ming GL, Song H, Waga C, Enomoto A, Kaibuchi K, et al. NMDA receptor regulates migration of newly generated neurons in the adult hippocampus via Disrupted-In-Schizophrenia 1 (DISC1) J Neurochem. 2011;118:34–44. doi: 10.1111/j.1471-4159.2011.07282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y, Beffert U, Ertunc M, Tang TS, Kavalali ET, Bezprozvanny I, et al. Reelin modulates NMDA receptor activity in cortical neurons. J Neurosci. 2005;25:8209–8216. doi: 10.1523/JNEUROSCI.1951-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.del Rio JA, Martinez A, Fonseca M, Auladell C, Soriano E. Glutamate-like immunoreactivity and fate of Cajal-Retzius cells in the murine cortex as identified with calretinin antibody. Cereb Cortex. 1995;5:13–21. doi: 10.1093/cercor/5.1.13. [DOI] [PubMed] [Google Scholar]
- 36.Manent JB, Demarque M, Jorquera I, Pellegrino C, Ben-Ari Y, Aniksztejn L, et al. A noncanonical release of GABA and glutamate modulates neuronal migration. J Neurosci. 2005;25:4755–4765. doi: 10.1523/JNEUROSCI.0553-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Behar TN, Scott CA, Greene CL, Wen X, Smith SV, Maric D, et al. Glutamate acting at NMDA receptors stimulates embryonic cortical neuronal migration. J Neurosci. 1999;19:4449–4461. doi: 10.1523/JNEUROSCI.19-11-04449.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirai K, Yoshioka H, Kihara M, Hasegawa K, Sakamoto T, Sawada T, et al. Inhibiting neuronal migration by blocking NMDA receptors in the embryonic rat cerebral cortex: a tissue culture study. Brain research. 1999;114:63–67. doi: 10.1016/s0165-3806(99)00019-x. [DOI] [PubMed] [Google Scholar]
- 39.Simonian SX, Herbison AE. Regulation of gonadotropin-releasing hormone (GnRH) gene expression during GnRH neuron migration in the mouse. Neuroendocrinology. 2001;73:149–156. doi: 10.1159/000054631. [DOI] [PubMed] [Google Scholar]
- 40.Kihara M, Yoshioka H, Hirai K, Hasegawa K, Kizaki Z, Sawada T. Stimulation of N-methyl-D-aspartate (NMDA) receptors inhibits neuronal migration in embryonic cerebral cortex: a tissue culture study. Brain research. 2002;138:195–198. doi: 10.1016/s0165-3806(02)00490-x. [DOI] [PubMed] [Google Scholar]
- 41.Reiprich P, Kilb W, Luhmann HJ. Neonatal NMDA receptor blockade disturbs neuronal migration in rat somatosensory cortex in vivo. Cereb Cortex. 2005;15:349–358. doi: 10.1093/cercor/bhh137. [DOI] [PubMed] [Google Scholar]
- 42.Lee LJ, Lo FS, Erzurumlu RS. NMDA receptor-dependent regulation of axonal and dendritic branching. J Neurosci. 2005;25:2304–2311. doi: 10.1523/JNEUROSCI.4902-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Datwani A, Iwasato T, Itohara S, Erzurumlu RS. NMDA receptor-dependent pattern transfer from afferents to postsynaptic cells and dendritic differentiation in the barrel cortex. Molecular and cellular neurosciences. 2002;21:477–492. doi: 10.1006/mcne.2002.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iwasato T, Datwani A, Wolf AM, Nishiyama H, Taguchi Y, Tonegawa S, et al. Cortex-restricted disruption of NMDAR1 impairs neuronal patterns in the barrel cortex. Nature. 2000;406:726–731. doi: 10.1038/35021059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Charych EI, Akum BF, Goldberg JS, Jornsten RJ, Rongo C, Zheng JQ, et al. Activity-independent regulation of dendrite patterning by postsynaptic density protein PSD-95. J Neurosci. 2006;26:10164–10176. doi: 10.1523/JNEUROSCI.2379-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma XM, Wang Y, Ferraro F, Mains RE, Eipper BA. Kalirin-7 is an essential component of both shaft and spine excitatory synapses in hippocampal interneurons. J Neurosci. 2008;28:711–724. doi: 10.1523/JNEUROSCI.5283-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Proepper C, Johannsen S, Liebau S, Dahl J, Vaida B, Bockmann J, et al. Abelson interacting protein 1 (Abi-1) is essential for dendrite morphogenesis and synapse formation. The EMBO journal. 2007;26:1397–1409. doi: 10.1038/sj.emboj.7601569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cuppini R, Sartini S, Ambrogini P, Falcieri E, Maltarello MC, Gallo G. Control of neuron outgrowth by NMDA receptors. Journal of submicroscopic cytology and pathology. 1999;31:31–40. [PubMed] [Google Scholar]
- 49.Baker RE, Wolters P, van Pelt J. Chronic blockade of glutamate-mediated bioelectric activity in long-term organotypic neocortical explants differentially effects pyramidal/non-pyramidal dendritic morphology. Brain research. 1997;104:31–39. doi: 10.1016/s0165-3806(97)00133-8. [DOI] [PubMed] [Google Scholar]
- 50.Black JE, Kodish IM, Grossman AW, Klintsova AY, Orlovskaya D, Vostrikov V, et al. Pathology of layer V pyramidal neurons in the prefrontal cortex of patients with schizophrenia. The American journal of psychiatry. 2004;161:742–744. doi: 10.1176/appi.ajp.161.4.742. [DOI] [PubMed] [Google Scholar]
- 51.Glantz LA, Lewis DA. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Archives of general psychiatry. 2000;57:65–73. doi: 10.1001/archpsyc.57.1.65. [DOI] [PubMed] [Google Scholar]
- 52.Kalus P, Muller TJ, Zuschratter W, Senitz D. The dendritic architecture of prefrontal pyramidal neurons in schizophrenic patients. Neuroreport. 2000;11:3621–3625. doi: 10.1097/00001756-200011090-00044. [DOI] [PubMed] [Google Scholar]
- 53.Akbarian S, Bunney WE, Jr, Potkin SG, Wigal SB, Hagman JO, Sandman CA, et al. Altered distribution of nicotinamide-adenine dinucleotide phosphate-diaphorase cells in frontal lobe of schizophrenics implies disturbances of cortical development. Archives of General Psychiatry. 1993;50:169–177. doi: 10.1001/archpsyc.1993.01820150007001. [DOI] [PubMed] [Google Scholar]
- 54.Falkai P, Schneider-Axmann T, Honer WG. Entorhinal cortex pre-alpha cell clusters in schizophrenia: quantitative evidence of a developmental abnormality. Biological Psychiatry. 2000;47:937–943. doi: 10.1016/s0006-3223(99)00250-4. [DOI] [PubMed] [Google Scholar]
- 55.Yang Y, Fung SJ, Rothwell A, Tianmei S, Weickert CS. Increased interstitial white matter neuron density in the dorsolateral prefrontal cortex of people with schizophrenia. Biol Psychiatry. 2011;69:63–70. doi: 10.1016/j.biopsych.2010.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dyck BA, Skoblenick KJ, Castellano JM, Ki K, Thomas N, Mishra RK. Synapsin II knockout mice show sensorimotor gating and behavioural abnormalities similar to those in the phencyclidine-induced preclinical animal model of schizophrenia. Schizophr Res. 2007;97:292–293. doi: 10.1016/j.schres.2007.08.026. [DOI] [PubMed] [Google Scholar]
- 57.Dyck BA, Skoblenick KJ, Castellano JM, Ki K, Thomas N, Mishra RK. Behavioral abnormalities in synapsin II knockout mice implicate a causal factor in schizophrenia. Synapse. 2009;63:662–672. doi: 10.1002/syn.20643. [DOI] [PubMed] [Google Scholar]
- 58.Porton B, Rodriguiz RM, Phillips LE, Gilbert JWt, Feng J, Greengard P, et al. Mice lacking synapsin III show abnormalities in explicit memory and conditioned fear. Genes Brain Behav. 2010;9:257–268. doi: 10.1111/j.1601-183X.2009.00555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guest KA, Dyck BA, Shethwala S, Mishra RK. Atypical antipsychotic drugs upregulate synapsin II in the prefrontal cortex of post-mortem samples obtained from patients with schizophrenia. Schizophr Res. 2010;120:229–231. doi: 10.1016/j.schres.2010.03.029. [DOI] [PubMed] [Google Scholar]
- 60.Tan ML, Dyck BA, Gabriele J, Daya RP, Thomas N, Sookram C, et al. Synapsin II gene expression in the dorsolateral prefrontal cortex of brain specimens from patients with schizophrenia and bipolar disorder: effect of lifetime intake of antipsychotic drugs. Pharmacogenomics J. 2014;14:63–69. doi: 10.1038/tpj.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pickering C, Ericson M, Soderpalm B. Chronic phencyclidine increases synapsin-1 and synaptic adaptation proteins in the medial prefrontal cortex. ISRN Psychiatry. 2013;2013:620361. doi: 10.1155/2013/620361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Porton B, Wetsel WC. Reduction of synapsin III in the prefrontal cortex of individuals with schizophrenia. Schizophr Res. 2007;94:366–370. doi: 10.1016/j.schres.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 63.Vawter MP, Thatcher L, Usen N, Hyde TM, Kleinman JE, Freed WJ. Reduction of synapsin in the hippocampus of patients with bipolar disorder and schizophrenia. Mol Psychiatry. 2002;7:571–578. doi: 10.1038/sj.mp.4001158. [DOI] [PubMed] [Google Scholar]
- 64.Yu GI, Kim SK, Park HJ, Kim JW, Chung JH, Shin DH. The C allele of synonymous SNP (rs1142636, Asn170Asn) in SYN1 is a risk factor for the susceptibility of Korean female schizophrenia. Synapse. 2012;66:979–983. doi: 10.1002/syn.21583. [DOI] [PubMed] [Google Scholar]
- 65.Porton B, Ferreira A, DeLisi LE, Kao HT. A rare polymorphism affects a mitogen-activated protein kinase site in synapsin III: possible relationship to schizophrenia. Biol Psychiatry. 2004;55:118–125. doi: 10.1016/j.biopsych.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 66.Chen Q, He G, Wang XY, Chen QY, Liu XM, Gu ZZ, et al. Positive association between synapsin II and schizophrenia. Biol Psychiatry. 2004;56:177–181. doi: 10.1016/j.biopsych.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 67.Chen Q, He G, Qin W, Chen QY, Zhao XZ, Duan SW, et al. Family-based association study of synapsin II and schizophrenia. Am J Hum Genet. 2004;75:873–877. doi: 10.1086/425588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lachman HM, Stopkova P, Rafael MA, Saito T. Association of schizophrenia in African Americans to polymorphism in synapsin III gene. Psychiatr Genet. 2005;15:127–132. doi: 10.1097/00041444-200506000-00009. [DOI] [PubMed] [Google Scholar]
- 69.Lee HJ, Song JY, Kim JW, Jin SY, Hong MS, Park JK, et al. Association study of polymorphisms in synaptic vesicle-associated genes, SYN2 and CPLX2, with schizophrenia. Behav Brain Funct. 2005;1:15. doi: 10.1186/1744-9081-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saviouk V, Moreau MP, Tereshchenko IV, Brzustowicz LM. Association of synapsin 2 with schizophrenia in families of Northern European ancestry. Schizophr Res. 2007;96:100–111. doi: 10.1016/j.schres.2007.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen SY, Huang PH, Cheng HJ. Disrupted-in-Schizophrenia 1-mediated axon guidance involves TRIO-RAC-PAK small GTPase pathway signaling. Proc Natl Acad Sci U S A. 2011;108:5861–5866. doi: 10.1073/pnas.1018128108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Waltereit R, Leimer U, von Bohlen Und Halbach O, Panke J, Holter SM, Garrett L, et al. Srgap3(−)/(−) mice present a neurodevelopmental disorder with schizophrenia-related intermediate phenotypes. FASEB J. 2012;26:4418–4428. doi: 10.1096/fj.11-202317. [DOI] [PubMed] [Google Scholar]
- 73.Valtorta F, Pozzi D, Benfenati F, Fornasiero EF. The synapsins: multitask modulators of neuronal development. Semin Cell Dev Biol. 2011;22:378–386. doi: 10.1016/j.semcdb.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 74.Kawauchi T, Chihama K, Nabeshima Y, Hoshino M. The in vivo roles of STEF/Tiam1, Rac1 and JNK in cortical neuronal migration. EMBO J. 2003;22:4190–4201. doi: 10.1093/emboj/cdg413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li LL, Ginet V, Liu X, Vergun O, Tuittila M, Mathieu M, et al. The nNOSp38MAPK pathway is mediated by NOS1AP during neuronal death. J Neurosci. 2013;33:8185–8201. doi: 10.1523/JNEUROSCI.4578-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]