Abstract
Calorie restriction (CR), a non-genetic intervention that promotes longevity in animals, may exert anti-aging effects by modulating mitochondrial function. Based on our prior mitochondrial proteome analysis, we focused on the potential roles of cytochrome c oxidase (Cox or Complex IV) subunit 6b1 on formation of mitochondrial supercomplexes comprised of Complex I, III, and IV. Blue native polyacrylamide gel electrophoresis followed by immunoblotting showed that the amount of Cox6b1 and the proportion of high molecular weight supercomplexes (SCs) comprised of Complexes I, III, and IV were increased in the liver of mice subjected to 30 % CR, compared with the liver of mice fed ad libitum. In in vitro experiments, in Cox6b1-overexpressing NIH3T3 (Cox6b1-3T3) cells, Cox6b1 was increased in the SC, III2IV1, and III2IV2 complexes and Cox was concomitantly recruited abundantly into the SC, compared with control (Con)-3T3 cells. The proportions of III2IV1, and III2IV2, relative to IV monomer were also increased in Cox6b1-3T3 cells. Cox6b1-3T3 cells showed increased oxygen consumption rates, Cox activity, and intracellular ATP concentrations, indicating enhanced mitochondrial respiration, compared with Con-3T3 cells. Despite the increased basal level of mitochondrial reactive oxygen species (ROS), cell viability after inducing oxidative stress was greater in Cox6b1-3T3 cells than in Con-3T3 cells, probably because of prompt activation of protective mechanisms, such as nuclear translocation of nuclear factor E2-related factor-2. These in vivo and in vitro studies show that Cox6b1 is involved in regulation of mitochondrial function by promoting the formation of SC, suggesting that Cox6b1 contributes to the anti-aging effects of CR.
Electronic supplementary material
The online version of this article (doi:10.1007/s11357-015-9787-8) contains supplementary material, which is available to authorized users.
Keywords: Calorie restriction, Cytochrome c oxidase, Cox6b1, Supercomplex
Introduction
Modest restriction of caloric intake with adequate essential nutrients, referred to as calorie restriction (CR), increases the life span of a range of organisms, including the replicative life span of yeasts (Mair et al. 2008). The effects of CR have also been tested in non-human primates, and the results to date suggest that CR extends the disease-free life span in monkeys but may not increase overall survival (Colman et al. 2009; Mattison et al. 2012). This implies that we could apply the CR paradigm to prolong a healthy life span in humans by preventing age-related disorders. Although our knowledge of the mechanisms underlying the effects of CR is increasing, our understanding of the process remains incomplete, particularly in mammals.
Mitochondria are dynamic organelles that play critical roles in energy production, generation of reactive oxygen species (ROS), apoptosis, and intracellular signaling (Finley et al. 2009). CR may affect mitochondrial bioenergetics by modulating the composition of the mitochondrial respiratory chains. CR is thought to increase mitochondrial biogenesis in rat hepatocytes and human skeletal muscle (Lopez-Lluch et al. 2006; Civitarese et al. 2007). However, Hancock et al. (2011) reported that 30 % CR for 3 months did not significantly affect the protein expression of mitochondrial respiratory complexes in multiple tissues in rats. Our mitochondrial proteome analysis of the rat liver demonstrated that CR altered the expression levels of multiple subunits of respiratory chain complexes I, IV, and V (Dani et al. 2010). Complex IV (cytochrome c oxidase, Cox) is thought to be a rate-limiting component of respiratory chain activity in vivo (Kadenbach 2003). CR decreased the expression of two subunits of Cox, namely, Cox2 and Cox5a, while modestly increasing the expression of one subunit, Cox6b1 (Dani et al. 2010). In the present study, we focused on Cox6b1, a non-transmembrane subunit of Cox that faces the mitochondrial intermembrane space and seems to stabilize Cox dimerization along with Cox6a (Tsukihara et al. 1996; Yoshikawa et al. 1998). In mammalian mitochondria, up to four copies of Cox are assembled into supercomplexes consisting of complexes I and III (Schägger and Pfeiffer 2000). It is thought that the formation of supercomplexes is associated with structural stabilization, increased electron transport, and sequestration of reactive intermediates, and prevents excess ROS generation (Genova et al. 2014). Although an earlier study suggested that removing Cox6b1 from the purified bovine complex substantially increased Cox activity (Weishaupt et al. 1992), a more recent study showed that mutations of the Cox6b1 gene cause severe infantile encephalopathy because of inadequate Cox activity (Massa et al. 2008). Therefore, Cox6b1 may play a role in CR-specific regulation of mitochondrial bioenergetics via the assembly of respiratory supercomplexes rather than through altered biogenesis.
In the present study, we assessed the formation of mitochondrial supercomplexes along with elevation of Cox6b1 in CR mouse liver. We also examined whether upregulation of Cox6b1 in cells in vitro modulates mitochondrial supercomplexes and then assessed effects of Cox6b1 on mitochondrial functions and stress resistance, because stress resistance is an important feature of CR (Masoro 2003).
Materials and methods
The animal care and experimental protocols of the present study were approved by the Ethics Review Committee for Animal Experimentation at Nagasaki University.
Materials
The following monoclonal or polyclonal antibodies (mAbs or pAbs) were used for immunoblotting: Total OXPHOS Rodent WB Antibody Cocktail (ab110413) containing five anti-mouse mAbs against 20-kDa subunit of complex I (NDUFB8), complex II 30-kDa subunit (SDHB), complex III Core protein 2 (Core2), complex IV subunit 1 (Cox1) and complex V alpha subunit (ATP5A), mouse anti-Cox6b1 mAb (ab110266), rabbit anti-Cox6b1 mAb (ab137089), mouse anti-Cox4 mAb (ab33985), rabbit anti-human nuclear factor E2-related factor-2 (Nrf2) mAb, mouse anti-human NAD(P)H:quinone oxidoreductase 1 (NQO1) mAb (ab28947), rabbit anti-VDAC/Porin pAb (ab34726), rabbit anti-mouse lamin B1 pAb (ab16048), rabbit anti-GAPDH conjugated with HRP pAb (ab9385), and rabbit anti-β-actin pAb (ab13822, Abcam, Cambridge, UK). Goat anti-human heme oxygenase 1 (HO-1) pAb (sc-1796) was purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Horseradish-peroxidase-conjugated anti-rabbit or anti-mouse IgG mAbs (GE Healthcare Lifescience, Piscataway, NJ, USA) and donkey anti-goat IgG mAb (sc-2033, Santa Cruz biotechnology) were used to visualize the proteins in the immunoblots. G418 antibiotics were purchased from Calbiochem (Merck, Darmstadt, Germany).
Mouse tissues
Liver tissues were obtained from 12-month-old male C57BL/6 J mice either fed ad libitum (AL) throughout the experiments or fed 30 % CR diets from 12 weeks of age, and were stored at −80 °C until use. The tissues were used to determine the mitochondrial protein and messenger RNA (mRNA) expression levels in vivo. The mitochondrial fractions of fresh liver tissues of 12-month-old mice were also used in blue native polyacrylamide gel electrophoresis (BN-PAGE) and two-dimensional sodium dodecyl polyacrylamide gel electrophoresis (2D SDS-PAGE). Animal care and the 30 % CR feeding regimen were carried out as described in our previous report (Yamaza et al. 2010).
Quantitative real-time RT-PCR analysis
Total RNA was extracted from murine tissues (40 mg) and cultured cells using Qiazol (Qiagen, Leusden, Netherlands). Any genomic DNA contaminating the sample was digested with DNase I (Qiagen). Further purification was performed using the RNeasy Cleanup Kit (Qiagen). One microgram of total RNA was reverse-transcribed using the High Capacity RNA-to-cDNA Kit for RT-PCR (Applied Biosystems, Foster City, CA, USA). Quantitative PCR was performed using Thunderbird SYBR qPCR Mix (Toyobo, Osaka, Japan) or Platinum qPCR SuperMix-UDG with ROX (Life Technologies). Gene-specific primer sets were obtained from SYBR green gene expression assays for Cox6b1 (MA035402), Cox5a (MA025060), Cox7c (MA135797) (Takara Bio, Tokyo, Japan), Cox2 (forward, AAC CGA GTC GTT CTG AT; reverse, CTA GGG AGG GGA CTG CTC AT), and Cox5b (forward, GAT GAG GAG CAG GCT ACT GG; reverse, CAG CCA AAC CAG ATG ATA), NQO1 (MA086424), HO-1 (MA089154), γGCS (MA058173), GST-α1 (MA085055), and SOD2 (MA031414) (Takara Bio). Gene-specific primer sets were obtained from TaqMan gene expression assays for Cox1 (Mm04225243_g1) and Cox4i1 (Cox4 isoform 1; Mm0150094_m1) (Applied Biosystems). Each sample was analyzed in duplicate. Amplification and real-time detection were performed using an ABI PRISM 7900HT (Applied Biosystems). Each sample was analyzed in duplicate. The expression levels of the target genes were normalized to those of mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or β-actin (ACTB) as an endogenous control (Takara Bio).
BN-PAGE and 2D SDS-PAGE
Mitochondrial fractions for BN-PAGE were prepared from tissues or cells using a Qproteome mitochondrial isolation kit (Qiagen) according to the manufacturer’s instructions. BN-PAGE analysis was performed using a NativePAGE™ Novex® Bis-Tris Gel System (Life Technologies) according to the manufacturer’s instructions. Mitochondrial fractions were solubilized in 2–2.5 % digitonin and were prepared using a NativePAGE Sample Prep Kit (BN2008; Life Technologies) and separated on 3–12 % NativePAGE Novex Bis-Tris Gels (BN1001BOX; Life Technologies). Bovine Heart Mitochondria WB Control (458322; Life Technologies) and NativeMark Unstained Protein Standard (LC0725; Life Technologies) were loaded as molecular mass standards. After electrophoresis, one-dimensional (1D) gels were stained with colloidal Coomassie brilliant blue G250 or were transferred to polyvinylidene difluoride (PVDF) membranes for immunoblotting. A wet transfer system (Trans-Blot Protein Transfer System, Bio-Rad) was used to transfer high molecular weight proteins and protein complexes to the PVDF membrane at 90 V for 4 h, in modified transfer buffer (25 mM Tris, 192 mM glycine, 20 % methanol, 0.01–0.2 % SDS). After protein transfer, the membranes were soaked in 8 % acetic acid for 5 min and washed with distilled water for 5 min. The membranes were washed in methanol for 5 min to remove the Coomassie stain and then immunoblotted with antibodies.
To separate proteins in two dimensions (2D), the lane cut from BN-PAGE gel was equilibrated in NuPAGE® LDS Sample Buffer (Life Technologies) according to the manufacturer’s instructions. The equilibrated gel strip was then applied to a Novex® 4–12 % Bis-Tris ZOOM® Protein Gel (NP0330BOX; Life Technologies) and separated by standard sodium dodecyl sulfate (SDS)-PAGE using standard transfer and immunoblotting procedures. Signal intensities were analyzed using Multi Gauge software (Fujifilm, Tokyo, Japan).
Plasmid construction
The sequences encoding Cox6b1 were amplified from mouse complementary DNA (cDNA) libraries generated from mouse skin samples. The forward and reverse primers for Cox6b1 were 5′-ACCATGGCTGAAGACATCAAGACT-3′ and 5′-TCAGATCTTCCCAGGAAATG-3′, respectively. Amplification was performed for 30 cycles with annealing at 55 °C for 30 s, extension at 68 °C for 1 min, and a final extension for 5 min at 68 °C. The PCR products were separated by electrophoresis on a 1 % agarose gel and were stained with SYBR Gold nucleic acid gel stain (Life Technologies, Carlsbad, CA, USA) to determine the Cox6b1 PCR product size (264 bp). The cDNA fragment of Cox6b1 was cloned into a pcDNA3.3-TOPO vector (Life Technologies). The plasmid sequences were verified by automated sequencing.
Establishment of stable NIH-3T3 cells overexpressing murine Cox6b1
NIH-3T3 cells (1.5 × 105 cells/cm2) were cultured in Dulbecco’s modified Eagle’s medium (Life Technologies) containing 10 % fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin, overnight at 37 °C under 5 % CO2. The seeded cells were counted using the Trypan blue exclusion method on a Countess Automated Cell Counter (Life Technologies). The cells were then transfected with the pcDNA3.3-Cox6b1 plasmid using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions. After 24 h, and every 48 h thereafter for 2 weeks, the culture medium was replaced with fresh medium containing 400 μg/ml of G418 (Wako, Tokyo, Japan). Between passages 3 and 20, four pools of clones that were stably transfected with the pcDNA3.3-Cox6b1 expression vector or a control vector (pcDNA3.3-TOPO) were collected for further studies. The intensity of transfection was confirmed by quantitative real-time PCR and immunoblotting.
Confocal microscopy
For confocal microscopy, Cox6b1-transfected NIH3T3 cells were stained with 100-nM Mitotracker Red (Life Technologies) for 15 min and were fixed in 4 % paraformaldehyde at room temperature for 15 min. For immunostaining, the cells were blocked with protein block serum-free reagent (Dako, Tokyo, Japan) for 1 h. The cells were immunostained with mouse Cox6b1 mAb at room temperature for 1 h. After three washes in PBS, the cells were stained with goat anti-mouse Alexa-Fluor 488-labeled secondary IgG Ab (Life Technologies) at room temperature for 1 h. The nuclei were stained with TO-PRO-3 (Life Technologies). Immunofluorescence images were obtained using a confocal laser scanning microscope (LSM 510 META; Carl Zeiss, Jena, Germany), and the images were processed with Zen software (Carl Zeiss).
Extracellular flux assays
Oxygen consumption rates (OCRs) were measured using the XFe96 Extracellular Flux Analyzer (Seahorse Bioscience, North Billerica, MA, USA). Seeded cells (3.0 × 105 cells/well) were incubated overnight in growth medium. At least 1 h before measurement, the cells were incubated with XF medium (non-buffered RPMI 1460 containing 25 mM glucose, 2 mM L-glutamine, and 1 mM sodium pyruvate) at 37 °C in a CO2-free incubator. The OCRs were detected under basal conditions and after the application of 1 μM oligomycin, 1 μM carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP), and 1 μM rotenone + 1 μM antimycin A (XF Cell Mito Stress Test Kit; Seahorse Bioscience) to measure basal respiration, coupled respiration (respiration linked to ATP production), maximal respiration, and uncoupled respiration (proton leak), respectively. Each sample was analyzed six times.
Measurement of Cox and citrate synthase activity
Mitochondrial fractions were prepared from cells using a mitochondrial isolation kit (Pierce Biochemicals/Thermo Fisher Scientific, Waltham, MA, USA). Mitochondrial pellets were suspended in 200 μl of buffer (10 mM HEPES pH 7.5, 250 mM sucrose, 1 mM ATP, 80 μM ADP, 5 mM sodium succinate, 2 mM potassium phosphate, and 1 mM dithiothreitol) and were stored at −70 °C until use. The protein concentrations of the mitochondrial preparations were determined using a bicinchoninic acid (BCA) protein assay kit (Pierce Biochemicals/Thermo Fisher Scientific). Cox activity in mitochondrial preparations was determined using a Complex IV assay kit (CYTOCOX1; Sigma-Aldrich, St. Louis, MO, USA). The Complex IV assay is a colorimetric assay that measures the decrease in absorbance at 550 nm corresponding to the oxidation of ferrocytochrome c to ferricytochrome c by cytochrome c oxidase. The reaction rate was measured for the first 45 s of the reaction. Cox activity was normalized for citrate synthase activity, which was measured in mitochondrial preparations using a citrate synthase activity assay kit (CS0720; Sigma-Aldrich) in accordance with the manufacturer’s instructions.
Measurement of intracellular ATP concentrations
The intracellular ATP concentrations were measured using an ATP Assay Kit (A22066; Life Technologies). Cells were grown in a 60-mm dish to approximately 80 % confluence and were then harvested. The background luminescence of 100 μl of the standard reaction solution was measured using a luminometer (FLUOstar Optima; BMG Labtech GmbH, Ortenberg, Germany), and a standard curve was generated. The standard ATP solutions were freshly prepared to measure the ATP concentration in each sample. The protein concentration of each extract was determined using the BCA protein assay kit (Pierce Biochemical/Thermo Fisher Scientific).
Determination of mitochondrial superoxide
Mitochondrial superoxide production was measured using Mitosox Red (Life Technologies). Cells were rinsed two times with phenol red-free Hanks’ balanced salt solution (HBSS; Life Technologies). Next, 3 μM Mitosox was added to the cells, which were then incubated in HBSS for 20 min at 37 °C. The stained cells were washed three times in HBSS, harvested by trypsinization and centrifugation, and resuspended in HBSS. Fluorescence was analyzed using a BD FACS Canto II flow cytometer (BD Biosciences, San Jose, CA, USA). Signals from 10,000 cells were recorded for each sample, and data were analyzed using BD FACS Diva software (BD Biosciences). Fluorescence values were standardized according to the fluorescence values of unstained control cells.
WST-1 cell viability assay
Cells were subcultured in 96-well plates. Confluent cells were deprived of serum for 24 h and were then treated with hydrogen peroxide (500 or 700 μM) for 18 h. Viable adherent cells were stained with Proliferation Reagent WST-1 (Roche Diagnostics GmbH, Penzberg, Germany) for 1 h at 37 °C under 5 % CO2. The WST-1 assay is based on the reduction of WST-1 by viable cells and is suitable for measuring cell proliferation, cell viability, and cytotoxicity. The reaction produces a soluble formazan salt. Absorbance was assayed at 570 nm using a microplate reader (Sunrise; Tecan, Männedorf, Switzerland).
Knock down of Cox6b1 using siRNA
For siRNA treatment, NIH3T3 cells (1.5 × 105 cells/cm2) were cultured in DMEM (Life Technologies) containing 10 % FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin, at 37 °C under 5 % CO2 overnight. The cells were transfected with Stealth RNAi Negative Control Medium GC Duplex #2 (Life Technologies) or Stealth Cox6b1 siRNA (Cox6b1-MSS272848, Life Technologies) using Lipofectamine2000 (Life Technologies) according to the manufacturer’s instructions, and the cells were stimulated or harvested 48–72 h after transfection.
Protein extraction and immunoblotting for Nrf2 and antioxidant enzymes
Cells were subcultured in six-well plates and grown to confluence. To examine Nrf2 nuclear translocation, the medium was replaced with serum-free medium for 24 h. The cells were then treated with 25 μM tert-butylhydroquinone (tBHQ) for 0, 60, or 120 min. The nuclear fraction was obtained using NE-PER nuclear and cytoplasmic extraction reagents (Thermo Fisher Scientific). To determine the expression of NQO1 and HO-1, proteins upregulated by Nrf2, the medium was replaced with serum-free medium for 24 h, and the cells were treated with vehicle (dimethyl sulfoxide), or with 25 μM tBHQ for 18 h. M-PER reagent (Life Technologies), containing protease and dephosphorylation inhibitors (Thermo Fisher Scientific), was added to the harvested cells. Debris in the supernatant was removed by centrifugation at 12,000 × g for 15 min. Protein samples (5 μg) were separated on 4–12 % NuPAGE Novex Bis-Tris gels (Life Technologies), transferred to a PVDF membrane, and blotted using standard methods. Protein bands were visualized using 5-bromo-4-chloro-3-indolyl phosphate/4-nitroblue tetrazolium (Wako) or an enhanced chemiluminescence system (Thermo Fisher Scientific). Data were analyzed using Multi Gauge software (Fujifilm, Tokyo, Japan).
Data analysis
Unpaired Student’s t test was used to determine the significance of differences between the treatment groups. Statistical significance was accepted at values of p < 0.05.
Results
Long-term CR promotes supercomplex formation
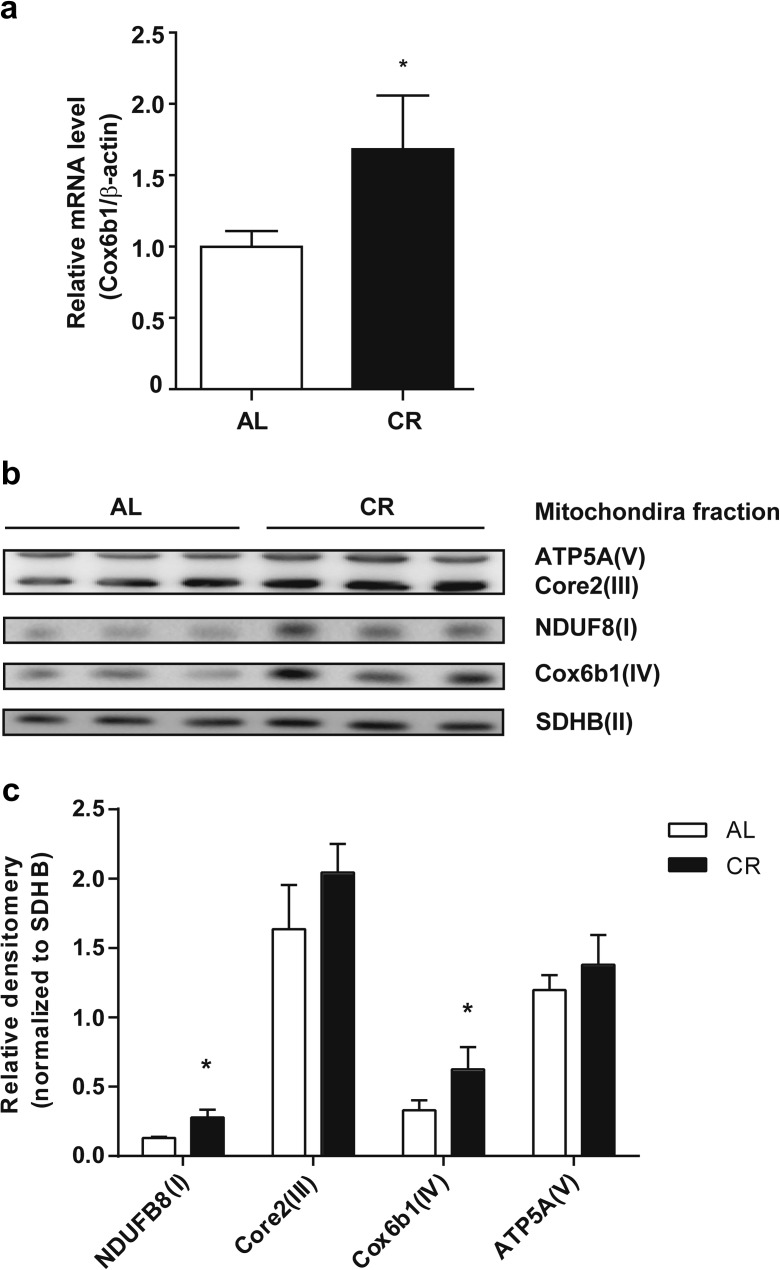
Some controversial findings have been reported regarding the effects of CR on mitochondrial biogenesis (Lopez-Lluch et al. 2006; Civitarese et al. 2007; Hancock et al. 2011). Our previous proteome analysis demonstrated that CR affects the expression of multiple subunits of Complexes I, IV, and V (Dani et al. 2010). In the present study, we reassessed mitochondrial respiratory complexes and subunits in liver tissues obtained from 12-month old CR C57BL/6 J male mice. First, we confirmed that the mRNA and protein expression levels of Cox6b1 were upregulated by CR in the mouse liver (Fig. 1a–c). The abundance of NDUFB8 protein, a marker of Complex I, was also significantly greater in CR mice than in AL mice (Fig. 1b, c). However, the protein expression levels of ATP5A and Core2, markers for Complex V and III, respectively, were not significantly different between CR and AL mice.
Fig. 1.
Effect of CR on the expression of Cox6b1 mRNA and mitochondrial proteins. a Cox6b1 mRNA expression in the liver of 12-month-old mice fed AL or 30 % CR was determined by qPCR. The mRNA expression of β-actin was used as an internal control. The results are presented as means ± SEM (n = 6, *p < 0.05). b Immunoblotting analysis of mitochondrial subunits using the indicated antibodies. c Densitometry of the immunoblots. The complex II subunit (30 kDa), SDHB, was immunodetected as a loading control. The results are presented as means ± SEM (n = 3, *p < 0.05)
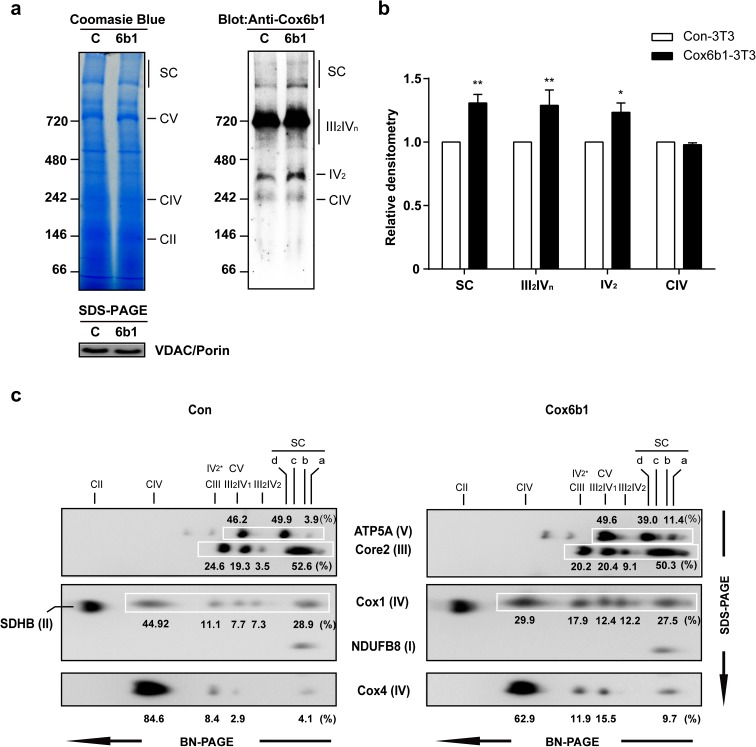
Previous studies have indicated that the formation or stabilization of mitochondrial respiratory supercomplexes modulates mitochondrial bioenergetics (Schägger 2001). A recent study also revealed that ischemic preconditioning in the heart, which minimizes the area of myocardial infarction, increased the abundance of Cox6b1 in high molecular weight supercomplexes (Wong et al. 2010). These findings imply a role of Cox6b1 in ischemic preconditioning, which is one of the hormetic responses potentially implicated in mechanisms underlying the effects of CR (Ristow et al. 2010). To examine the effects of CR on supercomplex formation, we performed 1D BN-PAGE and subsequent immunoblotting using the mitochondrial fraction of liver tissues. Coomassie blue-stained BN-PAGE gel showed that the abundances of the supercomplex (SC) and individual complexes (i.e., Complex V (CV), CIII, CIV, and CII) were similar between AL and CR mice (Fig. 2a, left panel). SDS-PAGE and immunoblotting of the sample with the anti-voltage-dependent anion channel (VDAC/Porin) antibody also confirmed similar loading concentrations of the mitochondria fraction between AL and CR mice. (Fig. 2a, left panel). However, immunoblotting for Cox6b1 showed an increased abundance of Cox6b1 in the SC region, and in the regions of complexes III2IV2 and III2IV1, but not significantly for IV2, in CR mice compared to AL mice (Fig. 2a, right panel, and Fig. 2b). 2D BN-PAGE and SDS-PAGE and subsequent immunoblotting studies with a cocktail of multiple antibodies were performed. The representative image (Fig. 2c) shows that the proportions of signal intensities in the region of high molecular weight SC, i.e., I/III2/IVn (n = 1-4) detected using the Core2, Cox1, NDUFB8, and Cox6b1 antibodies, were greater in CR samples than in AL samples. In contrast, the proportions of signal intensities at the positions of CIII and III2IVn detected by the Core2 antibody, and those of CIV and III2IVn detected by the Cox1 antibody, were lower in CR samples compared to AL samples. The proportion of signal intensities at the position of CIV detected by the Cox6b1 antibody was also lower in CR samples. These findings suggest that CR promotes the assembly of supercomplexes comprised of Complexes I, III, and IV, rather than the forms consisting of CIII, CIV, III2IV1, or III2IV2. In addition, the proportion of signal intensities at the position of CV detected by the ATP5A antibody was greater in CR samples than in AL samples; the proportion of signal intensities at the position of V2 was lower in CR samples. These findings suggest that CR inhibits the formation of V2.
Fig. 2.
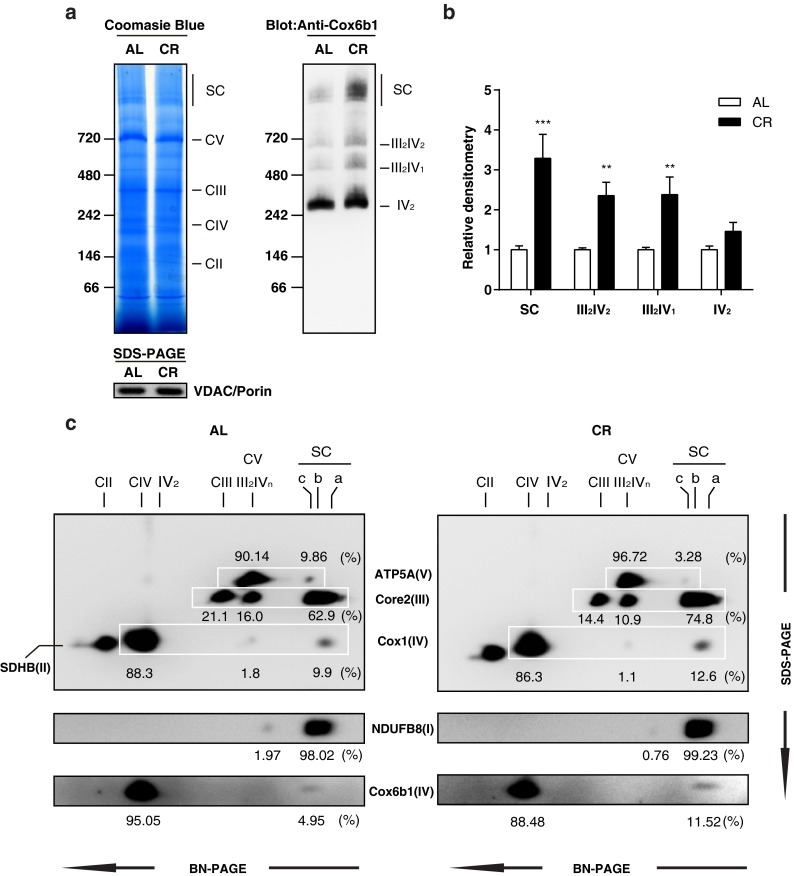
CR promotes the assembly of mitochondrial supercomplexes. a Blue native polyacrylamide gel electrophoresis (BN-PAGE) of liver tissue samples from 12-month-old mice fed AL or 30 % CR. Coomassie blue staining of 1D BN-PAGE gel (left panel) and subsequent immunoblotting with anti-Cox6b1 antibody (right panel). SDS-PAGE and immunoblotting with anti-VDAC/Porin antibody was also performed to confirm equivalent protein loading of mitochondria lysates (lower panel). b Densitometric analysis of immunoblots. The results are presented as mean ± SD (n = 6, **p < 0.01, ***p < 0.001, vs. AL group). c 2D BN/SDS-PAGE of AL and CR liver samples. Immunoblotting was performed using the anti-ATP5A antibody for Complex V, the anti-Core2 antibody for Complex III, the anti-SDHB antibody for Complex II, and the anti-Cox1 and the anti-Cox6b1 antibodies for Complex IV. The position SC-a corresponds to the mitochondrial supercomplexes containing I/III2/IVn (n = 1, 2, 3, or 4); the position SC-b corresponds to I/III2; The position SC-c corresponds to Complex V dimers. The positions of Complex V monomer (CV), Complex III monomer (CIII), Complex III dimers + Complex IV monomers or dimers (III2IVn, n = 1 or 2), Complex IV monomer or dimers (CIV or IV2), and Complex II monomers (CII) are also indicated. CV and III2IVn spots are overlapping. The values represent the proportion of signal intensity in each spot in a row detected by a specific antibody
Establishment of Cox6b1-overexpressing cells
To identify the role of Cox6b1 protein in regulation of mitochondrial functions and the formation of SC, we established stable Cox6b1-overexpressing cells. Of the clones of NIH-3T3 cells showing elevated Cox6b1 protein expression, we selected one clone in which the Cox6b1 mRNA expression level was modestly greater (1.8-fold) in Cox6b1-3T3 cells than in control cells (Con-3T3) transfected with the control vector (pcDNA3.3-TOPO) (Fig. 3a). Cox6b1 overexpression did not significantly affect the expression of the other Cox subunit genes selected in the present study (Fig. 3a). Cox6b1 protein was mostly localized in mitochondria (Fig. 3b). The expression of Cox6b1 protein in the mitochondrial fraction was 2.6 times greater in Cox6b1-3T3 cells than in Con-3T3 cells (Fig. 3b, c). The total amount of Cox1 and Cox4 was not significantly different between Con-3T3 and Cox6b1-3T3 cells (Fig. 3b). Confocal microscopic images confirmed that Cox6b1 immunofluorescence mostly overlapped with that of Mitotracker Red, which specifically labels mitochondria (Fig. 3d). We also obtained preliminary evidence that the overexpressed Cox6b1 labeled with green fluorescent protein was localized to the mitochondria (Supplemental Fig. S1).
Fig. 3.
Establishment of Cox6b1-3T3 cells. a Quantitative PCR analysis of the mRNA expression levels of Cox subunits, normalized for β-actin mRNA, in control NIH-3T3 cells (Con-3T3) and Cox6b1-overexpressing NIH-3T3 (Cox6b1-3T3) cells. The results are presented as mean ± SEM (n = 6, *p < 0.05 vs. Con-3T3). b Immunoblotting for Cox1, Cox4, and Cox6b1 in the cytosolic and mitochondrial fractions of Con-3T3 and Cox6b1-3T3 cells. β-actin and GAPDH were used as cytosolic markers and VDAC/Porin was used as a mitochondrial marker. c Relative expression of Cox6b1 as determined by densitometry of the immunoblots. Results are presented as mean ± SEM (n = 4, ***p < 0.001 vs. Con-3T3). The values were normalized to those of VDAC/Porin. d Localization of Cox6b1 and MitoTracker Red signals in Cox6b1-3T3 cells. Confocal microscopic images of Cox6b1-3T3 cells stained with MitoTracker Red (mitochondria) and TO-PRO-3 (nuclei). Microscopic images of MitoTracker Red signals, Cox6b1 cells, and the merged signals (TO-PRO-3 and MitoTracker Red) are shown in the left, middle, and right panels, respectively
Cox6b1 overexpression enhances the recruitment of Cox into mitochondrial supercomplexes
We investigated the contribution of Cox6b1 overexpression to supercomplex assembly. Using 1D BN-PAGE and subsequent immunoblotting, we found that the abundance of Cox6b1 was increased in the SC, III2IV1 or III2IV2, and IV2 regions, but not in the CIV region (Fig. 4a, b). We also confirmed that Cox (identified using anti-Cox4 antibody) was increased in the SC, indicating that Cox is integrated into the SC (Supplemental Fig. S2a). Moreover, siRNA-mediated knockdown of Cox6b1 reduced the abundance of Cox in the supercomplexes (Supplemental Fig. S2b). The 2D BN-PAGE and SDS-PAGE and immunoblotting studies revealed that the proportions of signal intensities of III2IV1 and III2IV2 in each row (which was detected using Core2, Cox1, or Cox4 antibody) were increased in Cox6b1-3T3 cells compared to Con-3T3 cells (Fig. 4c). For example, the proportions of signal intensities of Core2 at the position of III2IV2 were 3.5 and 9.1 % in Con-3T3 and Cox6b1-3T3 cells, respectively. The proportions of signal intensities of Cox1 in the position of III2IV2 were 7.3 % in the Con-3T3 and 12.2 % in the Cox6b1-3T3 cells. Similarly, the proportions of Cox4 signal intensities were 2.9 and 15.5 % in Con-3T3 and Cox6b1-3T3 cells, respectively (Fig. 4c). In the region of SC, the proportion of Cox4 signal intensities was greater in Cox6b1-3T3 cells than in Con-3T3 cells, although the proportions of Core2 and Cox1 signal intensities were not increased in Cox6b1-3T3 cells. In contrast, the proportions of Cox1 and Cox4 signal intensities in the position of CIV were decreased in Cox6b1-3T3 cells compared to Con-3T3 cells. Taken together, these findings suggest that Cox6b1 promotes the assembly of Cox into the high molecular weight supercomplexes (e.g., I/III2/IVn, n = 1–4) and their intermediate forms (III2IV1 and III2IV2). However, it should be noted that the 2D BN-PAGE and subsequent immunoblot images in vitro (Fig. 4) were different from those in vivo, particularly the proportions of Core2 and Cox1 signal intensities in the region of SC (Figs. 2c and 4c). In addition, the proportion of ATP5A signal intensities at the position of V3 or V4 was greater in Cox6b1-3T3 cells than in the Con-3T3 cells; in contrast, the proportion of ATP5A signals at the position of V2 (SC-d in Fig. 4c) was lower in Cox6b1-3T3 cells. These findings suggest that Cox6b1 overexpression promotes the formation of V3 or V4.
Fig. 4.
Effects of Cox6b1 overexpression on the recruitment of Cox6b1 to mitochondrial supercomplexes. BN-PAGE and immunoblotting of mitochondrial fractions from Con-3T3 and Cox6b1-3T3 cells. a 1D gels were stained with Coomassie R-250 (left panel) or immunoblotted with the anti-Cox6b1 antibody. SDS-PAGE and immunoblotting with the anti-VDAC/Porin antibody was also performed to confirm equivalent protein loading of mitochondria lysates (lower panel). Gels were transferred for the high molecular weight supercomplexes (right panel). b Densitometric analysis of immunoblots. The results are presented as mean ± SD (n = 4, *p < 0.05, **p < 0.01, vs. Con-3T3). c 2D SDS-PAGE of mitochondria fractions from Con-3T3 and Cox6b1-3T3 cells. Immunoblotting was performed with anti-ATP5A antibody for Complex V, anti-Core2 antibody for Complex III, anti-SDHB antibody for Complex II, and anti-Cox1 and anti-Cox4 antibodies for Complex IV. The positions corresponding to the mitochondrial supercomplexes are as follows: SC-a, Complex V trimer or tetramer (Vn, n = 3 or 4); SC-b, I/III2/IVn (n = 1, 2, 3, or 4); SC-c, Complex I + Complex III (I/III2); SC-d, Complex V dimer (V2). The positions of Complex V monomers (CV), Complex III dimers + Complex IV monomers or dimers (III2IVn, n = 1 or 2), predicted Complex IV dimers (IV2 *), Complex IV monomers (CIV), and Complex II monomers (CII) are shown. The values represent the proportion of signal intensity in each spot in a row detected by a specific antibody
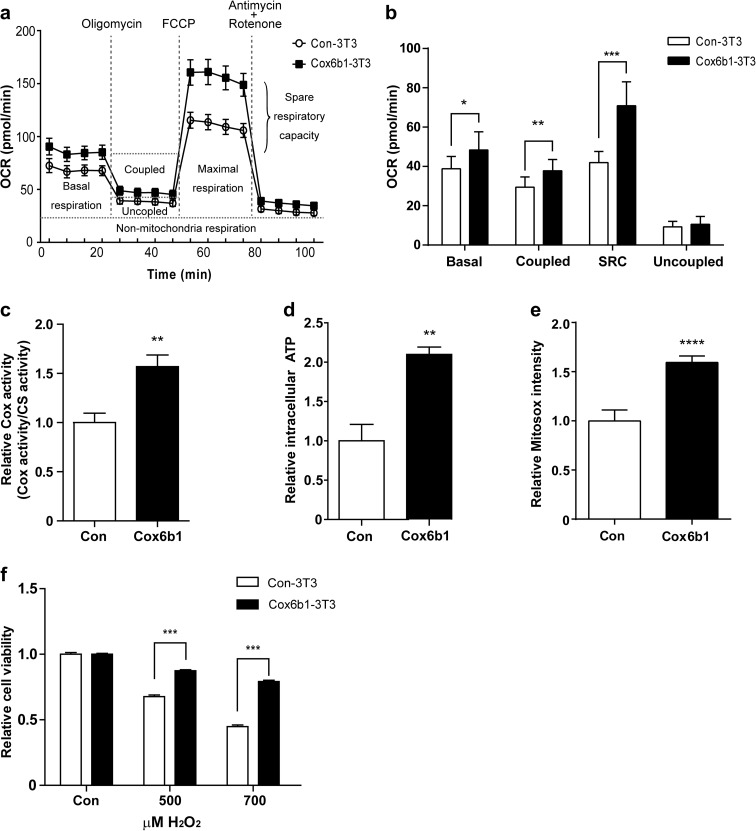
Overexpression of Cox6b1 induces mitochondrial respiration and stress resistance
Mitochondrial functions are tightly linked with the regulation of cellular energy metabolism, apoptosis, and ROS production. In the present study, we investigated mitochondrial respiration in Cox6b1-3T3 cells, by assessing the OCR. We found that the basal OCR was slightly higher in Cox6b1-3T3 cells than in Con-3T3 cells (Fig. 5a, b). Coupled respiration (i.e., OCR linked to ATP generation) was assessed by administration of oligomycin A, an inhibitor of ATP synthase (Fig. 5a). The OCR linked to ATP generation was calculated by subtracting OCR after oligomycin administration from basal OCR. Coupled respiration was increased 1.3-fold in Cox6b1-3T3 cells compared to Con-3T3 cells (Fig. 5b). Spare respiratory capacity (SRC), which is the additional mitochondrial capacity available in response to increased workload or stress, was evaluated by administration of FCCP, an uncoupling agent. SRC was also increased in Cox6b1-3T3 cells compared with Con-3T3 cells (Fig. 5a, b). Uncoupled respiration, which indicates proton leak state of respiration, was analyzed by administration of rotenone and antimycin A, inhibitors of Complex I and III respectively. Uncoupled respiration was not significantly different between Cox6b1-3T3 and Con-3T3 cells. Cox activity and cellular ATP concentrations were greater in Cox6b1-3T3 cells than in Con-3T3 cells (Fig. 5c, d). These findings indicate that oxidative phosphorylation is elevated in Cox6b1-3T3 cells. The basal level of mitochondrial superoxide, detected by Mitosox, was also greater in Cox6b1-3T3 cells than in Con-3 T3 cells (Fig. 5e). However, the viability of Cox6b1-3T3 cells following the induction of oxidative stress using hydrogen peroxide was also greater than that of Con-3T3 cells (Fig. 5f). In these experiments, we confirmed that cell viability prior to treatment with hydrogen peroxide did not differ between Con-3T3 cells and Cox6b103T3 cells (Supplemental Fig. S3).
Fig. 5.
Effect of Cox6b1 overexpression on mitochondrial function. a Oxygen consumption rate (OCR) was measured in Con-3T3 and Cox6b1-3T3 cells under basal conditions and after exposure to the indicated compounds using a XFe96 Extracellular Flux Analyzer. b Basal respiration (Basal), coupled respiration (Coupled), spare respiration capacity (SRC), and uncoupled respiration (Uncoupled) were calculated from the original XFe96 Extracellular Flux Analyzer data shown in Fig. 5a. The results are presented as mean ± SD (n = 5; *p < 0.05, **p < 0.01, ***p < 0.001 vs. Con-3T3). c Cytochrome c oxidase activity in isolated mitochondria from Con-3T3 and Cox6b1-3T3 cells. The results are presented as mean ± SEM (n = 6; **p < 0.01). Citrate synthase activity was used as an internal control. d Relative ATP concentrations were measured using a luminescence assay and were normalized for the protein concentration in each sample. Results are presented as mean ± SEM (n = 6; **p < 0.01). f Relative cellular viability following exposure to hydrogen peroxide (H2O2). Cell viability was measured using a WST-1 assay. Cells were exposed to 500 or 700 μM H2O2 for 24 h. The results are presented as mean ± SEM (n = 12; ***p < 0.001)
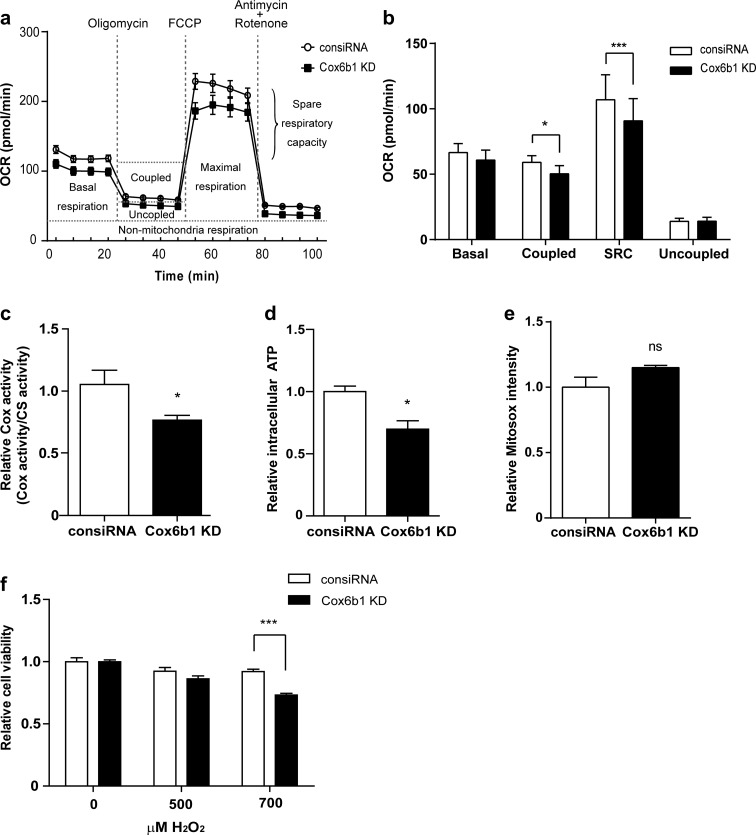
To verify the functional roles for Cox6b1, we also conducted knockdown (KD) experiments for Coxb61 in vitro. Silencing Cox6b1 expression using siRNA did not affect the mRNA expression levels of selected subunits (Supplemental Fig. S4a) including Cox4. However, Cox6b1 KD decreased the protein abundance not only of Cox6b1 but also of Cox4 (Supplemental Fig. S4b). We speculate that Cox6b1 KD confounds maturation of Complex IV and affects the stability of subunits of Complex IV, including Cox4. Thus, Cox6b1 KD may affect protein abundance of multiple subunits of Cox. In this condition, coupled respiration and SRC were slightly but significantly decreased in Cox6b1 KD cells compared with control siRNA-transfected (consiRNA) cells (Fig. 6a, b); there was no difference in basal respiration or uncoupled respiration. Cox activity and ATP levels were also reduced in Cox6b1 KD cells compared to consiRNA cells (Fig. 6c, d). Mitosox-detectable ROS levels did not significantly differ between Cox6b1 KD and consiRNA cells (Fig. 6e). The resistance of the cells to oxidative stress was attenuated in Cox6b1 KD cells (Fig. 6f). Thus, the reduction of Cox6b1 expression did not significantly affect mitochondrial superoxide levels, although mitochondrial oxidative phosphorylation was decreased.
Fig. 6.
The effect of Cox6b1 knockdown (KD) on mitochondrial function. a OCR was measured in control siRNA (consiRNA) and Cox6b1 KD cells under basal conditions and after exposure to the indicated compounds using a XFe96 Extracellular Flux Analyzer. b Basal respiration (Basal), coupled respiration (Coupled), spare respiration capacity (SRC), and uncoupled respiration (Uncoupled) were calculated from the original XFe96 Extracellular Flux Analyzer data in Fig. 6a. The results are presented as mean ± SD (n = 5; *p < 0.05, ***p < 0.001 vs. consiRNA). c Cytochrome c oxidase activity measured in isolated mitochondria from consiRNA and Cox6b1 KD cells. Results are presented as mean ± SEM, n = 6 (*p < 0.05; citrate synthase activity (CS) was used as internal control). d Relative ATP production was detected using a luminescence assay. Results are presented as mean ± SEM, n = 6 (*p < 0.05; each sample was normalized by protein concentration). e The relative cell viability in response to hydrogen peroxide was measured by WST-1 assay. The cells were exposed to different concentrations of H2O2 (500, 700 μM) for 24 h. Data represent mean ± SEM (n = 12, ***p < 0.001)
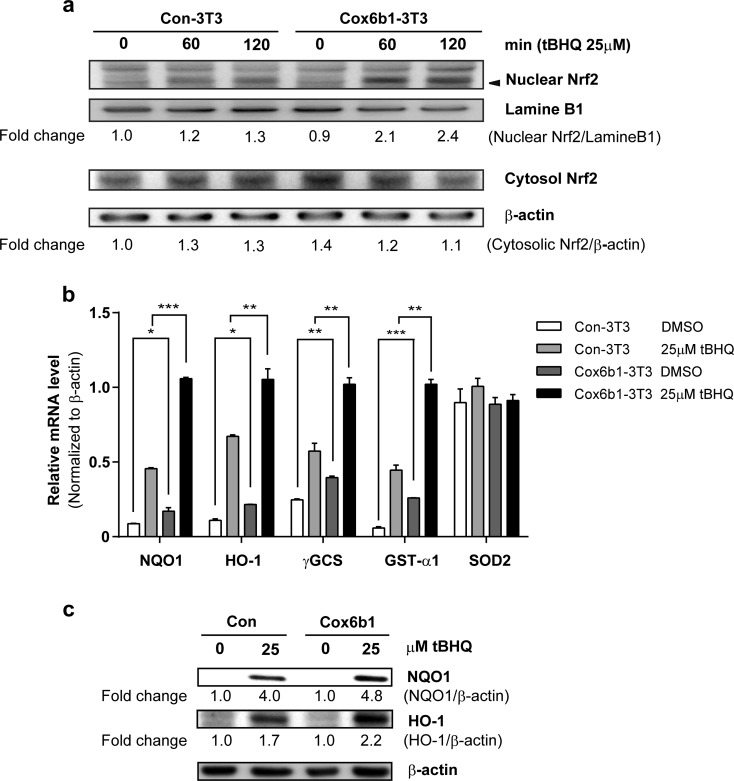
Overexpression of Cox6b1 enhances Nrf2-mediated stress response in cells
Nrf2 is thought to play important roles in CR-mediated stress responses in vivo (Pearson et al. 2008). Because the Cox6b1-3T3 cells displayed increased ROS generation but enhanced stress resistance, we evaluated the activation of Nrf2 in these cells. Subcellular fractionation and immunoblot analyses revealed that Nrf2 protein expression was significantly increased in the nuclear fraction of Cox6b1-3T3 cells. This effect occurred within 60 min after induction with tBHQ, a phase II enzyme inducer (Fig. 7a). The basal nuclear Nrf2 level was similar in Con-3T3 and Cox6b1-3T3 cells. We also confirmed that four of five target genes of Nrf2 were significantly upregulated in Cox6b1-3T3 cells in response to tBHQ, when compared to Con-3T3 cells (Fig. 7b). The protein abundance of NQO1 and HO-1, which are encoded by Nrf2 target genes, was significantly increased in Cox6b1-3T3 cells compared with Con-3T3 cells (Fig. 7c). KD of Nrf2 by siRNA did not affect the expression of Cox6b1-mRNA (Supplemental Fig. S5a, b), although Nrf2 KD attenuated the upregulation of HO-1 mRNA expression in response to tBHQ (Supplemental Fig. S5c). These findings suggest that Cox6b1 is not directly regulated by Nrf2.
Fig. 7.
Cox6b1 overexpression induces Nrf2 nuclear translocation and upregulates antioxidant enzymes. a Nuclear and cytosolic levels of Nrf2 in Con-3T3 and Cox6b1-3T3 cells. Cells were exposed to 25 μM of tBHQ for 60 or 120 min, and nuclear extracts were used for immunoblotting analysis. Lamin B1 and β-actin were used as internal controls for nuclear and cytosolic proteins, respectively. Fold changes of nuclear Nrf2 and cytosolic Nrf2, relative to those of Con-3T3 cells at 0 min, are indicated. b Quantitative PCR analysis of the mRNA expression levels of Nrf2-target genes, those are associated with oxidative stress responses (NQO1, HO-1, γGCS, GST-α1, SOD2) in control NIH-3T3 cells (Con-3T3) and Cox6b1-overexpressing NIH-3T3 cells (Cox6b1-3T3) in response to vehicle (DMSO only) or 25 μM of tBHQ for 6 h. β-actin was used as a control housekeeping gene. The results are presented as mean ± SEM (n = 4, *p < 0.05, **p < 0.01, ***p < 0.001). c Nrf2-dependent antioxidant enzyme expression is increased in Cox6b1-3T3 cells. Con-3T3 and Cox6b1-3T3 were incubated with 25 tBHQ for 18 h, and the total cell lysates were subjected to immunoblotting for NQO1 and HO-1. β-actin was used as an internal control. Fold changes of NQO-1 and HO-1, relative to those of Con-3T3 at 0 μM, are indicated
Discussion
In the first part of the present study, we examined whether CR promotes respiratory supercomplex assembly in vivo. BN-PAGE and SDS-PAGE and subsequent immunoblotting confirmed that the proportion of supercomplexes comprising Complexes I, III, and IV in the mitochondrial protein fraction was increased in CR mouse liver compared with control AL liver. However, the expression of NDUFB8 of Complex I, not just Cox6b1, was increased in the CR liver. In vivo, two studies have shown that CR upregulates mitochondrial biogenesis (Lopez-Lluch et al. 2006; Civitarese et al. 2007). Therefore, the effects of CR are not limited to Cox6b1. In fact, CR seems to influence the expression of several respiratory complexes or subunits, which promote the formation of functional supercomplexes.
Miwa et al. (2014) recently reported that CR reduces some matrix arm subunits of Complex I in mouse tissues, which is associated with improved Complex I assembly, higher Complex I-linked state 3 oxygen consumption rates, and decreased superoxide generation from purified mitochondria from tissues. They presume that incompletely assembled, and thus surplus, matrix arm subunits of Complex I can still use substrate and produce superoxide, but without proton pumping or electron transport (Miwa et al. 2014). In our previous study using rat liver tissues (Dani et al. 2010), NDUFA12 and NDUFA10 subunits, belonging to the peripheral arm of Complex I (Janssen et al. 2006), were also reduced with a 1.7-fold decrease of the abundance of monomeric Complex I in the CR group compared with the AL group. We speculate that CR modified assembly of Complex I, affecting formation of SC and thus mitochondrial respiration and generation of ROS. Thus, formation of SC with Complex I could be intrinsic to the mechanisms of CR.
In the present study, we assessed whether Cox6b1 overexpression promotes assembly of Cox into the mitochondrial SC. When compared to the effect of CR, the immunoblot images followed by BN-PAGE and 2D SDS-PAGE in Cox6b1-3T3 cells were similar in some, but not all, aspects. Nonetheless, the present analysis indicates promotion of Cox assembly into the SC and their intermediate forms (III2IV1 and III2IV2). As discussed above, CR in vivo affects multiple sites in the mitochondrial respiratory chain and thus leads to intricate effects. Upregulation of a single gene could not completely mimic the effect of CR on mitochondria.
In the present study, we also tested whether upregulation of Cox6b1, a subunit of Cox, affects mitochondrial respiration and also investigated its potential contribution to the anti-aging effects of CR. Upregulation of Cox6b1 increased mitochondrial respiration and ATP production but also increased mitochondrial ROS levels. However, despite the increase in ROS levels, Cox6b1 overexpression actually enhanced cell viability under oxidative stress conditions. Regarding the mechanism involved in these protective effects against oxidative stress, we found that Nrf2 expression was enhanced in Cox6b1 cells. It has been reported that Nrf2 mediates the oxidative stress response and the anti-neoplastic effects of CR (Pearson et al. 2008). These findings are consistent with the hypothesis that mitochondrial hormesis is a mechanism for CR (Ristow et al. 2010). CR was originally thought to slow aging by reducing mitochondrial ROS generation. However, recent studies have revealed that mitochondrial respiration and ROS are not necessarily decreased but are actually increased by CR (Ristow et al. 2010). In Caenorhabditis elegans, pretreatment with a ROS scavenger abolished the stress-resisting and life-extending effects of glucose restriction (Schultz et al. 2007), supporting the mitochondrial hormesis hypothesis of CR. In our previous study, hepatic mitochondrial ROS generation fluctuated during the fed–fasted cycle of the CR regimen (Hayashida et al. 2011). The ROS level decreased in the fed phase in CR rats but increased in the fasted phase to a level similar to that in AL rats. Thus, CR does not always decrease ROS generation in rodents. Although we used an in vitro cellular model in the present study, the ROS level was constantly elevated, and intermittent increases in mitochondrial ROS during the feeding cycle in vivo may induce a hormetic response. In fact, mitochondrial glutathione and oxidized glutathione levels were increased in the fasted phase relative to the fed phase of the CR regimen (Hayashida et al. 2011). Reversible fluctuations in ROS levels and the efficient buffering by the GSH:GSSG redox couple under a CR regimen have been proposed by Dani et al. (2010) as analogous to aging but a putatively hermetic response to CR.
In ischemic preconditioned heart models, ROS are required for the cardioprotective signal (Chen et al. 1995; Baines et al. 1997). In the preconditioned heart, the amount of Cox6b1 is increased in mitochondrial supercomplexes comprising Complexes I, III, and IV (Wong et al. 2010). In the present study, we also showed that upregulation of Cox6b1 promotes the assembly of Cox with Cox6b1 into respiratory supercomplexes. Therefore, the present in vitro experiments suggest that Cox6b1 regulates mitochondrial respiration and oxidative stress response by promoting the assembly of mitochondrial respiratory supercomplexes and contributes to the anti-aging effects of CR.
In summary, the present study provides evidence that mitochondrial supercomplex assembly rather than mitochondrial biogenesis may explain at least some of the anti-aging effects of CR.
Electronic supplementary material
(DOC 4710 kb)
Acknowledgments
We are grateful to the staff at the Laboratory Animal Center for Biomedical Research at the Center for Frontier Life Sciences, Nagasaki University for their animal care and technical assistance, and Yutaka Araki, Yuko Moriyama, Rieko Tahara, and Chika Matsumoto for their excellent technical assistance. The study was supported by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (I.S., nos. 22390042) and the JSPS Asian CORE Program (I.S., nos. FY2010-5). We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.
References
- Baines CP, Goto M, Downey JM. Oxygen radicals released during ischemic preconditioning contribute to cardioprotection in the rabbit myocardium. J Mol Cell Cardiol. 1997;29:207–216. doi: 10.1006/jmcc.1996.0265. [DOI] [PubMed] [Google Scholar]
- Chen W, Gabel S, Steenbergen C, Murphy E. A redox-based mechanism for cardioprotection induced by ischemic preconditioning in perfused rat heart. Circ Res. 1995;77:424–429. doi: 10.1161/01.RES.77.2.424. [DOI] [PubMed] [Google Scholar]
- Civitarese AE, Carling S, Heilbronn LK, Hulver MH, Ukropcova B, et al. Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med. 2007;4:e76. doi: 10.1371/journal.pmed.0040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman RJ, Anderson RM, Johnson SC, Kastman EK, Kosmatka KJ, et al. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–204. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani D, Shimokawa I, Komatsu T, Higami Y, Warnken U, et al. Modulation of oxidative phosphorylation machinery signifies a prime mode of anti-ageing mechanism of calorie restriction in male rat liver mitochondria. Biogerontology. 2010;11:321–334. doi: 10.1007/s10522-009-9254-y. [DOI] [PubMed] [Google Scholar]
- Finley LW, Haigis MC. The coordination of nuclear and mitochondrial communication during aging and calorie restriction. Ageing Res Rev. 2009;8:173–188. doi: 10.1016/j.arr.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genova ML, Lenaz G. Functional role of mitochondrial respiratory supercomplexes. Biochim Biophys Acta. 2014;1837:427–443. doi: 10.1016/j.bbabio.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Hancock CR, Han DH, Higashida K, Kim SH, Holloszy JO. Does calorie restriction induce mitochondrial biogenesis? a reevaluation. FASEB J. 2011;25:785–791. doi: 10.1096/fj.10-170415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashida T, Komatsu T, Henmi Y, Yanagihara-Ota K, Kim AR, Chiba T, et al. Modest inhibition of the growth hormone axis does not affect mitochondrial reactive oxygen species generation or redox state, unlike calorie restriction. Geriatr Gerontol Int. 2011;11:496–503. doi: 10.1111/j.1447-0594.2011.00692.x. [DOI] [PubMed] [Google Scholar]
- Janssen RJ, Nijtmans LG, van den Heuvel LP, Smeitink JAM. Mitochondrial complex I: structure, function and pathology. mitochondrial complex I: structure, function and pathology. J Inherit Metab Dis. 2006;29:499–515. doi: 10.1007/s10545-006-0362-4. [DOI] [PubMed] [Google Scholar]
- Kadenbach B. Intrinsic and extrinsic uncoupling of oxidative phosphorylation. Biochim Biophys Acta. 2003;1604:77–94. doi: 10.1016/S0005-2728(03)00027-6. [DOI] [PubMed] [Google Scholar]
- Lopez-Lluch G, Hunt N, Jones B, Zhu M, Jamieson H, Hilmer S. Calorie restriction induces mitochondrial biogenesis and bioenergetic efficiency. Proc Natl Acad Sci U S A. 2006;103:1768–1773. doi: 10.1073/pnas.0510452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annu Rev Biochem. 2008;77:727–754. doi: 10.1146/annurev.biochem.77.061206.171059. [DOI] [PubMed] [Google Scholar]
- Masoro EJ. Subfield history: caloric restriction, slowing aging, and extending life. Sci Aging Know Environ. 2003;2003:RE2. doi: 10.1126/sageke.2003.8.re2. [DOI] [PubMed] [Google Scholar]
- Massa V, et al. Severe infantile encephalomyopathy caused by a mutation in COX6B1, a nucleus-encoded subunit of cytochrome c oxidase. Am J Hum Genet. 2008;82:1281–1289. doi: 10.1016/j.ajhg.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison JA, Roth GS, Beasley TM, Tilmont EM, Handy AM, et al. Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature. 2012;489:318–321. doi: 10.1038/nature11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa S, Jow H, Baty K, Johnson A, Czapiewski R, Saretzki G, Treumann A, von Zglinicki T. Low abundance of the matrix arm of complex I in mitochondria predicts longevity in mice. Nat Comm. 2014;5:3837. doi: 10.1038/ncomms4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson KJ, Lewis KN, Price NL, Chang JW, Perez E, Cascajo MV, Tamashiro KL, et al. Nrf2 mediates cancer protection but not prolongevity induced by caloric restriction. Proc Natl Acad Sci U S A. 2008;105(7):2325–2330. doi: 10.1073/pnas.0712162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristow M, Zarse K. How increased oxidative stress promotes longevity and metabolic health: the concept of mitochondrial hormesis (mitohormesis) Exp Gerontol. 2010;45:410–418. doi: 10.1016/j.exger.2010.03.014. [DOI] [PubMed] [Google Scholar]
- Schägger H. Respiratory chain supercomplexes. IUBMB Life. 2001;52:119–128. doi: 10.1080/15216540152845911. [DOI] [PubMed] [Google Scholar]
- Schägger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, et al. Glucose restriction extends caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6:280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H. The whole structure of the 13-subunit oxidized cytochrome C oxidase at 2.8 a. Science (New York, NY) 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- Weishaupt A, Kadenbach B. Selective removal of subunit VIb increases the activity of cytochrome c oxidase. Biochemistry. 1992;31:11477–11481. doi: 10.1021/bi00161a028. [DOI] [PubMed] [Google Scholar]
- Wong R, Aponte AM, Steenbergen C, Murphy E. Cardioprotection leads to novel changes in the mitochondrial proteome. Am J Physiol Heart Circ Physiol. 2010;298:H75–H91. doi: 10.1152/ajpheart.00515.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaza H, Komatsu T, Wakita S, Kijogi C, et al. FoxO1 is involved in the antineoplastic effect of calorie restriction. Aging Cell. 2010;9(3):372–382. doi: 10.1111/j.1474-9726.2010.00563.x. [DOI] [PubMed] [Google Scholar]
- Yoshikawa S, Shinzawa-Itoh K, Tsukihara T (1998) Crystal structure of bovine heart cyto [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 4710 kb)