Abstract
PepBuild, a web server, will aid in designing and building a capped or uncapped peptide/protein with known secondary and tertiary structure. The user can build a peptide/protein by choosing the required amino acid residue with regular secondary structure. The torsional angles can be supplied by the user, if desired. The server also allows the user to add relevant protecting groups at the N- and/or C-terminal of the peptide. The amino acid side chains of the designed peptide are optimized using rotameric libraries. Finally, the server provides the option of displaying the result or downloading the complete file in PDB (Protein Data Bank) format. This PDB file can later be used as an input for various molecular simulation programs or for graphical display. The web server is available at http://www.imtech.res.in/bvs/pepbuild/.
INTRODUCTION
Peptide and protein structures provide important information in structural genomics. The three-dimensional (3D) models of these structures may be deduced either by such techniques as X-ray diffraction and NMR (nuclear magnetic resonance) or by theoretical prediction methods. Theoretical prediction methods, including knowledge-based methods, can quickly and efficiently predict the structure and also provide information about the 3D disposition of each and every atom in the peptide or protein. The required structure can also be designed using prior knowledge of either the cartesian or internal coordinates of the peptide or protein. Such structural data need to be built or designed in order to carry out molecular modeling (energy minimization and/or molecular dynamics) or docking studies, especially for small peptides. Some public domain software/programs from independent groups providing limited access, or commercial software such as InsightII/DS Modeling (http://www.accelrys.com/insight/ and http://www.accelrys.com/dstudio/ds_modeling/) and Sybyl (http://www.tripos.com/sciTech/inSilicoDisc/moleculeModeling/sybase.html), can aid in building the structure of the peptide/protein. Although the public domain software is able to build the structure, it lacks the ability to optimize the conformation of side chains of amino acid residues with respect to the backbone. Such un-optimized structures are likely to have high van der Waals energies and thus, may not allow minimization of the energy of the structure before the removal of bumpy overlaps of atoms of the structure. At present, there is no web service available that can build an optimized structure data file based on secondary structure information supplied. The web server PepBuild is intended to provide help in designing peptide/protein structures by generating structure data files in PDB (Protein Data Bank) format with optimized side chains for essential amino acid residues.
METHOD
Standard geometry (bond lengths and bond angles) is used for the amino acid residues and protecting groups. Realistic values for the torsional angles of the regular secondary structures are adopted (1,2). The overlaps between atoms of side chains developed while building the peptides need to be taken care of to avoid van der Waals contacts. SCRWL is used to predict the conformation of the side chains of the structure thus built. It employs a simple energy function based on a backbone-dependent rotameric library and a linear repulsive steric energy (3). The appropriate side chain torsional angles are incorporated into the initial model, based on this energy function. The final structure contains an optimized side chain conformation of the amino acid residues, in PDB format. Finally, protons are added to the PDB file to satisfy the valency requirements of the atoms. The algorithm for this web server has been developed and implemented in C++ and Java on the Solaris platform.
Input to the web server
The server constructs a peptide by taking residue-by-residue input from the user. It offers three different sets of choices for building the peptide:
secondary structure (extended, right- or left-handed α-helix, 310 helix, π-helix, β-sheet conformations) corresponding to each amino acid;
protonated or unprotonated amino acids;
different protecting groups ACE/BOC and NHE/NME for N- and C-terminals, respectively, if required.
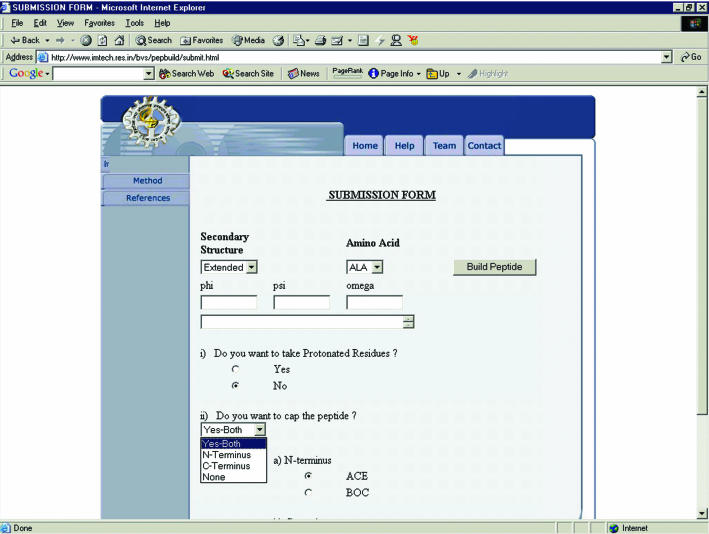
The submission form on the web server is shown in Figure 1. The torsional angles (φ, ψ and ω) for a non-regular secondary structure can also be supplied for an amino acid. There is an option to choose either protonated or unprotonated amino acid residues. Further, the peptide can optionally be capped by protecting groups such as ACE/BOC and NHE (for NH2)/NME at the N- and C-terminals, respectively. The user can also cap either the N-terminus or the C-terminus of the peptide. The user can build a peptide up to a maximum length of a 100 amino acid residues.
Figure 1.
PepBuild submission form.
RESULTS
Output of the web server
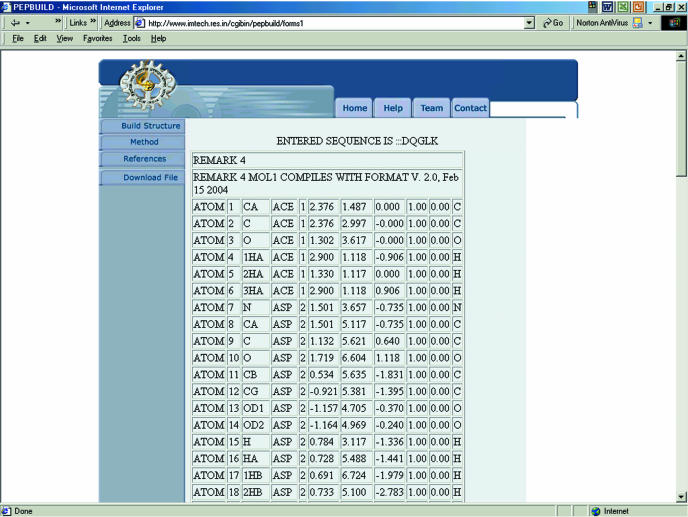
After submission of the input data, the peptide structure data file is built and displayed according to the input of the user. The structure data file, thus built contains an optimized conformation of the side chains of the amino acid residues, in PDB format. The user can download this file for further analysis and simulations. The optimized PDB file generated contains the positions of all the atoms including protons. The optimized structure does not have van der Waals overlaps of the side chains of the amino acid residues, and such a structure is in a low-energy state. This structure can be used by various molecular simulation programs for further studies. The complete optimization or exploration of the conformational space of the structure can be carried out using molecular dynamics approaches. PepBuild provides only a side-chain-optimized (with respect to backbone torsional angles) initial model of the peptide/protein. The conformation of the modeled peptide needs to be further compared with other possible low-energy conformations. An example PDB file of a heptapeptide (ACE–DQGLK–NME) generated using this web server is shown in Figure 2.
Figure 2.
A PDB file generated for a heptapeptide, ACE–DQGLK–NME.
PepBuild displays the information through a web-based graphical user interface that is very user-friendly and fast. PepBuild can be browsed via Netscape 6 and Internet Explorer version 5 or higher only. The user community involved in designing peptides and employing molecular modeling/simulation software such as AMBER (4) and CHARMm (5) can make use of this web server to build optimized peptide structures. However, PepBuild itself does not perform molecular dynamics.
Acknowledgments
ACKNOWLEDGEMENTS
The programming for the web server carried out by Ms Tejinder Kaur, Project Assistant, is duly acknowledged. The author is grateful to Prof. R. L. Dunbrack Jr for allowing usage of SCRWL for optimizing the side chains of the amino acid residues.
REFERENCES
- 1.Flory P.J. (1969) Statistical Mechanics of Chain Molecules. John Wiley, NY. [Google Scholar]
- 2.IUPAC-IUB Commission on Biochemical Nomenclature (1970) IUPAC-IUB Commission on Biochemical Nomenclature, abbreviations and symbols for the description of the conformation of polypeptide chains. Tentative rules (1969). Biochemistry, 9, 3471–3479. [DOI] [PubMed] [Google Scholar]
- 3.Canutescu A.A., Shelenkov,A.A. and Dunbrack,R.L.,Jr (2003) A graph theory algorithm for protein side-chain prediction. Prot. Sci., 12, 2001–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Case D.A., Pearlman,D.A., Caldwell,J.W., Cheatham,T.E.,III, Wang,J., Ross,W.S., Simmerling,C.L., Darden,T.A., Merz,K.M., Stanton,R.V. et al. (2002), AMBER 7. University of California, San Francisco.
- 5.Brooks B.R., Bruccoleri,R.E., Olafson,B.D., States,D.J., Swaminathan,S. and Karplus,M. (1983) CHARMm: a program for macromolecular energy minimization, and dynamics calculations, J. Comp. Chem. 4, 187–217. [Google Scholar]